Abstract
Roux-en-Y gastric bypass (RYGB) results in rapid weight loss, reduced adiposity, and improved glucose metabolism. These effects are not simply attributable to decreased caloric intake or absorption, but the mechanisms linking rearrangement of the gastrointestinal tract to these metabolic outcomes are largely unknown. Studies in humans and rats have shown that RYGB restructures the gut microbiota, prompting the hypothesis that some of the effects of RYGB are caused by altered host-microbial interactions. To test this hypothesis, we used a mouse model of RYGB that recapitulates many of the metabolic outcomes in humans. 16S ribosomal RNA gene sequencing of murine fecal samples collected after RYGB surgery, sham surgery, or sham surgery coupled to caloric restriction revealed that alterations to the gut microbiota after RYGB are conserved among humans, rats, and mice, resulting in a rapid and sustained increase in the relative abundance of Gammaproteobacteria (Escherichia) and Verrucomicrobia (Akkermansia). These changes were independent of weight change and caloric restriction, were detectable throughout the length of the gastrointestinal tract, and were most evident in the distal gut, downstream of the surgical manipulation site. Transfer of the gut microbiota from RYGB-treated mice to nonoperated, germ-free mice resulted in weight loss and decreased fat mass in the recipient animals relative to recipients of microbiota induced by sham surgery, potentially due to altered microbial production of short-chain fatty acids. These findings provide the first empirical support for the claim that changes in the gut microbiota contribute to reduced host weight and adiposity after RYGB surgery.
INTRODUCTION
Roux-en-Y gastric bypass (RYGB) is a highly effective treatment for severe obesity and type 2 diabetes, characterized by a marked and sustained loss of ~65 to 75% of excess body weight and fat mass (1). Initially thought to be a mechanical procedure causing restriction and calorie malabsorption, recent evidence suggests that RYGB alters the basic physiology of energy balance and metabolism in weight-dependent and weight-independent ways (2). These changes include marked improvement in glucose homeostasis before weight loss (3), increased secretion of gut hormones (4, 5), and alterations in energy expenditure (6–8); however, the mechanisms that drive these outcomes remain incompletely understood. It is likely that anatomical rearrangement of the gastrointestinal tract alters the composition of the luminal milieu and its interactions with the intestinal epithelium, which in turn affects downstream signaling pathways regulating host energy balance and metabolism (9). A particular luminal factor, however, has yet to be identified.
Of the several potential luminal components known to mediate energy balance after gastric bypass surgery, including dietary nutrients, biliopancreatic secretions, and the gut microbiota, we focused on the potential contribution of the microbiota to RYGB outcomes, particularly due to its increasingly recognized role in regulating energy balance and metabolic function. The gut microbiota has been reported to differ in community structure, gene content, and metabolic network organization between obese and lean individuals (10–13). Diets differing in fat and sugar content can also affect this structure (14–17), favoring the growth of bacteria whose secreted factors or structural components contribute to the development of adiposity, insulin resistance, and other metabolic derangements (18, 19). Furthermore, colonization of germ-free mice with a gut microbiota from conventionally raised mice has provided functional evidence that the gut microbiota can modulate host phenotype by decreasing food intake and increasing adiposity (10, 20). These phenotypes are further enhanced in germ-free animals colonized with the cecal microbiota from obese donors (10, 17). Because the gut microbiota influences some of the same metabolic parameters as RYGB, we hypothesized that changes to the gut microbiota after RYGB may contribute to some of the metabolic benefits of this procedure.
Recently, RYGB has been reported to cause marked shifts in fecal microbial profiles in both humans and rats (9, 21, 22), but it remains unclear to what degree these changes are caused by alterations in the gastrointestinal tract, by the surgical process itself, or by decreased weight and/or caloric intake induced by this procedure. Furthermore, it is unknown how rapidly these changes in microbial ecology occur, how stable they are over time within a given individual, and if they are limited to the distal gut (for example, feces). The microbial taxonomic groups that are enriched after RYGB have been associated with decreased adiposity and leptin levels in humans (21) and with changes in fecal and urinary metabolites in rats (9). To date, however, there has been no empirical data to support the hypothesis that these RYGB-altered microbial communities have a direct effect on improved metabolic outcomes.
To address these questions, we turned to a recently developed murine model of RYGB (23, 24) to (i) demonstrate that changes in the gut microbiota after gastric bypass surgery are conserved among humans, rats, and mice; (ii) demonstrate that the underlying cause of much of the microbial response to surgery is due to the reconfiguration of the gastrointestinal tract; (iii) characterize the temporal and spatial changes in the gut microbiota after this operation; and (iv) through transplantation of the RYGB-associated gut microbiota into germ-free mice, show that this altered gut microbiota is sufficient to trigger decreased host weight and adiposity (see Fig. 1, A to C, for experimental design).
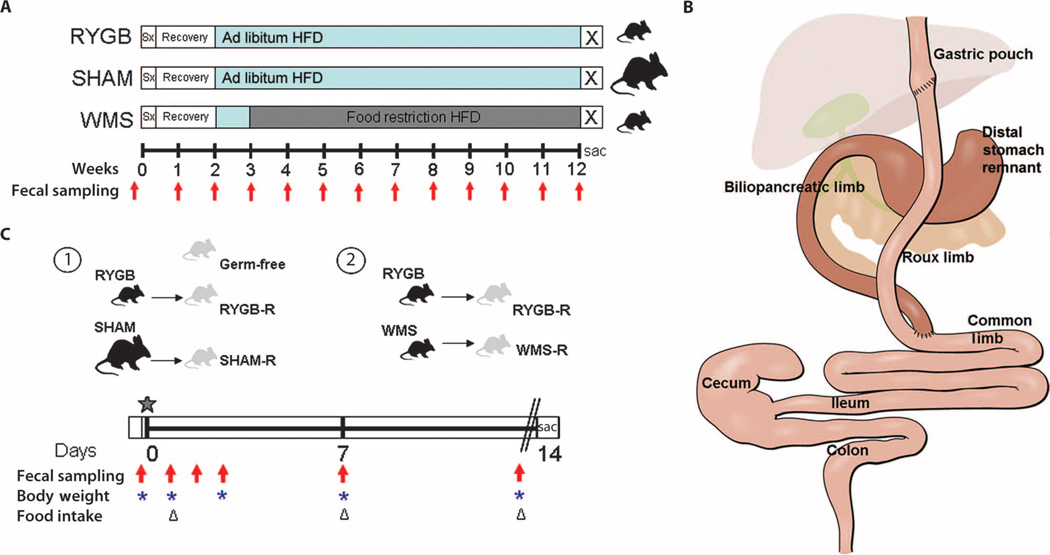
Fig. 1.
Schematic of experimental design. (A) DIO C57BL/6J mice fed a 60% HFD underwent either RYGB or sham operations and 2-week recovery on liquid diet before returning to HFD (blue bars). Sham animals that had successfully regained body weight within 3 weeks after surgery were divided into an ad libitum–fed SHAM group or food-restricted to match the weight of the RYGB animals (WMS). Fresh fecal samples were collected preoperatively and weekly for 12 weeks after surgery for microbiota analysis (red arrows). (B) Graphic of the RYGB anatomy and segments collected for luminal content and mucosal scrapings along the length of the gastrointestinal tract. Segments representative of the RYGB anatomy were also collected in SHAM and WMS animals. (C) Design of microbiota transfer experiments of the cecal contents from a representative donor animal from each group, depicted in (A), into germ-free mice (star), indicating timing of collection of fecal samples for microbiota analysis (red arrows), body weights (blue asterisks), and food intake (triangles). At the end of the colonization period, animals were fasted overnight (double lines), and final body weights, serum metabolic parameters, and adiposity scores were obtained.
RESULTS
The effect of RYGB on adiposity in the mouse model
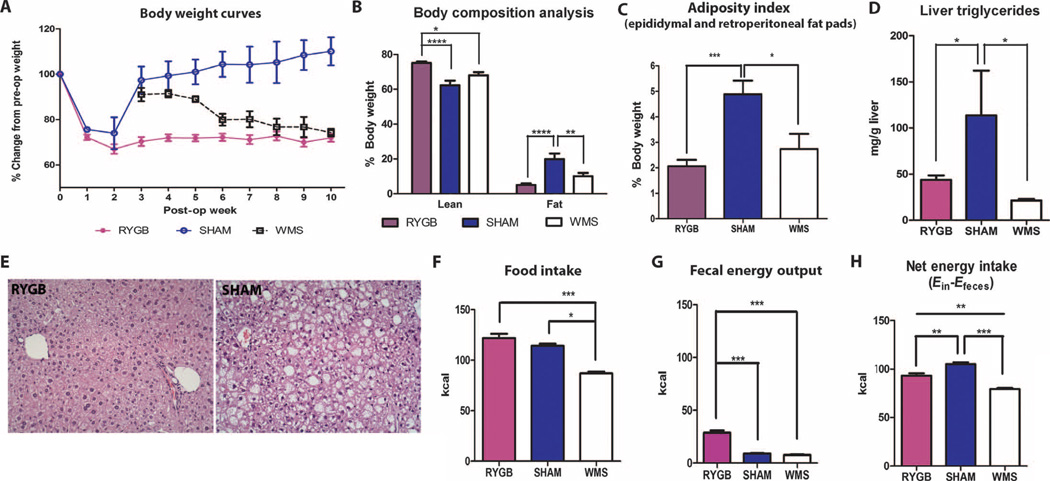
Diet-induced obese (DIO) C57BL/6J mice fed a high-fat diet (HFD) commonly develop obesity-related metabolic derangements, including glucose intolerance, hyperglycemia, hyperinsulinemia, and insulin resistance (25). Within 3 weeks after RYGB, male C57BL/6J mice lost 29 ± 1.9% of their initial body weight and remained at this lower weight throughout the study period. In contrast, ad libitum–fed sham-operated (SHAM) animals regained their body weight within 2 to 3 weeks after surgery (Fig. 2A; 32.8 ± 0.5 g versus 42.2 ± 2.1 g after 5 weeks; P = 0.007, Student’s t test). Weight-matched sham-operated (WMS) animals were fed ~25% fewer calories to match the weight of the RYGB animals. As we have seen previously (23, 24), the decreased body weight after RYGB reflected a preferential loss of fat mass and a preservation of lean mass compared to sham controls (Fig. 2B). Epididymal and retroperitoneal fat pad weight, the degree of hepatic steatosis, and liver triglyceride content were all reduced in both RYGB and WMS animals relative to SHAM animals (Fig. 2, C to E). Food intake was not different between RYGB and SHAM controls (Fig. 2F); however, taking into account the greater energy lost in the feces of RYGB animals (Fig. 2G), the net energy intake in RYGB animals was intermediate to that of SHAM and WMS controls (Fig. 2H and table S1). This finding indicates that the weight loss in response to RYGB is due both to a decrease in net energy intake and to an increase in total energy expenditure. Increased energy expenditure after RYGB has been previously demonstrated in both rat and mouse models (6–8).
Fig. 2.
Phenotypic data from the RYGB mouse model. Characteristics of DIO C57BL/6J mice undergoing either RYGB (n = 11 to 17), sham operation (SHAM; n = 4 to 6), or sham operation with weight matching to the RYGB group by food restriction (WMS; n = 5 to 6). (A) Body weight curves after surgery. (B) Body composition analysis. (C) Adiposity index calculated from epididymal and retroperitoneal fat pad weights. (D) Liver triglyceride content. (E) Liver histology (×20). (F to H) One-week cumulative (F) food intake, (G) fecal energy output, and (H) net energy intake in RYGB (n = 14), SHAM (n = 11), and WMS (n = 6) animals. Measurements for (B) to (E) were taken at the end of study, 15 weeks after surgery. Food intake and energy output studies were performed 4 to 6 weeks after surgery. *P < 0.05, **P < 0.01, ***P < 0.001, one-way analysis of variance (ANOVA) and post hoc Tukey test. Values represent means ± SEM.
RYGB animals maintained on HFD exhibited improved plasma glucose and insulin levels at 15 weeks after surgery, compared to SHAM animals, mirroring the metabolic phenotype of normal chow (NC)–fed lean mice (table S2). Although weight loss by food restriction yielded similar improvements in fasting plasma glucose and insulin levels, greater improvements in glucose tolerance and insulin sensitivity were seen in RYGB versus WMS animals (fig. S1), supporting human evidence that RYGB has weight-independent effects on glucose metabolism (26).
RYGB rapidly alters the distal gut microbiota
To determine how RYGB affects the distal gut microbiota, we performed 16S ribosomal RNA (rRNA) gene sequencing on fecal samples collected before and weekly for 3 months after intervention in the RYGB, SHAM, and WMS groups (Fig. 1A). Overall, we analyzed 166 samples from 23 animals (109,015 ± 5842 16S rRNA gene sequences per sample; table S3).
RYGB markedly altered the composition of the distal gut microbiota as early as 1 week after surgery, a change that progressed over time and stabilized after 5 weeks (Fig. 3 and fig. S2). The sham procedure also affected the fecal microbial communities but to a substantially lesser extent than RYGB; furthermore, the differences in microbial ecology between the SHAM and WMS groups were minimal (Fig. 3 and fig. S2). These observations suggest that rearrangement of the gastrointestinal tract by RYGB had a substantially greater effect on the fecal microbiota than either food restriction–mediated weight loss or the limited intestinal disruption caused by the sham procedure.
Fig. 3.
RYGB causes marked, rapid, and sustained changes in gut microbial ecology that are independent of weight and diet. (A) Heat map of pairwise Spearman rank correlations between species-level OTUs from fecal samples, ordered by treatment, individual, and time. Within-individual rank correlations in RYGB, SHAM, and WMS mice (n = 4 to 5 animals per group) compare weekly postoperative samples to a preoperative sample. Rank correlations between endpoint samples taken from unoperated DIO, unoperated NC-fed (NC), and RYGB-operated mice maintained on NC (NC-RYGB) are also included (n = 2 to 4 per group). Each correlation is colored from dark blue (no correlation) to dark red (perfect positive correlation). (B) Temporal effects of gastric bypass on overall community membership among fecal samples from RYGB (pink circles), SHAM (orange squares), and WMS (olive triangles) animals [first principal coordinate from an unweighted UniFrac-based analysis over time]. Includes endpoint fecal samples from age-matched DIO (purple circle), NC (green triangle), and NC-RYGB (blue diamond) mice. Values represent means ± SEM.
Additionally, we analyzed fecal samples at a single time point from unoperated DIO, unoperated lean NC-fed mice, and RYGB-operated mice maintained on NC (NC-RYGB). As expected, the abundance of species-level operational taxonomic units (OTUs) in samples from DIO and lean animals was significantly different (Fig. 3A; Spearman correlation range was 0.58 to 0.84 and 0.16 to 0.24 for within-group versus between-group comparisons, respectively; P < 10−7, Student’s t test). However, the effect of RYGB on the gut microbiota was similar regardless of whether mice were fed NC or HFD (Fig. 3), suggesting that the impact of surgery can overwhelm even the well-recognized effect of dietary composition on the gut microbiota (14).
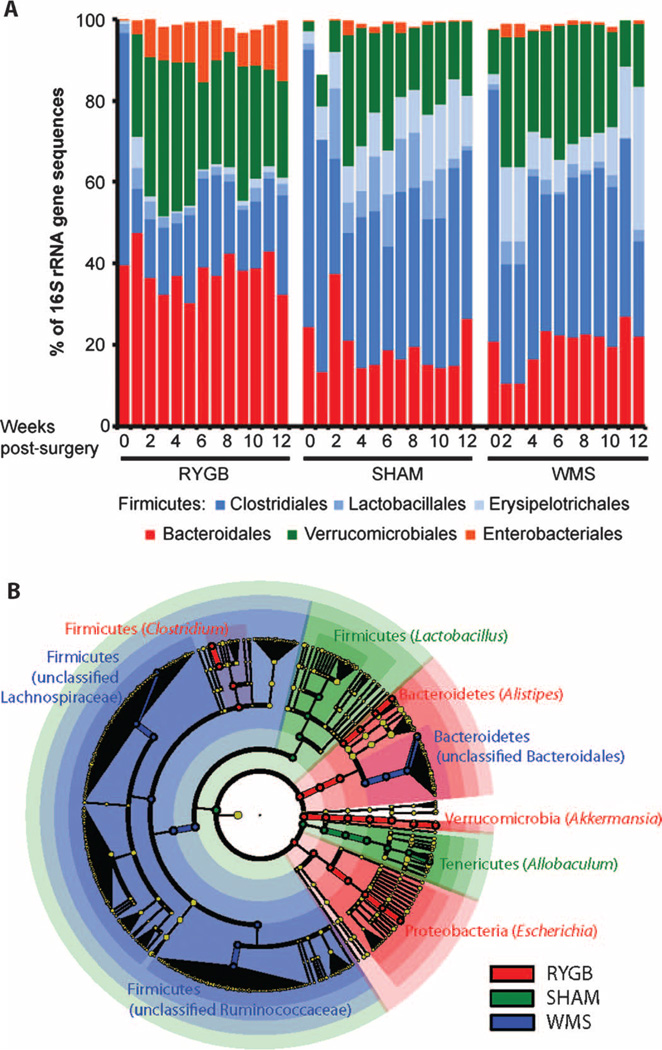
Weight loss after dietary restriction or RYGB was accompanied by similar changes in the abundance of taxonomic groups within the Firmicutes phylum as a proportion of the total sequences observed in the phylum. Compared to preoperative levels, both the RYGB and WMS microbiota were dominated by the order Clostridiales (78.9 ± 2.7% and 72.9 ± 2.7% of Firmicutes 16S rRNA gene sequences, respectively), whereas the SHAM microbiota had a significantly greater abundance of the orders Lactobacillales and Erysipelotrichales [42.5 ± 4.0% (SHAM), 27.1 ± 3.8% (WMS), and 21.1 ± 2.7% (RYGB); P < 0.01, SHAM versus WMS and SHAM versus RYGB, Student’s t test; Fig. 4A and table S4]. In contrast, the relative abundance of Bacteroidales remained stable when comparing pre- and postoperative abundances across all treatment groups over time (Fig. 4A).
Fig. 4.
Bacterial taxonomic groups that discriminate among RYGB-, SHAM-, and WMS-derived samples. (A) Average relative abundance of bacterial orders in RYGB, SHAM, and WMS mice before and up to 12 weeks after surgery. (B) LEfSe-derived (27) phylogenetic tree depicting nodes within the bacterial taxonomic hierarchy that are significantly enriched in fecal samples from RYGB (red), SHAM (green), and WMS (blue) mice. Significant phyla are labeled, with the genera in parentheses. LEfSe was used with the default parameters (n = 5000 sequences per sample; OTUs with <10 sequences and preoperation samples removed).
RYGB induced several specific changes in the gut microbial ecology including an increased abundance in the Enterobacteriales within the first 2 weeks after surgery; there was no change in the Enterobacteriales in either the SHAM or WMS groups (Fig. 4A and fig. S3A). There was also a significantly greater increase in Verrucomicrobiales within the first 2 weeks after RYGB (10,000-fold on average; P < 0.05, Student’s t test) compared to preoperative levels that was more marked than that observed after either SHAM or WMS treatment (3-fold, P < 0.05 for both; Fig. 4A). Using the linear discriminant analysis (LDA) effect size (LEfSe) (27) method, we identified more specific bacterial taxa and species-level phylotypes whose relative abundance varied significantly among fecal samples taken from the RYGB, SHAM, and WMS treatment groups. This analysis revealed 50 discriminative features (LDA score >2; Fig. 4B and table S4). RYGB microbiotas were enriched for three distinct taxonomic groups, evident from phylum level—Bacteroidetes, Verrucomicrobia, and Proteobacteria—to genus level—Alistipes, Akkermansia, and Escherichia, respectively (Fig. 4B). The relative abundance of Archaea in fecal samples, as determined by quantitative polymerase chain reaction (PCR), was also found to be greater in RYGB animals relative to SHAM animals (fig. S4, A and B). However, we were unable to detect the presence of methanogens using two previously validated primers targeting methyl coenzyme M reductase subunit A, a conserved enzyme in the methanogenesis pathway (fig. S4, C and D) (28, 29).
To determine how gastric bypass affects the spatial structure of the gut microbiota, we sampled the luminal and mucosal adherent microbial communities at eight sites spanning the stomach, small intestine, cecum, and colon of mice 15 weeks after RYGB, SHAM, or WMS treatments (n = 3 to 6 samples per site per treatment; 207 samples; 84,758 ± 3873 sequences per sample; Fig. 1B and table S5). Comparisons of microbial community membership using the unweighted UniFrac metric revealed that the distal stomach, ileal, cecal, and colonic microbiota were all strongly affected by RYGB (Fig. 5). As in the fecal samples, the aggregate RYGB microbiota had a significantly higher abundance of Enterobacteriales (16.7 ± 1.3%), Bacteroidales (28.7 ± 1.9%), and Verrucomicrobiales (17.6 ± 1.4%) compared to SHAM controls (5.0 ± 0.6%, 16.7 ± 1.8%, and 6.0 ± 1.0%, respectively; P < 10−5 for each, Student’s t test). The observed changes were qualitatively consistent throughout the gastrointestinal tract, with some notable exceptions (Fig. 5B). For example, the relative abundance of the Enterobacteriales increased to the greatest extent in the luminal contents of the distal gut, as well as in the intestinal segments not excluded from nutrient flow and in the mucosal adherent communities of the colon, ileum, and Roux limb (fig. S3B). Microbial communities in the mucosal and luminal content samples were similar across segments within both RYGB and SHAM animals (Fig. 5A); however, the luminal and mucosal adherent populations of WMS animals diverged throughout the small intestinal segments, including the biliopancreatic, Roux, and common limbs (Fig. 5). The latter result suggests that bacteria localized at the mucosal surface in the small intestine may be more responsive to food-restricted weight loss than to RYGB, where the effects are seen primarily in the distal intestine, cecum, and colon.
Fig. 5.
Relative abundance of bacterial taxa throughout the gastrointestinal tract of RYGB, SHAM, and WMS mice. (A) Spatial effects of gastric bypass. The first principal coordinate from an unweighted UniFrac-based analysis is shown for luminal and mucosal samples taken from the stomach, small intestine, cecum, and colon. Values represent means ± SEM. DS, distal stomach; GP, gastric pouch; BP, biliopancreatic limb. (B) The dominant bacterial orders are shown for samples from the stomach, small intestine, cecum, and colon. Mean values are shown for samples isolated from the lumen (blue squares) and mucosa (red squares).
To elucidate potential selection pressures that could be responsible for the observed changes in microbial ecology after RYGB, we analyzed fecal samples from RYGB, SHAM, and WMS animals for fecal fat and nitrogen content and pH (table S1). Fecal samples from RYGB mice had significantly lower pH and higher fat content than fecal samples from SHAM and WMS mice. In contrast, fecal nitrogen content was lower in both RYGB and WMS samples relative to SHAM. Furthermore, pH within the distal stomach was significantly increased in RYGB mice (3.8 ± 0.6; n = 8) relative to SHAM mice (1.9 ± 0.7; n = 5; P < 0.05, Mann-Whitney test), potentially due to the loss of direct stimulation of acid secretion by food. These changes underscore the presence of an altered luminal milieu after RYGB that could influence gut microbial ecology (14, 30).
Decreased host adiposity is transmissible through the microbiota
Given that previous studies have shown that host adiposity and glucose homeostasis can be affected by the gut microbiota (20, 31), we sought to determine whether any of the metabolic changes associated with RYGB could be directly influenced by RYGB-induced modulation of the gut microbiota. We tested this possibility by inoculating lean, germ-free mice with cecal contents from RYGB donors (RYGB recipient or RYGB-R mice) and comparing basal phenotypic outcomes to those of uninoculated germ-free mice and germ-free mice inoculated with cecal contents from SHAM or WMS donors (SHAM-R and WMS-R animals, respectively; Fig. 1C). During a 2-week colonization period, RYGB-R animals exhibited a significant decrease in body weight (−5.0 ± 1.8%; P < 0.05, one-sample t test), whereas both SHAM-R and germ-free control animals exhibited no significant weight change (0.2 ± 1.1% and 2.5 ± 1.4%, respectively; Fig. 6, A and B). Food intake in SHAM-R animals was significantly decreased compared to germ-free controls, similar to what has been seen previously [P < 0.01, SHAM-R versus germ-free, Student’s t test (20)], whereas food intake in RYGB-R animals was not affected (P = 0.39, RYGB-R versus germ-free, Student’s t test; Fig. 6C). A similar trend toward increased food intake in RYGB-R mice relative to SHAM-R mice (Fig. 6C) was also seen relative to WMS-R mice (fig. S5A). Both SHAM-R and WMS-R animals exhibited a similar increase in overall adiposity (fig. S6), including relative fat pad weights, at the end of the colonization period. Consolidating both sham groups together, the recipients of cecal transfer from sham donors had a significantly greater fat mass than either RYGB-R or uninoculated germ-free controls [P < 0.01 (germ-free versus SHAM-R) and P < 0.05 (RYGB-R versus SHAM-R), Student’s t test; Fig. 6D and fig. S5B], a difference that was also reflected in plasma leptin levels (P < 0.05, RYGB-R versus SHAM-R, Student’s t test; Fig. 6E and fig. 5C). Fat pad weight did not increase in the RYGB-R animals relative to the uninoculated germ-free controls (Fig. 6D). Liver weights were no different among groups, but there was a trended increase in liver triglyceride levels in the SHAM-R animals (Table 1).
Fig. 6.
Decreased weight and adiposity is transmissible via the gut microbiota. (A) Body weight curves for SHAM-R (n = 6) and RYGB-R (n = 10) mice, represented as change from initial body weight. (B) Change in body weights (BW) among the groups relative to baseline. (C) Cumulative food intake over the 13-day colonization period. (D and E) Visceral fat pad weights (D) and plasma leptin levels (E) in the RYGB-R (n = 10), uninoculated germ-free (n = 7), and consolidated SHAM-R (n = 10) groups. Values in (A) to (E) are means ± SEM. *P < 0.05, **P < 0.01, ANOVA post hoc test. Representative of three experiments. (F) Relative abundance of bacterial taxa in recipient animals after gavage with cecal contents from RYGB, SHAM, and WMS donors. Mean values across each time point (1 to 13 days after gavage) are shown (n = 3 to 15 samples per time point; 10,000 sequences per sample).
Table 1.
Fasting blood parameters of germ-free (GF) recipient animals receiving cecal contents from RYGB-R, SHAM-R, and GF mice. Values represent means ± SEM. Data annotated by different letters are significantly different (P < 0.05, ANOVA).
| RYGB-R | SHAM-R | GF | |
|---|---|---|---|
| n | 15 | 10 | 7 |
| Blood glucose (mg/dl) | 148 ± 5.5A | 147 ± 7.4A | 120 ± 3.9B |
| Insulin (ng/ml) | 0.46 ± 0.10A | 0.99 ± 0.19A | 0.73 ± 0.19A |
| HOMA-IR | 4.4 ± 0.98A | 9.8 ± 4.1A | 5.5 ± 1.5A |
| Serum triglyceride (mg/dl) | 77.8 ± 9.7A | 113 ± 8.5B | 73.5 ± 16.8A |
| Serum NEFA (meQ/liter) | 0.58 ± 0.06A | 0.65 ± 0.06A | 0.62 ± 0.05A |
| Liver weight (% body weight) | 4.9 ± 0.10A | 4.8 ± 0.20A | 4.8 ± 0.18A |
| Liver triglyceride (mg/g liver) | 22.9 ± 4.2A | 36.2 ± 9.1A | 21.6 ± 6.0A |
Although marked changes in serum glucose, insulin, and triglycerides in lean recipient mice fed a standard chow diet were not expected, there was a trend toward improved insulin sensitivity, estimated by homeostasis model assessment of insulin resistance (HOMA-IR), and significantly reduced fasting triglyceride levels in RYGB-R mice relative to SHAM-R mice (Table 1). Glucose tolerance and insulin tolerance in the recipients were not measured.
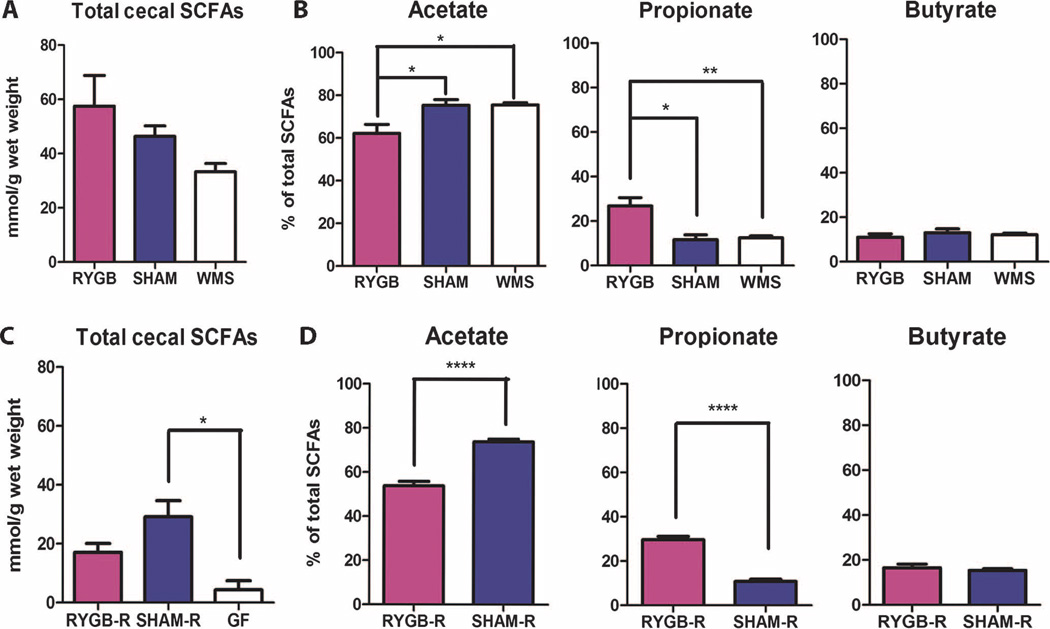
In an attempt to find potential microbiota-derived signals that could be directing the loss of body weight and adiposity in recipient animals, we examined both ileal fasting-induced adiposity factor (FIAF) gene expression and cecal short-chain fatty acid (SCFA) composition. Colonization with either SHAM-R or RYGB-R microbiota into germ-free animals caused a similar decrease in ileal FIAF expression (fig. S7), consistent with previously published results (20). Cecal SCFAs, however, differed among the germ-free, RYGB-R, and SHAM-R groups (Fig. 7, C and D). Microbial colonization of germ-free mice increased SCFA production, as indicated by the increase in cecal SCFA levels, with SHAM-R animals producing significantly more SCFAs relative to germ-free animals (P < 0.05, Student’s t test) and RYGB-R animals producing an intermediate quantity of cecal SCFAs (Fig. 7C). This difference is not as apparent in the operated donor groups (Fig. 7A); however, the relative proportion of acetate, propionate, and butyrate levels were maintained in both donor and recipient animals. The acetate/propionate/butyrate ratios in SHAM and WMS animals were 75:12:13, similar to those seen in SHAM-R animals (74:11:15); in contrast, the acetate/propionate/butyrate ratios in RYGB and RYGB-R animals were in the same range at 62:27:11 and 54:30:16, respectively (Fig. 7, B and D).
Fig. 7.
SCFA levels are consistent between donor and recipient animals. (A and B) Total cecal SCFAs (A) and percentage of total SCFAs (B) of acetate, propionate, and butyrate in RYGB-operated (n = 6), SHAM-operated (n = 4), and WMS-operated (n = 5) animals. (C) Total cecal SCFAs of RYGB-R (n = 5), SHAM-R (n = 6), and germ-free (GF; n = 4) mice 2 weeks after colonization of their respective operated donor cecal microbiota. (D) Percentage of total SCFA of acetate, propionate, and butyrate in RYGB-R and SHAM-R mice. Values represent means ± SEM. *P < 0.05, **P < 0.01, ****P < 0.001, one-way ANOVA post hoc Tukey test or Student’s t test.
To identify RYGB-specific microbial biomarkers that may contribute to the recipient phenotype, we collected fecal samples from RYGB-R, SHAM-R, and WMS-R animals at 1, 2, 3, 7, and 13 days after colonization with their respective donor microbiota and searched for bacterial taxonomic groups shared between the donor and recipient animals. 16S rRNA gene sequencing was performed on 91 total samples from 26 recipient animals (n = 5 to 6 mice per treatment group per experiment; 53,739 ± 2740 sequences per sample; table S6). All three treatment groups had high levels of Enterobacteriales during the first 2 days after colonization, whereas the Bacteroidales consistently became the dominant taxa by day 13. The levels of Verrucomicrobiales, however, remained significantly higher in RYGB-R samples (30.1 ± 1.6% of 16S rRNA gene sequences) than in either SHAM-R (9.3 ± 1.7%; P < 10−4) or WMS-R (14.6 ± 2.8%; P < 10−4, Student’s t test) controls (Fig. 6F). Analysis with LEfSe confirmed the enrichment for Akkermansia as well as Alistipes (Bacteroidetes phylum) in RYGB-R fecal samples, similar to what we observed in the donor animals (table S7).
DISCUSSION
Rodent models of RYGB assist in identifying potential mechanisms of weight loss, fat loss, and metabolic improvements in ways that would otherwise be limited in clinical studies. Here, we show that changes in microbial ecology after RYGB are due to gastrointestinal reconfiguration and not merely due to the associated changes in weight loss, diet, or intestinal transection. Furthermore, we demonstrate that this altered microbiota is sufficient to trigger a reduction in host body weight and adiposity.
Our study was designed to minimize potential interindividual variation by assessing within-individual changes in gut microbial ecology before and after surgery. Marked changes in this ecology were observed within 1 week after RYGB, with a pronounced increase in the abundance of the Verrucomicrobia (genus: Akkermansia) and Gammaproteobacteria (order: Enterobacteriales). These changes were similar to those observed in the fecal microbiota of human patients (21, 22) and in nonobese, NC-fed rats (9) that had undergone RYGB, and were observed in RYGB mice fed either a high-fat or standard chow diet. Several studies have demonstrated the role of dietary composition in modulating microbial ecology and diversity (14–16, 32); however, it is clear that RYGB produces unique selective pressures to modulate community structure that is not merely a reversion to a community seen in the lean, NC-fed state (14, 17). Together, many aspects of the microbial response to gastric bypass surgery are conserved among humans, rats, and mice, despite significant differences in the specific microbial strains and species found within their gastrointestinal tracts (13, 33).
Given the increase in Gammaproteo-bacteria in humans (21, 22), rats (9), and now our mouse model of RYGB, we reasoned that this population could be a key contributor to regulating host metabolic outcomes after surgery. Escherichia, the genus most highly enriched after RYGB in the mouse, includes several pathogenic strains associated with metabolic syndrome, obesity, and insulin resistance (19, 34) as well as nonpathogenic commensal species that have been used as probiotic agents to prevent gastrointestinal inflammatory conditions (35). It is possible that the specific Escherichia population enriched after RYGB may have some beneficial role in driving host metabolic improvements after surgery.
The Verrucomicrobium Akkermansia was also substantially increased in RYGB mice and was maintained after transfer to germ-free recipients. Given this observation, it is possible that Akkermansia may have a substantial role in regulating host adiposity and weight loss. Akkermansia can use mucus as a sole source of carbon and nitrogen in times of health and particularly in times of caloric restriction (36). This observation likely explains why Akkermansia is selectively increased in the mucus layer of WMS mice. The physiological relevance of the overall increase in Akkermansia throughout the RYGB-altered gastrointestinal tract, and how this may differ from the site-specific increases seen in WMS animals, remains to be determined. Additional studies are needed to elucidate whether increased foraging on host mucins or altered inflammation and insulin sensitivity in response to Akkermansia (37, 38) contribute to the metabolic outcomes after RYGB. In addition, we cannot discount the potential interactions among Enterobacteriales, Akkermansia, and other enriched bacterial groups (such as Alistipes) in the RYGB model and how they may work independently or interdependently to influence host metabolic improvements.
Microbial community structure is likely altered after RYGB through rerouting of nutrients and biliopancreatic secretions. Although we did see changes in fecal fat and pH profiles in the RYGB animal, these changes would be expected to promote the growth of Firmicutes (17, 30) rather than the observed decrease in Firmicutes after surgery. Therefore, the precise factors responsible for modulating microbial communities have yet to be defined.
Phenotypic characterization of germ-free recipient mice receiving a microbiota transplant from RYGB-operated donors highlights the potential functional and metabolic contributions of the RYGB microbiota. We found that RYGB-R animals had reduced body weight and decreased fat deposition compared to SHAM-R and uninoculated germ-free controls. WMS-R animals shared similar body weight and adiposity phenotypes as the SHAM-R animals (fig. S5), likely due to the similar cecal microbial profiles between the WMS and SHAM donors (Fig. 3C and fig. S3), suggesting that the effects of the RYGB microbiota on germ-free recipients are unique to this particular microbial profile. This was a surprising observation because inoculation of the entire complex microbial communities (14, 19, 20) or specific microorganisms (39) from either lean or obese mice has previously been shown to cause an increase in epididymal fat pad weight and overall adiposity in the face of decreased food intake (10, 20). Although the degree of weight loss by the RYGB microbiota into lean recipient mice is not as profound as the effect of RYGB in obese mice (5% versus 29% weight loss, respectively), these findings are consistent with the hypothesis that alterations in the gut microbiota after RYGB at least partially modulate body weight and adiposity.
A decrease in adiposity and body weight without a change in food intake suggests that the RYGB-associated microbiota may either reduce the ability to harvest energy from the diet or produce signals regulating energy expenditure and/or lipid metabolism. Previous studies had identified ileal FIAF as a microbiota-influenced mechanism promoting lipid uptake into adipose tissue (20, 31). Colonization of germ-free recipients with either a RYGB or SHAM microbiota resulted in equally reduced ileal FIAF expression despite the differences in fat pad weight, suggesting that the effect of the RYGB microbiota on metabolic outcomes is independent of FIAF.
SCFAs are by-products of microbial fermentation that affect host physiology in several ways, including serving as a primary energy source for colonocytes, substrates for lipid storage in adipose tissue, immune cell modulators, and molecules regulating lipid and glucose metabolism and appetitive drives via G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor (GPR41 and GPR43) activation and signaling (40, 41). Increased adiposity has been demonstrated in previously germ-free recipients of gut microbiota contents, likely due to increased availability of SCFAs (10, 17). Indeed, an increased production of SCFAs in SHAM-R animals relative to RYGB-R and germ-free animals may explain the differences in adiposity among the groups. However, these observations do not explain the physiological differences among the RYGB, SHAM, and WMS groups, which do not exhibit significant differences in total cecal SCFA content.
It is possible that the different SCFAs affect host metabolic physiology in different ways. Both RYGB and RYGB-R animals had relatively greater propionate and lower acetate production than the WMS, SHAM, and SHAM-R groups, likely a consequence of the different microbial profiles seen in the RYGB and RYGB-R animals (tables S4 and S7) (42). The decreased adiposity in RYGB-R animals relative to SHAM-R animals could be due to the reduced level of acetate available for lipogenesis by adipose tissue and peripheral tissues (43). Furthermore, propionate is known to inhibit acetate conversion into lipid in the liver (44) and adipose tissue (45), and may contribute to the lowered adiposity and serum triglyceride levels and a trend toward decreased hepatic triglyceride content. Propionate is also thought to inhibit food intake through GPR41- and GPR43-induced secretion of satiety-regulating gastrointestinal hormones (46), although this is less likely given the lack of change in food intake in the RYGB-R animals.
Given the well-established effect of RYGB to increase energy expenditure (7, 8), we cannot discount the possibility that a reduction in body weight without a change in food intake is due to altered microbial signaling that up-regulates host energy expenditure. Male mice lacking GPR41 have reduced energy expenditure and increased adiposity (47), suggesting that SCFAs regulate energy expenditure via GPR41. Among the known SCFAs, propionate has the highest affinity for GPR41, and its administration has been shown to increase sympathetic activity, resulting in elevated energy expenditure (48). Therefore, the increase in energy expenditure after RYGB could be mediated, in part, through enhanced propionate activation of the GPR41 pathway. Another potential mechanism is differential microbial modification of bile acids, which are also signaling molecules that can influence energy expenditure (49). Additional studies are necessary to further clarify the specific contributions of these various microbially influenced metabolites on changes in host energy balance and metabolism after RYGB.
It is clear from studies in our laboratory and others that RYGB improves glucose metabolism in animals and in people (50). However, the degree to which the RYGB-altered microbiota mediates those improvements when administered to germ-free mice remains uncertain. In these experiments, recipient mice were lean and maintained on NC. Although there were no changes in fasting glucose levels between RYGB-R and SHAM-R mice, the RYGB-R group exhibited a trend toward lower fasting insulin levels than either SHAM-R or germ-free mice. It is possible that, in this model system, the effect of the microbial community on host glucose metabolism is diet-dependent. Therefore, future microbiota transplantation studies in germ-free recipients fed a HFD are warranted.
In summary, these studies have given us preliminary insights to the potential contributions of the RYGB-associated gut microbiota, and particularly SCFAs, in regulating host physiology, energy balance, and metabolism (fig. S8). Future studies incorporating surgery, metagenomics, gnotobiotics, and genetic deletions for key metabolic signaling pathways promise to expand our understanding of the gut microbiota in shaping host energy balance and the metabolic response to surgical intervention.
MATERIALS AND METHODS
Animals
Male C57BL/6J DIO mice were purchased at 22 to 26 weeks of age (Jackson Laboratories) and maintained on a 60% HFD (Research Diets, D12492) until they reached a preoperative weight of 40 to 50 g. All surgically operated animals were individually housed in cages with wire floor throughout the study period. Male, age-matched (7 to 10 weeks old), germ-free Swiss Webster mice were obtained from Taconic. All animal studies were performed under protocols approved by the Massachusetts General Hospital and Harvard Medical School Institutional Animal Care and Use Committees.
Surgery, postoperative care, and diet
The surgical approach for RYGB mice is a modified version of that previously described (8, 23), the only difference being that the glandular and nonglandular portions of the stomach were double-sutured and transected to form the distal stomach and gastric pouch, respectively (Fig. 1B). With this approach, we achieved less than 20% mortality over the course of the experiment. Sham animals were treated in a manner similar to the RYGB animals, with a single transection just distal to the ligament of Treitz followed by reanastomosis to restore the preoperative intestinal anatomy.
Operated animals did not receive postoperative antibiotics or anti-inflammatory drugs, but were given buprenorphine (0.05 mg/kg, intramuscularly) for pain. Animals were maintained on a liquid diet (Vital HN, Abbott Laboratories) for 2 weeks until weaned back onto solid HFD. Body weights were monitored weekly. Three weeks after surgery, a group of weight-stabilized SHAM animals was randomly chosen and maintained on a restricted diet of about 75% of the calories consumed daily by RYGB-treated mice so as to match the weight of the RYGB animals (WMS). With constant adjustment of the daily food intake of these animals, weight matching with the RYGB group was stabilized by 6 weeks after surgery (3 weeks after start of food restriction).
Food intake and fecal analysis
A subgroup of animals was housed singly on wire floors, and food in the hopper and food spilled were weighed every 3 to 4 days. All fecal pellets were collected, weighed, and submitted to the University of Arkansas Central Analytical Laboratory (http://www.uark.edu/ua/cal/) for measurements of fecal calorie, crude fat, and protein (nitrogen) and pH. The net intake for each component (energy, fat, or protein) was calculated by subtracting the energy expelled in the stool from the total amount consumed during a 1-week period.
Fecal sampling, processing, and analysis
Fecal samples were collected weekly and stored at −80°C until processing. DNA was extracted with the PowerSoil bacterial DNA extraction kit (MO-BIO) and PCR-amplified with universal bacterial primers targeting variable region 4 of the 16S rRNA gene as described previously (51). Amplicons were sequenced with the Illumina HiSeq platform (52). Multivariable statistical analysis was used to compare microbial composition among the treatment groups. 16S rRNA gene sequences were analyzed with the QIIME [Quantitative Insights Into Microbial Ecology (53)] software package along with custom Perl scripts to analyze α (within-sample) and β (between-sample) diversity. The LEfSe package was used to identify taxonomic groups significantly associated with each treatment (27). Spearman rank correlations were calculated based on the number of sequences assigned to abundant species-level phylotypes in each fecal microbiota (phylotypes with ≥100 sequences across a combined data set were included; 10,000 randomly subsampled sequences were included per sample).
Tissue harvest and intestinal axis sampling
Mice were sacrificed between 12 and 15 weeks after surgery. Before sacrifice, animals were fasted for 2 hours and blood glucose was measured from the tail tip with a handheld glucometer (AlphaTRAK, Abbott Laboratories). The animals were then anesthetized with sodium pentobarbital (100 mg/kg, intraperitoneally) and euthanized by cardiac exsanguination. Plasma was isolated and stored at −80°C. The following intestinal sections were collected for bacterial DNA analysis: gastric pouch, distal stomach remnant, biliopancreatic limb, Roux limb, common limb, ileum, cecum, and colon. Sections representative of the intestinal segments comprising each limb were also collected in SHAM animals (Fig. 1B). Each segment was flushed with extraction buffer (200 mM NaCl, 200 mM tris, 20 mM EDTA, pH 8.0) to retrieve luminal bacterial contents. Each section was then cut longitudinally, and the mucosa was scraped off with slides for assessment of mucosal adherent bacterial populations. All contents and mucosal adherent samples were flash-frozen in liquid nitrogen and stored at −80°C until DNA extraction. In a separate group of animals, pH was measured in different segments of the gastrointestinal tract with a micro-pH electrode (Thermo Scientific). Epididymal and retroperitoneal fat pads were collected and weighed as a biomarker for visceral adiposity. Whole-body lean and fat masses were determined by time-domain nuclear magnetic resonance (Bruker TD Minispec).
Germ-free mouse experiments
Cecal contents from a donor animal representing each group (RYGB, WMS, and SHAM) were saved in reduced anaerobic phosphate-buffered saline and homogenized within an anaerobic chamber. The resultant slurry was administered by oral gavage to germ-free mice (five to six animals per group). Animals were housed one to two per cage on wire floors and fed autoclaved rodent breeder chow. Uninoculated germ-free mice were used as controls. Body weight and cumulative food intake were measured and recorded weekly. Fecal pellets were collected at days −1, 1, 2, 3, 7, and 13 days after gavage. On day 13, food was withheld from animals overnight. The next morning, fasting blood glucose was measured in blood collected from the tail tip. After euthanasia, a terminal blood collection was taken, tissue was harvested, the liver and visceral (retroperitoneal and epididymal) fat pads were weighed, and cecal contents were obtained for SCFA analysis.
Biochemical assays
Plasma insulin was measured with the mouse ultrasensitive insulin enzyme-linked immunosorbent assay (ELISA) (Alpco). Fasting triglyceride and nonesterified fatty acids (NEFAs) were assayed in serum with kits for these analytes (Wako Chemicals). Liver triglycerides were measured with free glycerol reagent (Sigma) after ethanolic KOH extraction. Plasma leptin was measured by ELISA (Crystal Chem). HOMA-IR, an estimate of insulin resistance, was calculated as follows: [fasting glucose (mg/dl) × fasting insulin (µU/ml)]/405. Cecal SCFA content was determined by gas chromatography.
Gene expression
Total RNA was extracted with TRIzol reagent (Invitrogen), and complementary DNA was produced with the SuperScript III First-Strand Synthesis System (Invitrogen). TaqMan quantitative PCR was performed with primers for FIAF (Mm00480431_m1, Applied Biosystems). Gene expression was normalized to the housekeeping gene β-actin.
Statistical analysis
For in vivo physiology experiments, all data are expressed as means ± SEM. Statistical analyses were performed with GraphPad Prism (v. 5). The threshold of statistical significance was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank C. Maurice for helpful advice; D. Gootenberg, S. Lajoie, and A. Pfalzer for technical assistance; N. Stylopoulos for contributions to surgical model development; C. Reardon and C. Daly of the Bauer Core Facility for sequencing; L. Kirby (University of Arkansas Central Analytical Laboratory) for fecal analysis; and V. Yeliseyev, M. Delaney, and L. Bry of the Harvard Digestive Diseases Center Gnotobiotic Core Facility for technical assistance, advice with the microbiota transfer studies, and SCFA analysis of cecal samples.
Funding: NIH DK088661 (L.M.K.), NIH P50 GM068763 (P.J.T.), F32 DK095561 (A.P.L.), P30DK034854 (Harvard Digestive Diseases Center), and Ethicon Surgical Care (L.M.K.).
Footnotes
Author contributions: A.P.L., P.J.T., and L.M.K. designed the experiments and wrote the paper; M.P. produced surgical mouse models; J.-M.L. performed 16S sequencing of samples; P.J.T. analyzed 16S sequencing data; A.P.L. collected and processed the samples, performed metabolic phenotyping experiments, and analyzed related data; S.M. provided technical assistance and critical reading and editing of the manuscript.
Competing interests: L.M.K. receives funding as a consultant for Ethicon Surgical Care. A.P.L., L.M.K., and P.J.T. are named inventors on a patent related to this work (serial 13/780,284). M.P., J.-M.L., and S.M. declare no competing financial interests.
Data and materials availability: 16S rRNA gene sequencing reads are deposited in MG-RAST (accession no. 1331).
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/5/178/178ra41/DC1
Materials and Methods
Fig. S1. Effect of RYGB on glucose metabolism.
Fig. S2. RYGB alters the gut microbiota independently of weight loss.
Fig. S3. Relative abundance of Enterobacteriales over time and space.
Fig. S4. Determination and quantification of Archaea and methanogens in fecal DNA samples in RYGB, SHAM, and WMS animals.
Fig. S5. Phenotypic changes between germ-free mice colonized with microbiota from either RYGB or WMS donors.
Fig. S6. Comparisons of body weight and adiposity markers in SHAM-R and WMS-R mice.
Fig. S7. Ileal FIAF gene expression in germ-free recipient mice.
Fig. S8. Potential mechanisms by which altered microbiota after RYGB affect host metabolic function.
Table S1. One-week food intake and fecal calorimetry analysis of a cohort of RYGB, SHAM, and WMS animals.
Table S2. Two-hour fasted blood concentrations of glucose, insulin, calculated HOMA-IR, triglycerides, and NEFAs in mice maintained on HFD.
Table S3. 16S rRNA gene sequencing metadata from fecal time series.
Table S4. Taxonomic groups with differential relative abundance between treatments.
Table S5. 16S rRNA gene sequencing metadata from intestinal axis sampling.
Table S6. 16S rRNA gene sequencing metadata from fecal transplants.
Table S7. Taxonomic groups with relative abundance between transplant recipients.
REFERENCES AND NOTES
- 1.Hatoum IJ, Greenawalt DM, Cotsapas C, Reitman ML, Daly MJ, Kaplan LM. Heritability of the weight loss response to gastric bypass surgery. J. Clin. Endocrinol. Metab. 2011;96:E1630–E1633. doi: 10.1210/jc.2011-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn SM, Pomp A, Rubino F. Metabolic surgery for type 2 diabetes. Ann. N. Y. Acad. Sci. 2010;1212:E37–E45. doi: 10.1111/j.1749-6632.2011.05984.x. [DOI] [PubMed] [Google Scholar]
- 3.Rubino F, Gagner M, Gentileschi P, Kini S, Fukuyama S, Feng J, Diamond E. The early effect of the Roux-en-Y gastric bypass on hormones involved in body weight regulation and glucose metabolism. Ann. Surg. 2004;240:236–242. doi: 10.1097/01.sla.0000133117.12646.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thaler JP, Cummings DE. Minireview: Hormonal and metabolic mechanisms of diabetes remission after gastrointestinal surgery. Endocrinology. 2009;150:2518–2525. doi: 10.1210/en.2009-0367. [DOI] [PubMed] [Google Scholar]
- 5.le Roux CW, Aylwin SJ, Batterham RL, Borg CM, Coyle F, Prasad V, Shurey S, Ghatei MA, Patel AG, Bloom SR. Gut hormone profiles following bariatric surgery favor an anorectic state, facilitate weight loss, and improve metabolic parameters. Ann. Surg. 2006;243:108–114. doi: 10.1097/01.sla.0000183349.16877.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stylopoulos N, Hoppin AG, Kaplan LM. Roux-en-Y gastric bypass enhances energy expenditure and extends lifespan in diet-induced obese rats. Obesity. 2009;17:1839–1847. doi: 10.1038/oby.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bueter M, Löwenstein C, Olbers T, Wang M, Cluny NL, Bloom SR, Sharkey KA, Lutz TA, le Roux CW. Gastric bypass increases energy expenditure in rats. Gastroenterology. 2010;138:1845–1853. doi: 10.1053/j.gastro.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Nestoridi E, Kvas S, Kucharczyk J, Stylopoulos N. Resting energy expenditure and energetic cost of feeding are augmented after Roux-en-Y gastric bypass in obese mice. Endocrinology. 2012;153:2234–2244. doi: 10.1210/en.2011-2041. [DOI] [PubMed] [Google Scholar]
- 9.Li JV, Ashrafian H, Bueter M, Kinross J, Sands C, le Roux CW, Bloom SR, Darzi A, Athanasiou T, Marchesi JR, Nicholson JK, Holmes E. Metabolic surgery profoundly influences gut microbial-host metabolic cross-talk. Gut. 2011;60:1214–1223. doi: 10.1136/gut.2010.234708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc. Natl. Acad. Sci. U.S.A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1 doi: 10.1126/scitranslmed.3000322. 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 19.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, Mariat D, Corthier G, Doré J, Henegar C, Rizkalla S, Clément K. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: Links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kucharczyk J, Nestoridi E, Kvas S, Andrews R, Stylopoulos N. Probing the mechanisms of the metabolic effects of weight loss surgery in humans using a novel mouse model system. J. Surg. Res. 2013;179:e91–e98. doi: 10.1016/j.jss.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 24.Hatoum IJ, Stylopoulos N, Vanhoose AM, Boyd KL, Yin DP, Ellacott KL, Ma LL, Blaszczyk K, Keogh JM, Cone RD, Farooqi IS, Kaplan LM. Melanocortin-4 receptor signaling is required for weight loss after gastric bypass surgery. J. Clin. Endocrinol. Metab. 2012;97:E1023–E1031. doi: 10.1210/jc.2011-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: Physiological and molecular characteristics. Physiol. Behav. 2004;81:243–248. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Bikman BT, Zheng D, Pories WJ, Chapman W, Pender JR, Bowden RC, Reed MA, Cortright RN, Tapscott EB, Houmard JA, Tanner CJ, Lee J, Dohm GL. Mechanism for improved insulin sensitivity after gastric bypass surgery. J. Clin. Endocrinol. Metab. 2008;93:4656–4663. doi: 10.1210/jc.2008-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl. 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009;11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 31.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U.S.A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3:279–288. doi: 10.4161/gmic.19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trebichavsky I, Splichal I, Rada V, Splichalova A. Modulation of natural immunity in the gut by Escherichia coli strain Nissle 1917. Nutr. Rev. 2010;68:459–464. doi: 10.1111/j.1753-4887.2010.00305.x. [DOI] [PubMed] [Google Scholar]
- 36.Belzer C, de Vos WM. Microbes inside—From diversity to function: The case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucindegrader Akkermansia muciniphila. Front. Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, Buschard K, Hansen AK. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55:2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 39.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 41.Layden BT, Angueira AR, Brodsky M, Durai V, Lowe WL., Jr. Short chain fatty acids and their receptors: New metabolic targets. Transl. Res. 2013;161:131–140. doi: 10.1016/j.trsl.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.sge_macfarlane. [DOI] [PubMed] [Google Scholar]
- 43.Hong YH, Nishimura Y, Hishikawa D, Tsuzuki H, Miyahara H, Gotoh C, Choi KC, Feng DD, Chen C, Lee HG, Katoh K, Roh SG, Sasaki S. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005;146:5092–5099. doi: 10.1210/en.2005-0545. [DOI] [PubMed] [Google Scholar]
- 44.Wolever TM, Spadafora P, Eshuis H. Interaction between colonic acetate and propionate in humans. Am. J. Clin. Nutr. 1991;53:681–687. doi: 10.1093/ajcn/53.3.681. [DOI] [PubMed] [Google Scholar]
- 45.Reshef L, Niv J, Shapiro B. Effect of propionate on lipogenesis in adipose tissue. J. Lipid Res. 1967;8:682–687. [PubMed] [Google Scholar]
- 46.Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta. 2010;1801:1175–1183. doi: 10.1016/j.bbalip.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Bellahcene M, O’Dowd JF, Wargent ET, Zaibi MS, Hislop DC, Ngala RA, Smith DM, Cawthorne MA, Stocker CJ, Arch JR. Male mice that lack the G-protein-coupled receptor GPR41 have low energy expenditure and increased body fat content. Br. J. Nutr. 2012:1–10. doi: 10.1017/S0007114512003923. [DOI] [PubMed] [Google Scholar]
- 48.Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, Kobayashi M, Hirasawa A, Tsujimoto G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc. Natl. Acad. Sci. U.S.A. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 50.Rubino F, Schauer PR, Kaplan LM, Cummings DE. Metabolic surgery to treat type 2 diabetes: Clinical outcomes and mechanisms of action. Annu. Rev. Med. 2010;61:393–411. doi: 10.1146/annurev.med.051308.105148. [DOI] [PubMed] [Google Scholar]
- 51.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl. 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8:79. doi: 10.1186/1471-2180-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deplancke B, Hristova KR, Oakley HA, McCracken VJ, Aminov R, Mackie RI, Gaskins HR. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 2000;66:2166–2174. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in //10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.