Abstract
Objective
Many patients following severe trauma have complicated recoveries due to the development of organ injury. Physiological and anatomical prognosticators have had limited success in predicting clinical trajectories. We report on the development and retrospective validation of a simple genomic composite score that can be rapidly used to predict clinical outcomes.
Design
Retrospective cohort study
Setting
Multi-institution level 1 trauma centers
Patients
Data was collected from 167 severely traumatized (ISS >15) adult (18–55 yo) patients
Methods
Microarray-derived genomic data obtained from 167 severely traumatized patients over 28 days were assessed for differences in mRNA abundance between individuals with different clinical trajectories. Once a set of genes was identified based on differences in expression over the entire study period, mRNA abundance from these subjects obtained in the first 24 hours was analyzed in a blinded fashion using a rapid multiplex platform, and genomic data reduced to a single metric.
Results
From the existing genomic data set, we identified 63 genes whose leukocyte expression differed between an uncomplicated and complicated clinical outcome over 28 days. Using a multiplex approach that can quantitate mRNA abundance in less than 12 hours (nanoString™), we reassessed total mRNA abundance from the first 24 hours after trauma, and reduced the genomic data to a single composite score using the difference from reference (DFR). This composite score showed good discriminatory capacity to distinguish patients with a complicated outcome (area under a receiver-operator curve, 0.811, p < 0.001). This was significantly better than the predictive power of either APACHE II or NISS scoring systems.
Conclusions
A rapid genomic composite score obtained in the first 24 hours after trauma can retrospectively identify trauma patients who are likely to develop a complicated clinical trajectories. A novel platform is described in which this genomic score can be obtained within 12 hours of blood collection, making it available for clinical decision making. (300 words; limit 300)
Keywords: nanostring, microarray, blunt trauma
Introduction
Traumatic injury remains the most common cause of death in individuals less than 45 years of age and is a significant healthcare burden (1, 2). Although risk factors such as increasing age, number of premorbid conditions, and injury severity are well known causes of increased mortality following traumatic injury (3, 4), predictive models based on physiologic and clinical parameters still lack the specificity and sensitivity to identify individual patients at risk of developing organ injury or sepsis.
Current evidence suggests that a systemic immunological dysregulation is central to the organ injury following trauma. While our understanding of the systemic consequences of severe injury and sepsis remains incomplete, as evidenced by the lack of success in translational and clinical trials of therapies aimed at modulating the inflammatory response (5, 6), the development of prognosticators based on immunological measures are plentiful (7–12). Unfortunately, these diagnostics and prognostics have not proved sufficiently successful to enter the clinical armamentarium.
To obtain a better global understanding of the complex immune response following severe injury in humans, we examined genome-wide expression patterns of blood leukocytes following severe injury, and observed what was termed a ‘genomic storm’, in which greater than 75% of the host leukocyte transciptome was altered following severe injury (13). More importantly, we demonstrated that genomic information collected in the first 12 hours of severe trauma contained information independent of anatomical and physiological prognosticators that was associated with different clinical outcomes (14). In a more recent study, we further demonstrated that differences in leukocyte genomic information obtained over the first four days was associated with patients who would die from organ failure or would have prolonged complicated courses (15). Such findings strongly suggest that genomic information contained in whole blood leukocytes in the immediate post-trauma period could be used to develop a prognostic tool to determine outcome, and theoretically, identify those patients most likely to benefit from interventions.
In this report, we propose a novel platform to conduct genomic measurements from whole blood leukocytes to identify severely traumatized patients who will have a complicated clinical recovery. Using existing microarray data from the ‘Inflammation and the Host Response to Injury’ Glue-Grant (GG) collaborative research program, we identify 63 genes from 167 severely injured patients whose leukocyte gene expression differs between patients with different clinical outcomes over 28 days. Using existing RNA samples from the collaborative research program obtained in the first 24 hours after trauma, we then conduct a proof of principal study to retrospectively validate the microarray data using a multiplex RNA quantification scheme based on fluorescently labeled codesets that can quantitate the 63 gene expression levels simultaneously within 12 hours of sample collection (nanoString nCounter™ Gene Analysis). The gene expression data were re-evaluated in all 167 subjects, and the resulting gene expression data was reduced to a single metric, as described previously (14).
Based on the findings reported here, we propose that such a genomic score can feasibly be utilized as a prognostic tool in the clinical setting to identify trauma patients at risk of developing organ injury and adverse outcomes, to guide clinical decisions regarding treatment, and should be validated prospectively in future studies.
Methods
Subject Recruitment and Affymetrix U133 GeneChip Data Analysis
One hundred, sixty three patients between the ages of 18 and 55 who suffered severe blunt injury were enrolled from six institutions between November 2003 and January 2005 and compared to 35 age and gender matched, healthy control subjects (13). Samples were collected with signed informed consent; each institution had their own IRB approval, and de-identified clinical data are now in the public domain (16). Blood samples were collected within 12 hours of injury and at 1, 4, 7, 14, 21, and 28 days after the injury while hospitalized in the intensive care unit, whole blood nucleated cells were isolated, and genome-wide expression analyses of 54,675 probe sets were performed using the Affymetrix U133 GeneChip™ (17, 18).
Statistical analysis to assess the overall gene expression patterns following trauma was performed (13). Briefly, trauma patients were divided into three cohorts based on their clinical trajectory being either “uncomplicated” (as defined by the absence or resolution of organ injury (modified Marshall MOF Score) in less than 5 days), “complicated” (resolution of organ injury in greater than 14 days or no recovery by 28 days, or death), or “intermediate”. We focused our initial analyses on the extremes of outcome, those patients who had a either a “complicated” trajectory versus those who had an uncomplicated clinical course. Using the EDGE software program, statistical analysis was performed to assess differences in gene expression over the entire 28 day sampling period between complicated and uncomplicated patients(19, 20). Using this approach, we were able to identify a set of genes that were differentially expressed using a false discovery rate (FDR) adjusted probability of <0.001. We then filtered that set to only include the genes that had at least a two-fold maximal difference in expression between the two clinical trajectories, which resulted in a set of 63 genes. Gene expression data from those 63 genes were reduced to a single metric that was then independently tested for its predictive ability at a single clinically-relevant time point, 24 hours, that evaluated the magnitude of the difference in expression from healthy, control subjects using a statistical tool that reduces multiplex genomic data to a single metric (DFR) (14). The DFR calculates the sum of the normalized differences in expression for each of the genes from the mean expression obtained from age, gender and ethnicity matched controls, using the equation (14):
where ei is the patient’ s expression level and Mi and Vi are the control group mean and variance for the ith probe set. Division by the control variance is a rescaling that prevents the DFR score from being dominated by genes that are inherently more variable or more highly expressed. The natural logarithm is applied to make the distribution of the resulting DFR more symmetric over the patient population (14). In essence, DFR calculates the distance between each patient’s gene expression profile and the profile obtained from the healthy reference subjects.
Validation of Affymetrix U133 GeneChip Analysis with nanoString nCounter™ Gene Expression Analysis
A custom-made nanoString nCounter™ codeset was used to perform a quantitative analysis that could retrospectively validate the Affymetrix U133 GeneChip™ microarray analysis, and included only the 63 genes that were found to be differentially expressed between complicated and uncomplicated patients, and seven variably expressed housekeeping genes. In order to assess the analytical variance of the nanoString™ platform to detect small differences in array expression, we performed an ex-vivo study using a human whole blood sample that was stimulated with lipopolysaccharide (E. coli O26:B6, 100 ngs/ml) for two hours at 37 °C and compared to unstimulated samples. One healthy, human subject provided signed informed consent, and the protocol had been approved by the University of Florida IRB. The sample was divided into 10 aliquots and five were stimulated with lipopolysaccharide and five were unstimulated, and processed immediately. RNA was isolated using identical protocols to those employed for the clinical samples (15, 18), and five replicates from both LPS-stimulated and unstimulated blood were analyzed using both the Affymetrix U133+ v2 GeneChip™ and the nanoString™ technology.
The nanoString™ technology employs two sequence specific complimentary probes with a unique code for each of the genes of interest (21). Probe pairs containing a reporter probe with a signal on its 5′ end, and a capture probe with biotin on the 3′ end with an individualized fluorescent barcode were hybridized to 100 ng of target mRNA for varying lengths of time (21). The excess probes were washed away using a magnetic bead-based purification on the nCounter™ Prep station. Data collection and quantitative analysis of the codeset was carried out using the nCounter™ Digital Analyzer. The raw data were collected and normalized for the expression of included house-keeping genes. The analysis was then repeated on the 167 patient samples and 35 controls at <12 and 24 hours using the nanoString™ technology and compared the analysis for the 63 predictive genes with the previously reported microarray analysis (15). Using the microarray data and the DFR (14), we then derived a metric that could be used to prospectively validate the genomic test.
Statistical Analysis
Continuous demographic variables were tested for normality, variance and was tested for significance via one-way ANOVAs via Dunn’s or Tukey post-hoc analysis. Categorical demographic variables were tested for significance using a χ2 test. Separate univariate logistic regression models were fit to assess the association between DFR, ISS, NISS, and APACHE II scores as continuous independent measurements and outcome variable (complicated versus uncomplicated clinical course). Receiver operating characteristic (ROC) curve analysis was performed by calculating sensitivity and specificity for each independent variable. We constructed separate multivariable logistic models using clinical independent predictors only (ISS and APACHE II scores) or combining clinical predictors with genomic score (DFR in addition to ISS and APACHE II). All independent predictors were entered in each model as continuous variables. For each model we performed model diagnostics to test for collinearity (using variance inflation factor and tolerance) and interactions. Model fit was evaluated using the Hosmer-Lemeshow goodness of fit test. Multiple ROC curve comparison analyses were performed to assess model predictive performance using AUC for each model. Area under the curve (AUC) was used to assess model fit. AUC comparisons were made using ROCCONTRAST option available in LOGISTIC procedure using a nonparametric method that exploits the mathematical equivalence of the AUC to the Mann-Whitney U-statistic. All analyses were performed using SAS (v.9.2, Cary, N.C.) and SigmaStat v11.
Results
Genome-wide expression of whole blood leukocytes was determined at 12 hours, 1, 4, 7, 14 and 28 days after severe blunt trauma in a cohort of 167 severe blunt trauma patients whose characteristics have been previously published (13). Using clinical parameters, the 167 enrolled patients were divided into three cohorts based on their time to organ recovery (TTR), a new metric of organ failure recovery recently described by our group, which is defined as the time at which a patient is free from organ failure for at least two days. Prolonged TTR is associated with advanced age, increased severity of injury, shock, and increased transfusion requirements, thus making it a comprehensive metric to monitor the complex response in trauma patients(22). This allowed us to divide the cohort into three distinct groups of patients who had either an uncomplicated, intermediate, or complicated recovery, as defined in the Materials and Methods (Table 1 and 2). As expected, patients who experienced intermediate and complicated clinical courses had significantly higher injury severity scores, ICU and hospital length of stay, noninfectious and infectious complications, and mortality compared to those patients who had uncomplicated clinical recoveries (p<0.001) (Table 1).
Table 1.
Separation of “Inflammation and Host Response to Injury” severe trauma cohort into tertiles.
| Patient outcome designation | ||
|---|---|---|
| Uncomplicated | Marshall | < 6 |
| TTR | <4 days | |
|
| ||
| Intermediate | Marshall | < or >6 |
| TTR | 4–14 days | |
|
| ||
| Complicated | Marshall | >6 |
| TTR | >14 days | |
Table 2.
Patient demographics and outcomes. Table summarizes the demographics and outcome variables in the 167 patients with severe blunt trauma. The trauma patients were assigned to three subgroups based on their extreme clinical trajectories: ‘Uncomplicated Recovery’ indicating those subjects who did not develop MOF (MOF <6) and had a TTR of less than 4 days, ‘Intermediate Recovery’ indicating those patients that had MOF scores < or > 6 and a TTR of 4 to 14 days, and ’Complicated Recovery’ where patients had MOF and a time to recovery in excess of 14 days. Values represent the mean ± S.E.M., with median and middle quartiles in parentheses. Demographic and outcome data were analyzed by one-way ANOVA with a Tukey’s or Kruskall-Wallis (‡) post hoc analysis, or Chi-square (†), where appropriate. Significance was designated at the p<0.05 level of confidence.
| Uncomplicated Recovery, n=55 | Intermediate Recovery, n=71 | Complicated Recovery, n=41 | Probability | |
|---|---|---|---|---|
|
| ||||
| Demographics | ||||
|
| ||||
| Age | 33 ± 2 (32, 21–43) | 35 ± 2 (34, 25–45) | 34 ± 2 (34, 26–42) | p=0.515‡ |
|
| ||||
| Sex (M/F) | 30/25 | 47/24 | 30/11 | p=0.151† |
|
| ||||
| APACHE II | 24.4 ± 0.8 (25, 21–29) | 28.4 ± 0.6 (29, 24–32) | 29.4 ± 0.8 (29, 26–33) | p<0.001 |
|
| ||||
| ISS | 26.2 ± 1.8 (24, 17–35) | 32.6 ± 1.5 (34, 22–42) | 35.7 ± 2.0 (38, 27–42) | p=0.001 |
|
| ||||
| NISS | 32.6 ± 1.8 (29, 22–40) | 36.9 ± 1.5 (34, 27–45) | 39.8 ± 1.9 (41, 29–44) | p=0.01‡ |
|
| ||||
| Total Transfusion, ml | 1705 ± 172 (1400, 700–2229) | 2782 ± 241 (2100, 1400–3787) | 2952 ± 423 (2150, 1050–3500) | p<0.001‡ |
|
| ||||
| Total Crystalloid, ml | 10544 ± 765 (9070, 7409–12163) | 13802 ± 808 (12000, 9025–17837) | 15226 ± 1530 (12935, 8728–18683) | p<0.001‡ |
|
| ||||
| Worst Base Deficit | −9.2 ± 0.4 (−8.9, −11.6–(−6.4)) | −8.7 ± 0.6 (−8.0, −10–(−5.9)) | −10.6 ± 0.8 (−10.3, −13.8–(−6.0)) | p=0.456‡ |
|
| ||||
| Lowest Systolic BP, mmHg | 92.3 ± 3.0 (88, 80–108) | 88.3 ± 2.1 (87, 78–100) | 86.6 ± 3.0 (84, 77–97) | p=0.233‡ |
|
| ||||
| Outcomes | ||||
|
| ||||
| Survival | 100 % | 94% (67/4) | 83% (34/7) | p=0.002 |
|
| ||||
| Maximum Marshall Score | 3.0 ± 0.1 (3, 2–4) | 5.5 ± 0.3 (5, 3–7) | 8.8 ± 0.4 (8, 7–10) | p<0.001‡ |
|
| ||||
| Hospital Length of Stay, days | 15.2 ± 1.7 (12, 9–18) | 26 ± 1.7 (22, 17–33) | 35.8 ± 3.6 (30, 23–42) | p<0.001‡ |
| ICU Length of Stay, Days | 4.8 ± 0.4 (5, 3–6) | 12.3 ±0.7 (11, 8–15) | 25.1 ± 2.3 (21, 18–30) | p<0.001‡ |
|
| ||||
| Time to Recovery, days | 2.9 ± 0.1 (3, 2–4) | 10.0 ± 1.0 (5, 3–14) | 22.0 ± 0.9 (20, 18–28) | p<0.001† |
|
| ||||
| Noninfectious Complications | 5.5 % (3/52) | 64.8% (46/25) | 90.2 % (37/4) | p<0.001† |
|
| ||||
| Nosocomial Infections | 20.0 % (11/44) | 64.8% (46/25) | 85.4 % (35/6) | p<0.001† |
|
| ||||
| Surgical Site Infections | 7.3 % (4/51) | 22.5% (16/55) | 41.5 % (17/24) | p<0.001† |
|
| ||||
| Ventilator Associated Pneumonia, Cases/1000 vent days | 3.8 | 21.6 | 25.3 | p<0.001† |
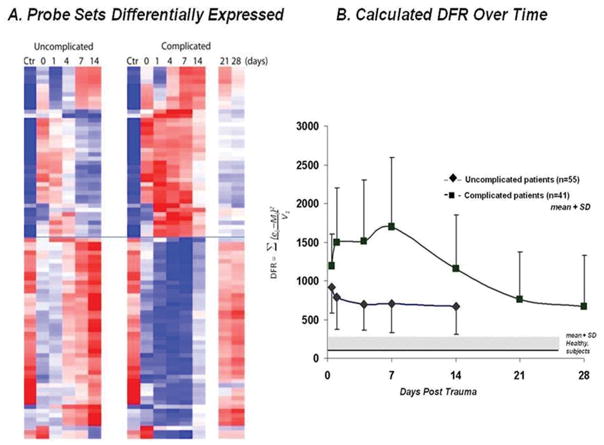
To explore if there were differences in the genomic response to severe blunt trauma that may be associated with clinical recovery, we focused initially on the two extremes of the study population, those patients designated as having an uncomplicated clinical recovery versus those patients designated as having a complicated recovery. We found that there were 3,681 probe sets representing 2,435 genes whose expression over 28 days was significantly different between patients with an uncomplicated and complicated recovery, using a FDR adjusted probability of Q < 0.001. These genes were then filtered to include only those genes whose expression differed by at least two fold. A total of 63 significant genes were found to be different between patients with a complicated or uncomplicated recovery (Figure 1A and Table 3). Of the 63 genes whose expression was found to be significantly different between trauma patients with complicated and uncomplicated clinical recoveries, greater than two thirds of these genes were directly related to protective immunity. In fact, the greatest difference between complicated and uncomplicated patients was with the magnitude and duration of suppression of genes involved in adaptive immunity pathways, most notably the suppression of genes involved in antigen presentation (HLA-DQ, HLA-DR, HLA-DM and CD74) and with interferon and interferon-inducible genes (IFIT 1,2,3,5, IFI44, IFI44L, IFI6).
Figure 1.
Heat map and calculated DFR for the 63 genes that distinguish clinical trajectory. Using a false discovery adjusted probability of <0.001 and a 2-fold difference in expression, the temporal pattern of expression of 63 genes that differed between patients with the two clinical outcomes is presented. A. Cluster analysis of the two cohorts B. Summary of the difference from reference (DFR) score calculated for each patient in the uncomplicated and complicated cohorts at each time point. Statistical analysis at each time point (0, 1, 4, 7, and 14 days) revealed significant differences in DFR between complicated and uncomplicated patients (p<0.05, Mann Whitney Rank analysis).
Table 3.
Sixty three genes identified as having a different expression between patients with a complicated or uncomplicated recovery.
| Gene ID | Official Name | Complicated vs. Uncomplicated Fold Change |
|---|---|---|
| AGFG1 | ArfGAP with FG repeats 1 | 2.03 |
| ANKRD55 | ankyrin repeat domain 55 | 3.29 |
| ATP6V1C1 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | 2.13 |
| CD24 | CD24 molecule | 2.38 |
| CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | −2.03 |
| CDK5RAP2 | CDK5 regulatory subunit associated protein 2 | 2.91 |
| CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non- specific cross reacting antigen) | 2.01 |
| CEACAM8 | carcinoembryonic antigen-related cell adhesion molecule 8, CD66b | 2.19 |
| CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial | −2.91 |
| DACH1 | dachshund homolog 1 (Drosophila) | 2.04 |
| EPSTI1 | epithelial stromal interaction 1 (breast) | −3.01 |
| FLJ39051 | hypothetical protein LOC399972 | 2.50 |
| FOLR3 | folate receptor 3 (gamma) | 2.01 |
| GALNT14 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N- acetylgalactosaminyltransferase 14 (GalNAc-T14) | 2.04 |
| GNLY | granulysin | −2.16 |
| GRB10 | growth factor receptor-bound protein 10 | 2.14 |
| HERC5 | hect domain and RLD 5 | −3.27 |
| HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 2.06 |
| HLA-DMA | major histocompatibility complex, class II, DM alpha | −2.25 |
| HLA-DMB | major histocompatibility complex, class II, DM beta | −2.25 |
| HLA-DPA1 | major histocompatibility complex, class II, DP alpha 1 | −2.57 |
| HLA-DPB1 | major histocompatibility complex, class II, DP beta 1 | −2.31 |
| HLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 | −2.41 |
| HLA-DQB1 | major histocompatibility complex, class II, DQ beta 1 | −2.68 |
| HLA-DRA | major histocompatibility complex, class II, DR alpha | −2.69 |
| HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 | −2.48 |
| IFI44 | interferon-induced protein 44 | −3.27 |
| IFI44L | interferon-induced protein 44-like | −4.53 |
| IFI6 | interferon, alpha-inducible protein 6 | −2.68 |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | −6.96 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | −2.99 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | −3.23 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 | −2.19 |
| IL1R1 | interleukin 1 receptor, type I | 2.06 |
| IL1R2 | interleukin 1 receptor, type II | 2.69 |
| ISG15 | ISG15 ubiquitin-like modifier | −2.48 |
| LCN2 | lipocalin 2 | 2.43 |
| LOC100127983 | hypothetical protein LOC100127983 | 2.03 |
| LRG1 | leucine-rich alpha-2-glycoprotein 1 | 2.20 |
| LTF | lactotransferrin | 2.23 |
| MIAT | myocardial infarction associated transcript (non-protein coding) | 2.35 |
| MMP8 | matrix metallopeptidase 8 (neutrophil collagenase) | 4.2 |
| MX1 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 | −2.68 |
| NAIP | NLR family, apoptosis inhibitory protein | 2.33 |
| NSUN7 | NOL1/NOP2/Sun domain family, member 7 | 3.03 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46kDa | −2.04 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71kDa | −2.11 |
| OAS3 | 2′-5′-oligoadenylate synthetase 3, 100kDa | −2.46 |
| OLAH | oleoyl-ACP hydrolase | 5.82 |
| OLFM4 | olfactomedin 4 | 3.61 |
| PCOLCE2 | procollagen C-endopeptidase enhancer 2 | 2.77 |
| PDGFC | platelet derived growth factor C | 2.04 |
| PMAIP1 | phorbol-12-myristate-13-acetate-induced protein 1 | −2.27 |
| PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | −2.64 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 | −4.47 |
| SIPA1L2 | signal-induced proliferation-associated 1 like 2 | 2.01 |
| SLC26A8 | solute carrier family 26, member 8 | 2.23 |
| TCN1 | transcobalamin I (vitamin B12 binding protein, R binder family) | 2.19 |
| TDRD9 | tudor domain containing 9 | 3.29 |
| TGFBI | transforming growth factor, beta-induced, 68kDa | −2.14 |
| THBS1 | thrombospondin 1 | 2.06 |
| VNN1 | vanin 1 | 3.14 |
| XAF1 | XIAP associated factor 1 | −2.58 |
Next, we wanted to distill the expression values of these 63 genes to an easily understandable and clinically applicable, single metric. To do this, we utilized the DFR score, which we have previously shown to describe the overall genomic response of a patient compared to the response from a reference population, in this case, age, gender and ethnicity matched healthy controls. For each of the 96 patients classified as complicated or uncomplicated, we calculated a DFR score based on the expression level of the 63 genes over time (Figure 1B) (14). The genomic response curves for these two patient populations diverge and separate by 24 hours extending for at least seven days; the differences in the genomic scores at <12, 24 hours and days 4, and 7 were statistically significant (p<0.05 by Mann-Whitney rank analysis). The data suggest that a composite genomic score or DFR could be generated to identify patients by 24 hours that will go on to experience an adverse outcome in response to trauma (Figure 1B).
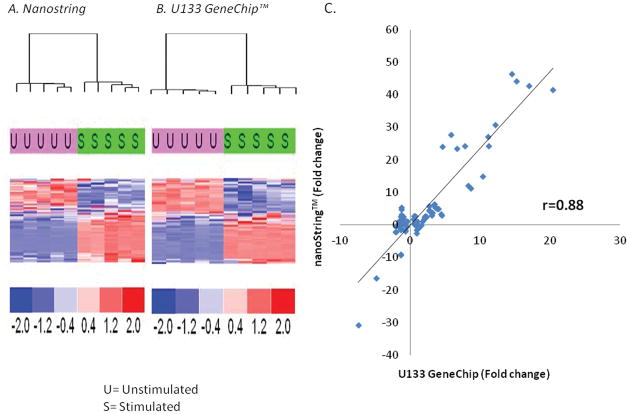
Unfortunately, neither deep sequencing nor microarrays are amenable to a rapid turnaround time required for clinical decision making in the critically-ill. In contrast, the nanoString™ nCounter multiplex array is a rapid (12–18 hours) assay that is capable of evaluating the expression of a large number of genes simultaneously (up to 800) for a patient and would offer a rapid turn-around alternative to microarray (21). By modifying the quantities of starting material, we could further reduce the analytical time to a manageable 6–10 hours. To validate analytical variance; whether the nanoString™ technology could reproducibly generate similar gene expression patterns, we prospectively compared the nanoString™ quantitation of gene expression with the Affymetrix U133+ v2 GeneChip™ data using ex vivo LPS stimulation of whole human blood. Five replicates from a single sample of unstimulated or LPS-stimulated healthy, human whole blood were assessed for expression of the 63 genes identified above. As demonstrated in Figure 2A, both nanoString™ and microarray platform yielded similar results for the 63 genes, and were able to segregate samples based on whether or not the sample was stimulated with LPS. In addition, the expression of these 63 genes were also correlated well between platforms (Figure 2B), r=0.88 (p<0.0001), suggesting that the nanoString™ platform was as reproducible as microarray, and could offer a rapid turn-around alternative (6–10 hrs) to lengthier microarray analyses, which can take upwards of 3–4 days.
Figure 2.
Comparison of gene expression patterns and variation in human whole blood ex vivo stimulated with bacterial lipopolysaccharide. A single sample was divided and one-half stimulated with 100 ng/ml of E. coli LPS for two hours at 37° C. Total RNA was isolated from the two preparations and divided into ten equal aliquots, five analyzed by microarray, and five analyzed with the nanoString™ technology. A and B. Heat maps of the hierarchical clustering of the gene expression data for the two technologies, revealing that both methodologies easily detected the change in gene expression changes induced by LPS (U=Unstimulated samples; S=Stimulated samples) C. Pearson correlation coefficients for the change in gene expression of the 63 genes produced by the LPS stimulation. Values represent fold change over control expression values.
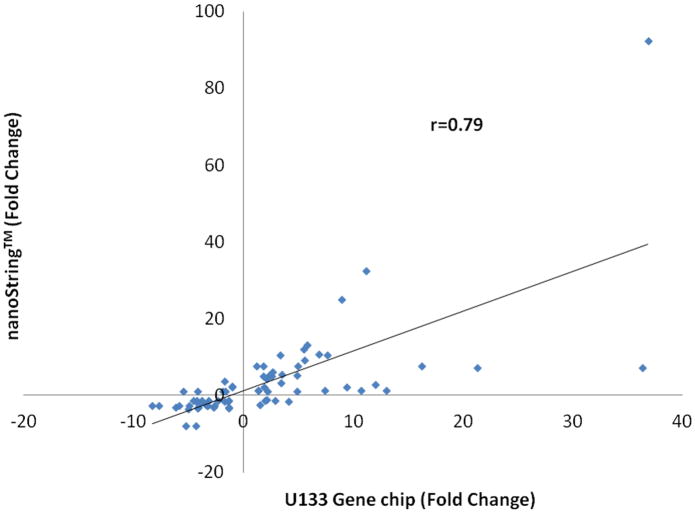
To further support and validate the nanoString™ platform in our trauma patient population, we evaluated the correlation of gene expression derived from both the microarray or nanoString™ platform of the 63 genes in the 167 trauma patients from samples taken at 24 hours after trauma. The expression of the 63 genes (Table 3) were well correlated with an r value of 0.79 (p<0.0001) (Figure 3) between both nanoString and microarray platforms, similar to the ex vivo LPS analyses above. These data further support the capability of using a nanoString™ based assay, in place of microarray, to rapidly identify the 63 genes within a trauma population.
Figure 3.
Pearson correlation between the microarray and nanoString™ expression level of the 63 genes found to be differentially regulated between uncomplicated and complicated patient cohorts. Values represent fold change over control expression values.
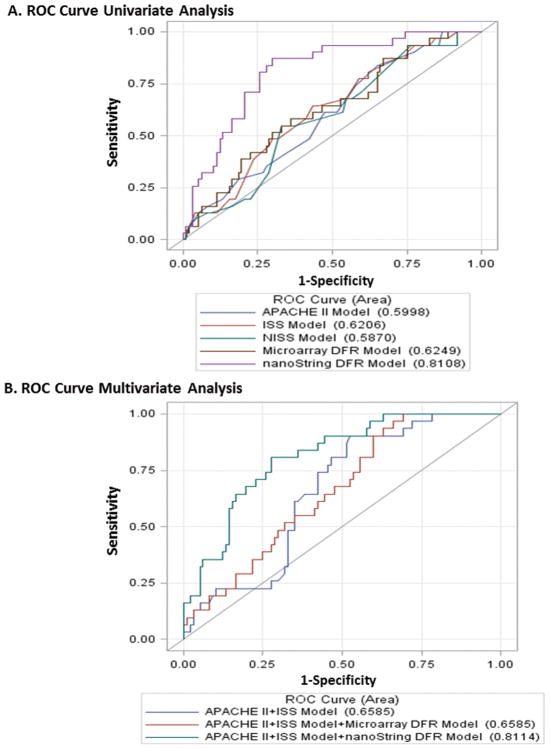
Next, we wanted to prospectively determine if a DFR score based on the nanoString™ platform obtained in the first 24 hours, could be used to discriminate between patients that experienced adverse clinical recoveries versus those that did not. DFR scores were calculated for each patient at the 24 hour period, a time point that would have clinical relevance to the subsequent management of this population (Figure 1B). Receiver operator curves were then generated to determine the ability of the nanoString™ based DFR to determine between either uncomplicated outcomes versus the other two groups, or complicated outcomes versus intermediate or uncomplicated outcomes. Interestingly, nanoString-based DFR scores could differentiate those patients who would experience a complicated recovery from those who would go on to have either an uncomplicated or an intermediate recovery markedly better than seen with microarray. AUC 0.625 vs 0.811 microarray vs nanoString™ respectively, p-value=0.0011 (Figure 4). Surprisingly, the nanoString™ AUC values were also better than anatomic and physiologic predictors of clinical course and recovery, such as new injury severity score (NISS), injury severity score (ISS), or acute physiology and chronic health evaluation (APACHE II) metrics (Table 4)collected for these patients with p-values of 0.005, 0.0009, and 0.0011 respectively (Figure 4) (14). As shown in Figure 4, addition of physiologic or anatomic scoring systems to the nanoString™ DFR resulted in only minimal and nonsignificant improvements in the AUC.
Figure 4.
Receiver operator curves (ROC) demonstrating the ability of nanoString™ versus microarray platforms to discriminate between clinical outcome. A. ROC curves for comparison of univariate analysis. B. ROC curves for comparison of multivariate analysis. AUC for nanoString DFR is significantly higher than AUC for ISS, NISS, and microarray models, with p=0.005, 0.0009, and 0.0011 respectively. Adding nanoSpring DFR to APACHE II and ISS model increases AUC significantly from 0.66 to 0.81 (p=0.0026). AUC for model with APACHE II, ISS, and nanoString DFR is significantly higher than AUC of the model with APACHE II, ISS, and mcicroarray DFR (p=0.0008).
Table 4.
Comparison of microarray and nanoString™ DFR to established clinical predictor models of outcome with corresponding area under the curves (AUC) and their 95% confidence interval values for each multivariable model.
|
Complicated/intermediate vs. uncomplicated
| |
|---|---|
| Variable | AUC (95% CI) |
| nanoString™ DFR | 0.784 (0.702, 0.867) |
| Microarray DFR | 0.801 (0.721, 0.88) |
| APACHE II | 0.729 (0.637, 0.82) |
| ISS | 0.65 (0.544, 0.756) |
| NISS | 0.638 (0.531, 0.744) |
| APACHE II+ISS Model | 0.75 (0.655, 0.844) |
| APACHE II+ISS Model+nanoString DFR Model | 0.819 (0.742, 0.896) |
|
Complicated vs. uncomplicated/intermediate
| |
|---|---|
| Variable | AUC (95% CI) |
| nanoString™ DFR | 0.811 (0.729, 0.893) |
| Microarray DFR | 0.625 (0.514, 0.736) |
| APACHE II | 0.6 (0.49, 0.709) |
| ISS | 0.621 (0.512, 0.729) |
| NISS | 0.587 (0.477, 0.697) |
| APACHE II+ISS Model | 0.659 (0.56,0.757) |
| APACHE II+ISS Model+nanoString DFR Model | 0.811 (0.731,0.892) |
Discussion
MOF and sepsis remains a significant cause of post traumatic injury morbidity and mortality (23, 24). Identifying severely injured patients early who will have a complicated clinical outcome has been difficult and remains a significant clinical and therapeutic hurdle. Unfortunately, clinical trials for the treatment of patients with severe trauma or sepsis have been plagued by the inability to identify patients prospectively who will or will not have a complicated recovery and might benefit from interventional therapies. Entry criteria are primarily physiologic and are limited to nonspecific inflammatory responses and overall organ injury, which may be too delayed to permit intervention to affect outcome. In addition, it is generally accepted that the well-established clinical criteria used to enter patients into severe trauma or sepsis clinical trials includes individuals who will either not benefit from the therapy or may actually be harmed (25). Thus being able to calculate a rapid (<24 hr) genomic score to exclude those patients who will recover and not require intervention, while identifying patients who will have a poor outcome will be a useful tool for future clinical trials.
Genome-wide expression analysis offers the advantage of surveying the entire transcriptome of a cell or tissue, and evaluating changes in expression without any pre-existing bias. Past studies conducted by the Inflammation and Host Response to Injury Large Scale Collaborative Research Program (Glue Grant) have strongly suggested that information contained in the leukocyte transcriptome early after trauma could be used to discriminate much later clinical outcomes (14, 15). Those studies identified families of genes involved in inflammation, antigen presentation and T cell responses as being discriminatory. Similarly, we were able to identify 63 genes whose expression was significantly different (FDR Q<0.001 and fold change >2) between trauma patients with an uncomplicated and complicated outcome (Table 3 and Figure 1).
Although a multiplex genomic approach to patient identification remains attractive, there are a number of technical challenges that must be overcome before a genomic test can be readily used in a critical care setting. There are three main hurdles. First, there must be techniques that can readily and easily isolate the cell populations of interest from critically ill patients, in this case total blood leukocytes. Second, microarray and sequencing approaches are both too time consuming and expensive to be used routinely and still suffer from a lack of clinical validation and reproducibility (26). Third, genomic data must be reduced to a single metric that can be effectively interpreted and applied quickly. In severely traumatized or septic patients, this all must be accomplished within a matter of hours, not days or weeks.
We have successfully addressed all three issues. An alternative simpler and more rapid approach is the use of microfluidics chambers that lyse erythrocytes and yield a pure leukocyte population(27). We have used these in the past in critically ill patients, and demonstrated that in ten minutes, we can obtain yields of total leukocytes that give genomic signals comparable to that seen with macroscale techniques (28). Second, the nanoString™ multiplex array was selected as an alternative to microarray because of its ability to rapidly assay for genomic expression, and when appropriately modified, can generate quantitative, reproducible results within 8–12 hours. In addition, because the hybridization technology does not require prior sample amplification, the mRNA concentration response curves should theoretically be more linear and result in less data compression than seen with microarrays. This increase in dynamic range offered by the nanoString™ platform may be one of the reasons why there is a difference in the discriminatory ability between the two platforms as shown in Figure 4. Regardless, we confirm that the genomic expression patterns for the 63 genes obtained by microarray correlates well with the expression values obtained by the using the nanoString™ platform both in vitro (Figure 2B), as well as in vivo in the original 167 consortium patients who experienced severe blunt trauma (Figure 3) (13). Finally, we used a genomic tool developed by the Program that consolidates all of the information in the 63 genes equally into a single metric (14). Importantly, when the gene expression profile of each patient was distilled to a single genomic score, these scores were able to discriminate patients that developed a poor clinical outcome from those who recovered (Figure 4). Additionally, the calculated genomic score is a linear, normally distributed value in this cohort, and it would be very easy to reset the thresh-hold to capture a less complicated clinical trajectory if one wished to do so. The precision of the assay decreases as the thresh-hold is reduced, but it can be used to capture a larger at-risk population.
There are, however, a number of important considerations. First, this is a retrospective analysis and the genes were selected by identifying those whose expression differed over the 28 day study period. The DFR generated to predict clinical trajectory over the first 24 hours is therefore not independently obtained, and must be validated prospectively before its value can be properly evaluated. Second, DFR scores are based on peripheral leukocyte expression from a relatively small cohort of 167 severely injured trauma patients without significant head injury. As a result, the ability to extend these results to other patient populations currently remains unknown. These caveats aside, the value and importance presented within this report rests with the ability to develop the infrastructure for integrating genomics into a rapid prognostic for the assessment of the critically ill patient.
It is generally accepted that past randomized controlled interventional trials have failed in this patient population in large part due to the heterogeneity of the patient population as well as the inability to select the sickest patient with the highest mortality who have been shown to be the ones to ultimately benefit from therapy (25). The clinical implications of this lie in the fact that this platform could ultimately be coupled to a therapeutic intervention that is aimed at the subset of patients predicted to have a complicated outcome, thus eliminating those patients expected to recover regardless of intervention.
Along the same lines, such a metric could actually be used to determine response to therapy. Of note, two thirds (42) of the 63 genes identified as being differentially expressed between uncomplicated and complicated trauma patients are genes known to be associated with adaptive immunity in general, and specifically are known to be associated with interferon signaling., We found that the expression of these genes was suppressed in trauma patients with complicated outcomes. Such findings imply that this battery of genes might not only be useful in identifying patients at risk of complications after trauma, but also who are responsive to biological response modifiers (29). In this manner, the metric may be useful at identifying those patients who might benefit from therapies that target adaptive immunity such as IL-7, IL-15, IFNγ, and GITR agonists, and also those patients who respond appropriately to the treatment modality.
Importantly, we describe the development of a novel rapid turn-around multiplex array that can be performed from blood sampling within a 12–18 hour time frame and integrated into a single metric or score that can predict long term outcome and identify patients in whom interventional therapies may be beneficial. Though these findings will need to be validated in a larger multicenter trial, a nanoString™ DFR score based on the expression of the 63 gene subset has better discriminatory ability compared to other established clinical predictors based on organ failure or injury (Table 4 and Figure 4). This would allow clinicians to identify patients who will be at risk for developing adverse clinical outcomes, thus allowing them to focus care on prevention of complications such as secondary infections and multisystem organ failure.
Acknowledgments
This work was supported by a contract (U54 GM-062119-10) awarded by the National Institute of General Medical Sciences (NIGMS). AGC and LFG were supported by a T32 training grant (T32 GM-008721-13) in burns and trauma from the NIGMS. AGC was also supported by an individual NRSA (F32 GM-093665-01) awarded by NIGMS, USPHS. AB was supported by award (K23 GM-087709-03) from NIGMS.
Footnotes
The authors have not disclosed any potential conflicts of interest
References
- 1.Probst C, Pape HC, Hildebrand F, et al. 30 years of polytrauma care: An analysis of the change in strategies and results of 4849 cases treated at a single institution. Injury. 2009;40(1):77–83. doi: 10.1016/j.injury.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Cothren CC, Moore EE, Hedegaard HB, et al. Epidemiology of urban trauma deaths: a comprehensive reassessment 10 years later. World J Surg. 2007;31(7):1507–1511. doi: 10.1007/s00268-007-9087-2. [DOI] [PubMed] [Google Scholar]
- 3.Bamvita JM, Bergeron E, Lavoie A, et al. The impact of premorbid conditions on temporal pattern and location of adult blunt trauma hospital deaths. J Trauma. 2007;63(1):135–141. doi: 10.1097/TA.0b013e318068651d. [DOI] [PubMed] [Google Scholar]
- 4.Demetriades D, Murray J, Charalambides K, et al. Trauma fatalities: time and location of hospital deaths. J Am Coll Surg. 2004;198(1):20–26. doi: 10.1016/j.jamcollsurg.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Bone RC, Balk RA, Fein AM, et al. A second large controlled clinical study of E5, a monoclonal antibody to endotoxin: results of a prospective, multicenter, randomized, controlled trial. The E5 Sepsis Study Group. Crit Care Med. 1995;23(6):994–1006. doi: 10.1097/00003246-199506000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Fisher CJ, Jr, Slotman GJ, Opal SM, et al. Initial evaluation of human recombinant interleukin-1 receptor antagonist in the treatment of sepsis syndrome: a randomized, open-label, placebo-controlled multicenter trial. Crit Care Med. 1994;22(1):12–21. doi: 10.1097/00003246-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Abraham E, Glauser MP, Butler T, et al. p55 Tumor necrosis factor receptor fusion protein in the treatment of patients with severe sepsis and septic shock. A randomized controlled multicenter trial. Ro 45-2081 Study Group. JAMA. 1997;277(19):1531–1538. [PubMed] [Google Scholar]
- 8.Abraham E, Laterre PF, Garbino J, et al. Lenercept (p55 tumor necrosis factor receptor fusion protein) in severe sepsis and early septic shock: a randomized, double-blind, placebo-controlled, multicenter phase III trial with 1,342 patients. Crit Care Med. 2001;29(3):503–510. doi: 10.1097/00003246-200103000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Oberholzer A, Souza SM, Tschoeke SK, et al. Plasma cytokine measurements augment prognostic scores as indicators of outcome in patients with severe sepsis. Shock. 2005;23(6):488–493. [PubMed] [Google Scholar]
- 10.de Pablo R, Monserrat J, Reyes E, et al. Mortality in patients with septic shock correlates with anti-inflammatory but not proinflammatory immunomodulatory molecules. Journal of intensive care medicine. 2011;26(2):125–132. doi: 10.1177/0885066610384465. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann U, Bertsch T, Dvortsak E, et al. Matrix-metalloproteinases and their inhibitors are elevated in severe sepsis: prognostic value of TIMP-1 in severe sepsis. Scand J Infect Dis. 2006;38(10):867–872. doi: 10.1080/00365540600702058. [DOI] [PubMed] [Google Scholar]
- 12.Pruitt JH, Welborn MB, Edwards PD, et al. Increased soluble interleukin-1 type II receptor concentrations in postoperative patients and in patients with sepsis syndrome. Blood. 1996;87(8):3282–3288. [PubMed] [Google Scholar]
- 13.Xiao W, Mindrinos MN, Seok J, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208(13):2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren HS, Elson CM, Hayden DL, et al. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15(7–8):220–227. doi: 10.2119/molmed.2009.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai KH, Tan CS, Leek JT, et al. Dissecting inflammatory complications in critically injured patients by within-patient gene expression changes: a longitudinal clinical genomics study. PLoS Med. 2011;8(9):e1001093. doi: 10.1371/journal.pmed.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minei JP, Cuschieri J, Sperry J, et al. The changing pattern and implications of multiple organ failure after blunt injury with hemorrhagic shock. Crit Care Med. 2011 doi: 10.1097/CCM.0b013e3182376e9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feezor RJ, Baker HV, Mindrinos M, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19(3):247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 18.Cobb JP, Mindrinos MN, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102(13):4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Xu W, Herndon D, et al. Analysis of factorial time-course microarrays with application to a clinical study of burn injury. Proc Natl Acad Sci U S A. 2010;107(22):9923–9928. doi: 10.1073/pnas.1002757107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leek JT, Monsen E, Dabney AR, et al. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22(4):507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 21.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 22.Cuschieri J, Johnson JL, Sperry J, et al. Benchmarking Outcomes in the Critically Injured Trauma Patient and the Effect of Implementing Standard Operating Procedures. Ann Surg. 2012;255(5):993–999. doi: 10.1097/SLA.0b013e31824f1ebc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciesla DJ, Moore EE, Johnson JL, et al. A 12-year prospective study of postinjury multiple organ failure: has anything changed? Arch Surg. 2005;140(5):432–438. doi: 10.1001/archsurg.140.5.432. discussion 438–440. [DOI] [PubMed] [Google Scholar]
- 24.Pfeifer R, Tarkin IS, Rocos B, et al. Patterns of mortality and causes of death in polytrauma patients--has anything changed? Injury. 2009;40(9):907–911. doi: 10.1016/j.injury.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Marshall JC. Such stuff as dreams are made on: mediator-directed therapy in sepsis. Nat Rev Drug Discov. 2003;2(5):391–405. doi: 10.1038/nrd1084. [DOI] [PubMed] [Google Scholar]
- 26.Frantz S. An array of problems. Nat Rev Drug Discov. 2005;4(5):362–363. doi: 10.1038/nrd1746. [DOI] [PubMed] [Google Scholar]
- 27.Sethu P, Anahtar M, Moldawer LL, et al. Continuous flow microfluidic device for rapid erythrocyte lysis. Anal Chem. 2004;76(21):6247–6253. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- 28.Russom A, Sethu P, Irimia D, et al. Microfluidic leukocyte isolation for gene expression analysis in critically ill hospitalized patients. Clin Chem. 2008;54(5):891–900. doi: 10.1373/clinchem.2007.099150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turina M, Dickinson A, Gardner S, et al. Monocyte HLA-DR and interferon-gamma treatment in severely injured patients--a critical reappraisal more than a decade later. J Am Coll Surg. 2006;203(1):73–81. doi: 10.1016/j.jamcollsurg.2006.03.010. [DOI] [PubMed] [Google Scholar]