Abstract
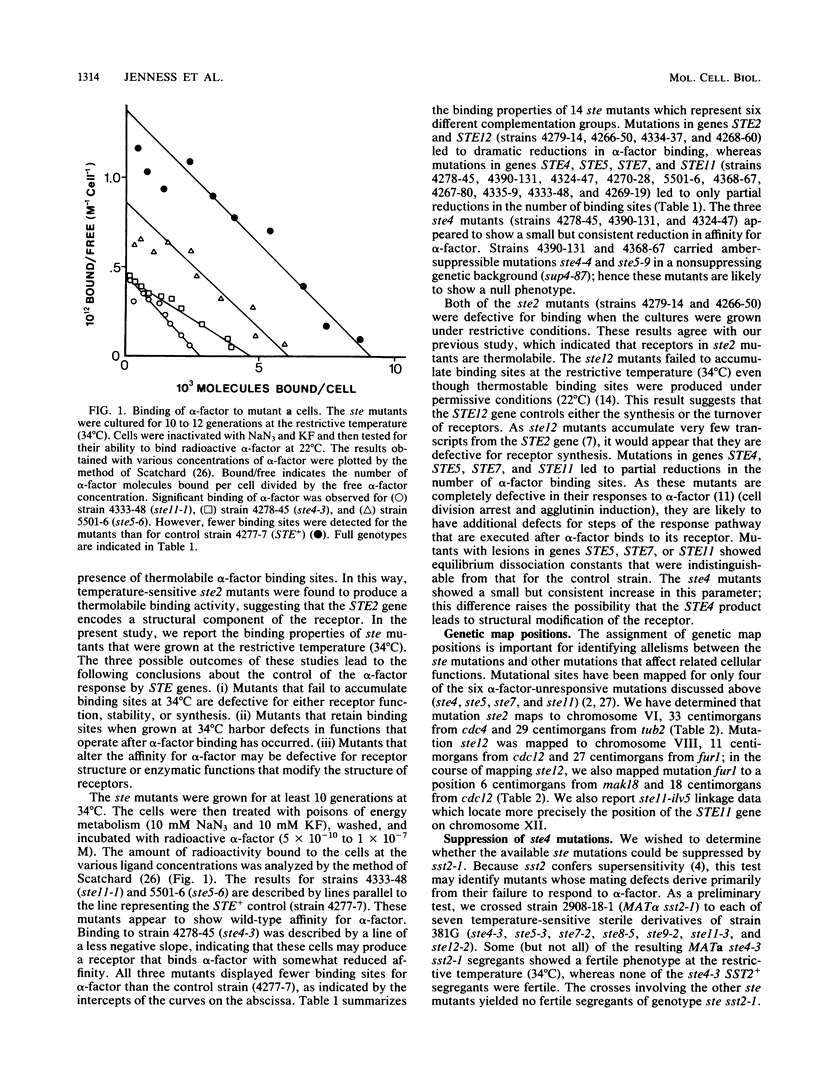
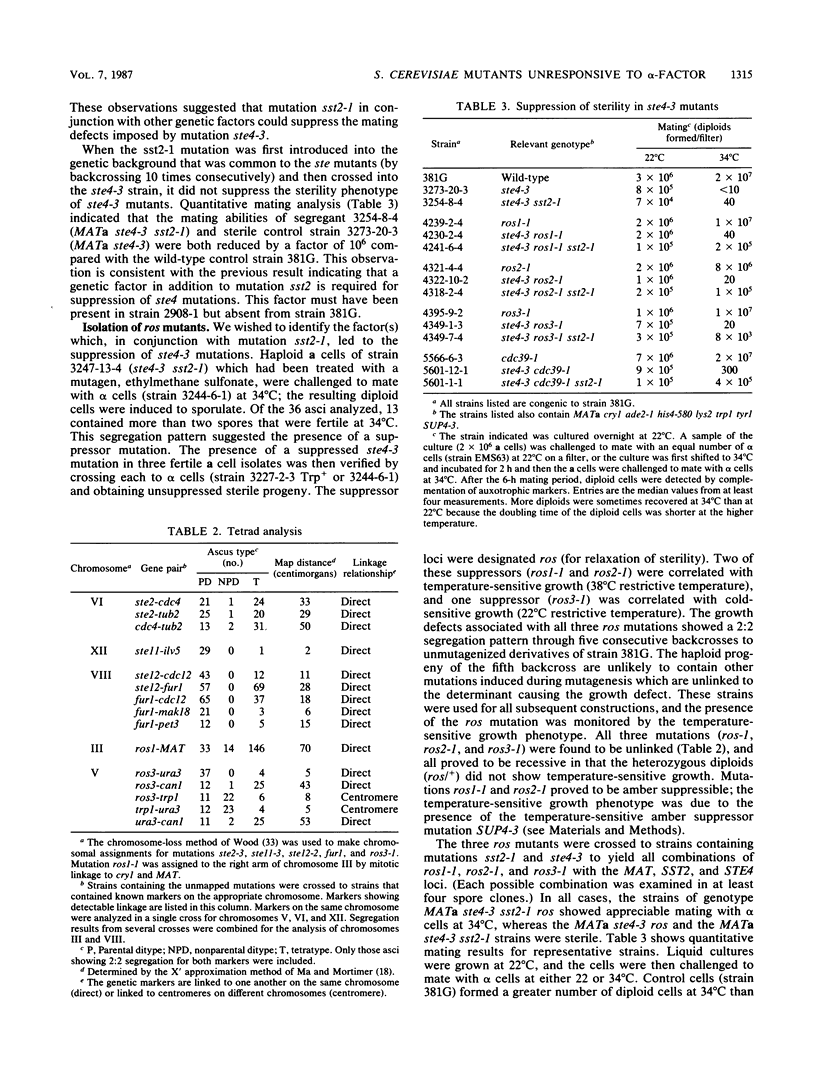
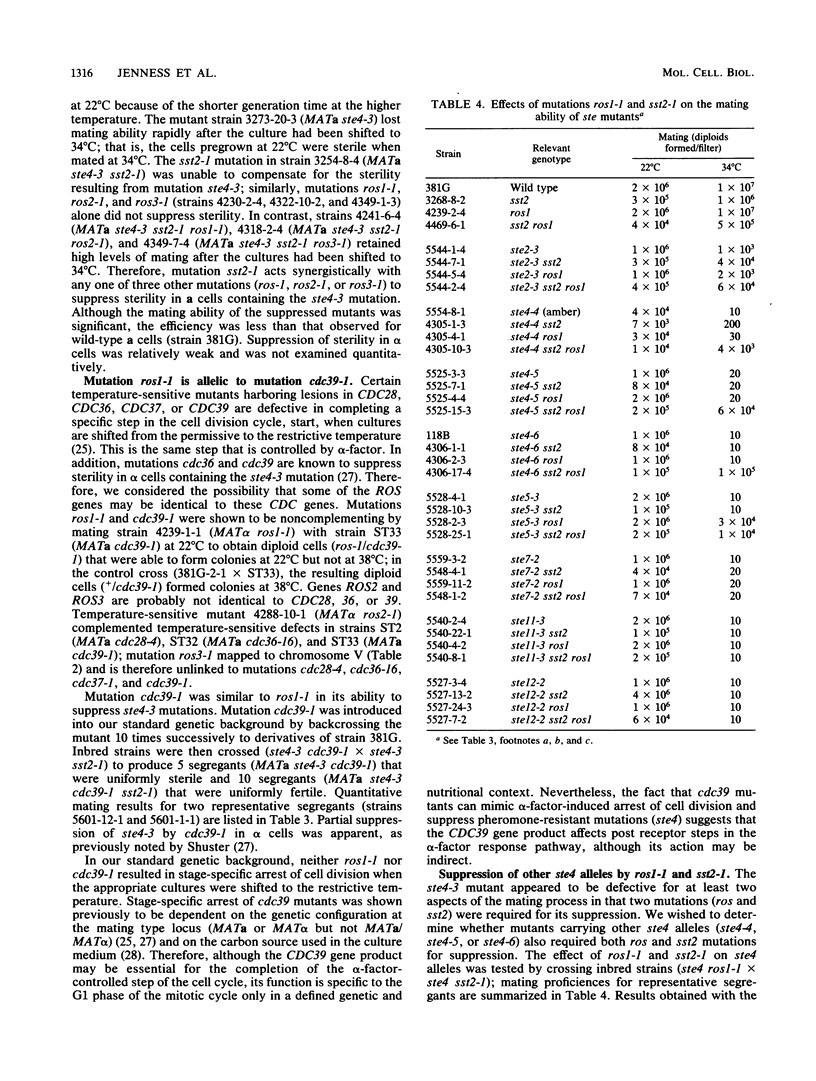
Mutations in six genes that eliminate responsiveness of Saccharomyces cerevisiae a cells to alpha-factor were examined by assaying the binding of radioactively labeled alpha-factor to determine whether their lack of responsiveness was due to the absence of alpha-factor receptors. The ste2 mutants, known to be defective in the structural gene for the receptor, were found to lack receptors when grown at the restrictive temperature; these mutations probably affect the assembly of active receptors. Mutations in STE12 known to block STE2 mRNA accumulation also resulted in an absence of receptors. Mutations in STE4, 5, 7, and 11 partially reduced the number of binding sites, but this reduction was not sufficient to explain the loss of responsiveness; the products of these genes appear to affect postreceptor steps of the response pathway. As a second method of distinguishing the roles of the various STE genes, we examined the sterile mutants for suppression. Mating of the ste2-3 mutant was apparently limited by its sensitivity to alpha-factor, as its sterility was suppressed by mutation sst2-1, which leads to enhanced alpha-factor sensitivity. Sterility resulting from each of four ste4 mutations was suppressed partially by mutation sst2-1 or by mutation bar1-1 when one of three other mutations (ros1-1, ros2-1, or ros3-1) was also present. Sterility of the ste5-3 mutant was suppressed by mutation ros1-1 but not by sst2-1. The ste7, 11, and 12 mutations were not suppressed by ros1 or sst2. Our working model is that STE genes control the response to alpha-factor at two distinct steps. Defects at one step (requiring the STE2 gene are suppressed (directly or indirectly) by mutation sst2-1, whereas defects at the other step (requiring the STE5 gene) are suppressed by the ros1-1 mutation. The ste4 mutants are defective for both steps. Mutation ros1-1 was found to be allelic to cdc39-1. Map positions for genes STE2, STE12, ROS3, and FUR1 were determined.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burkholder A. C., Hartwell L. H. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985 Dec 9;13(23):8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaleff D. T., Tatchell K. Molecular cloning and characterization of the STE7 and STE11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Aug;5(8):1878–1886. doi: 10.1128/mcb.5.8.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Otte C. A. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982 Jan;2(1):11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chvatchko Y., Howald I., Riezman H. Two yeast mutants defective in endocytosis are defective in pheromone response. Cell. 1986 Aug 1;46(3):355–364. doi: 10.1016/0092-8674(86)90656-2. [DOI] [PubMed] [Google Scholar]
- Ciejek E., Thorner J. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell. 1979 Nov;18(3):623–635. doi: 10.1016/0092-8674(79)90117-x. [DOI] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell. 1985 Oct;42(3):923–930. doi: 10.1016/0092-8674(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., McCaffrey G., Sprague G. F., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig A., Holly J., Saari G., MacKay V. L. Multiple regulation of STE2, a mating-type-specific gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2106–2114. doi: 10.1128/mcb.6.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp Cell Res. 1973 Jan;76(1):111–117. doi: 10.1016/0014-4827(73)90425-4. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Hartwell L. H. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974 Apr 15;84(3):445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to Saccharomyces cerevisiae a cells: dissociation constant and number of binding sites. Mol Cell Biol. 1986 Jan;6(1):318–320. doi: 10.1128/mcb.6.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to yeast a cells: chemical and genetic evidence for an alpha-factor receptor. Cell. 1983 Dec;35(2 Pt 1):521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Spatrick P. Down regulation of the alpha-factor pheromone receptor in S. cerevisiae. Cell. 1986 Aug 1;46(3):345–353. doi: 10.1016/0092-8674(86)90655-0. [DOI] [PubMed] [Google Scholar]
- Kurjan J. Alpha-factor structural gene mutations in Saccharomyces cerevisiae: effects on alpha-factor production and mating. Mol Cell Biol. 1985 Apr;5(4):787–796. doi: 10.1128/mcb.5.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Mortimer R. K. Empirical equation that can be used to determine genetic map distances from tetrad data. Mol Cell Biol. 1983 Oct;3(10):1886–1887. doi: 10.1128/mcb.3.10.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. A. Comparison of dose-response curves for alpha factor-induced cell division arrest, agglutination, and projection formation of yeast cells. Implication for the mechanism of alpha factor action. J Biol Chem. 1983 Nov 25;258(22):13849–13856. [PubMed] [Google Scholar]
- Moore S. A. Yeast cells recover from mating pheromone alpha factor-induced division arrest by desensitization in the absence of alpha factor destruction. J Biol Chem. 1984 Jan 25;259(2):1004–1010. [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse-Messenguy F., Fink G. R. Temperature-sensitive nonsense suppressors in yeast. Genetics. 1973 Nov;75(3):459–464. doi: 10.1093/genetics/75.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980 Jul;95(3):561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J. R. "Start" mutants of Saccharomyces cerevisiae are suppressed in carbon catabolite-derepressing medium. J Bacteriol. 1982 Aug;151(2):1059–1061. doi: 10.1128/jb.151.2.1059-1061.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J. R. Mating-defective ste mutations are suppressed by cell division cycle start mutations in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1052–1063. doi: 10.1128/mcb.2.9.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley D. R., Lefkowitz R. J. Molecular mechanisms of receptor desensitization using the beta-adrenergic receptor-coupled adenylate cyclase system as a model. Nature. 1985 Sep 12;317(6033):124–129. doi: 10.1038/317124a0. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Herskowitz I. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J Mol Biol. 1981 Dec 5;153(2):305–321. doi: 10.1016/0022-2836(81)90280-1. [DOI] [PubMed] [Google Scholar]
- Stoscheck C. M., Carpenter G. Down regulation of epidermal growth factor receptors: direct demonstration of receptor degradation in human fibroblasts. J Cell Biol. 1984 Mar;98(3):1048–1053. doi: 10.1083/jcb.98.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. S. Mitotic chromosome loss induced by methyl benzimidazole-2-yl-carbamate as a rapid mapping method in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1080–1087. doi: 10.1128/mcb.2.9.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]



