Abstract
In this issue of Science Translational Medicine, Tanas and colleagues describe a disease-defining genetic alteration for the vascular cancer epithelioid hemangioendothelioma (EHE). The resulting EHE-associated fusion gene encodes an aberrantly expressed putative transcription factor. This molecular information is the latest in a series of genetic discoveries that aid in cancer diagnosis and may pave the way to targeted therapeutic agents.
In the ancient Chinese text entitled The Art of War, author Sun Tzu writes that a key step to achieving victory is to “know thy enemy” (1). One of the challenges in the war on cancer has been in understanding the roots of this complex disease, which represents a multitude of neoplastic illnesses with different underlying genetic alterations. Cancer is commonly defined as a disease of the tissue of origin (for example, cancer of the colon, breast, or lung) and of specific cell types (carcinoma, sarcoma, etc.). Certain cancer subtypes have been classified by immunohistochemical labeling of marker proteins that are differentially present in selected tumors. However, conventional pathological analyses alone often cannot precisely identify the underlying tumor type, nor can they fully predict why individual patients respond to certain therapies or can display markedly different prognoses. Given the high complexity of this disease, one might expect that an intricate understanding of human cancer and, thus, disease management might only be possible when the underlying genetic changes are fully elucidated. A recent flurry of genome-wide sequencing analyses have revealed that tumors possess an underlying wealth of tumor-specific (somatic) genetic alterations that can be useful for improved tumor classification, appropriate therapeutic stratification, and disease monitoring.
In this issue of Science Translational Medicine, Tanas and colleagues describe the identification of a novel gene fusion between the WWTR1 and CAMTA1 genes in EHE (2). This study elegantly demonstrates that this gene fusion, resulting from a reciprocal t(1;3)(p36;q25) translocation, is present in virtually all EHEs but absent from other vascular neoplasms. This is an important finding because EHEs are difficult to distinguish clinically from other vascular tumors. Using this molecular translocation, the authors provide an immediately useful fluorescence in situ hybridization (FISH) test for accurate identification of EHE.
The WWTR1/CAMTA1 fusion gene consists of the vascular tissue–specific WWTR1 promoter and part of the gene that encodes the NH2 terminus of WWTR1 (a protein that is highly expressed in endothelial cells), fused in frame to the part of the CAMTA1 gene that encodes the COOH terminus of the brain-specific CAMTA1 transcription factor. Although the function of the resulting fusion protein is not known, it is likely that the aberrant expression of CAMTA1 in vascular tissue is crucial to the biology of this disease.
The observations by Tanas and colleagues (2) represent the latest in a series of recent discoveries of genomic alterations identified in human cancers. With the advent of unbiased approaches for massively parallel sequencing of tumor genomes and transcriptomes, a variety of previously unrecognized genes are now emerging as cancer gene “mountains,” or recurrently mutated genes in specific tumors (Table 1). One characteristic of these mountains that distinguishes them from common cancer genes, such as KRAS or TP53, is that they appear to be mutated predominantly in one or a few tumor types.
Table 1.
Cancer gene mountains identified through unbiased approaches.
| Altered gene | Tumors with specific gene alterations | References |
|---|---|---|
| ALK | Lung cancer, neuroblastoma | (16–21) |
| ARID1A | Ovarian clear cell carcinoma, endometrioid tumors | (7, 8) |
| ATRX | Pancreatic neuroendocrine tumors | (9) |
| DAXX | Pancreatic neuroendocrine tumors | (9) |
| DNMT3A | Acute myeloid leukemia | (14) |
| FOXL2 | Ovarian granulosa cell tumors | (32) |
| IDH1 | Gliomas, acute myeloid leukemia | (3–5) |
| IDH2 | Gliomas, acute myeloid leukemia | (3–5) |
| MLL2 | Medulloblastoma, non-Hodgkin lymphoma | (11, 12) |
| NOTCH1 | Head and neck cancer, chronic lymphocytic leukemia | (33–35) |
| PBRM1 | Clear cell renal cancer | (10) |
| PPP2R1A | Ovarian clear cell carcinoma, uterine carcinoma | (7, 36) |
| TMPRSS/ERG | Prostate cancer | (6) |
| WWTR1/CAMTA1 | Epithelioid hemangioendothelioma | (2) |
Analogous to the specificity of the WWTR1/CAMTA1 rearrangement for EHEs, mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 have been found in a significant fraction of gliomas and acute myeloid leukemias (AML) but rarely in cancers of epithelial or other origins (3–5). A similar pattern emerges for many of the genes in Table 1, including TMPRSS/ERG rearrangements in prostate cancer (6), mutations in ARID1A (which encodes a SWI/SNF transcriptional regulatory protein) in ovarian clear cell carcinoma (7, 8), alterations of the chromatin remodeling genes DAXX and ATRX genes in pancreatic neuroendocrine tumors (9), mutations in PBRM1 (which encodes part of the SWI/SNF chromatin-remodeling complex) in clear cell renal cancer (10), and alterations in the histone methyltransferase–encoding MLL2 gene in medulloblastomas and non-Hodgkin lymphoma (11, 12). The mechanism for this tumor specificity is unknown but may be related to the function of the products of altered genes in cell types that give rise to these tumors, such as high expression of WWTR1 in endothelial cells or the androgen-regulated spatial proximity of the TMPRSS and ERG genes in prostate cancer cells (2, 13).
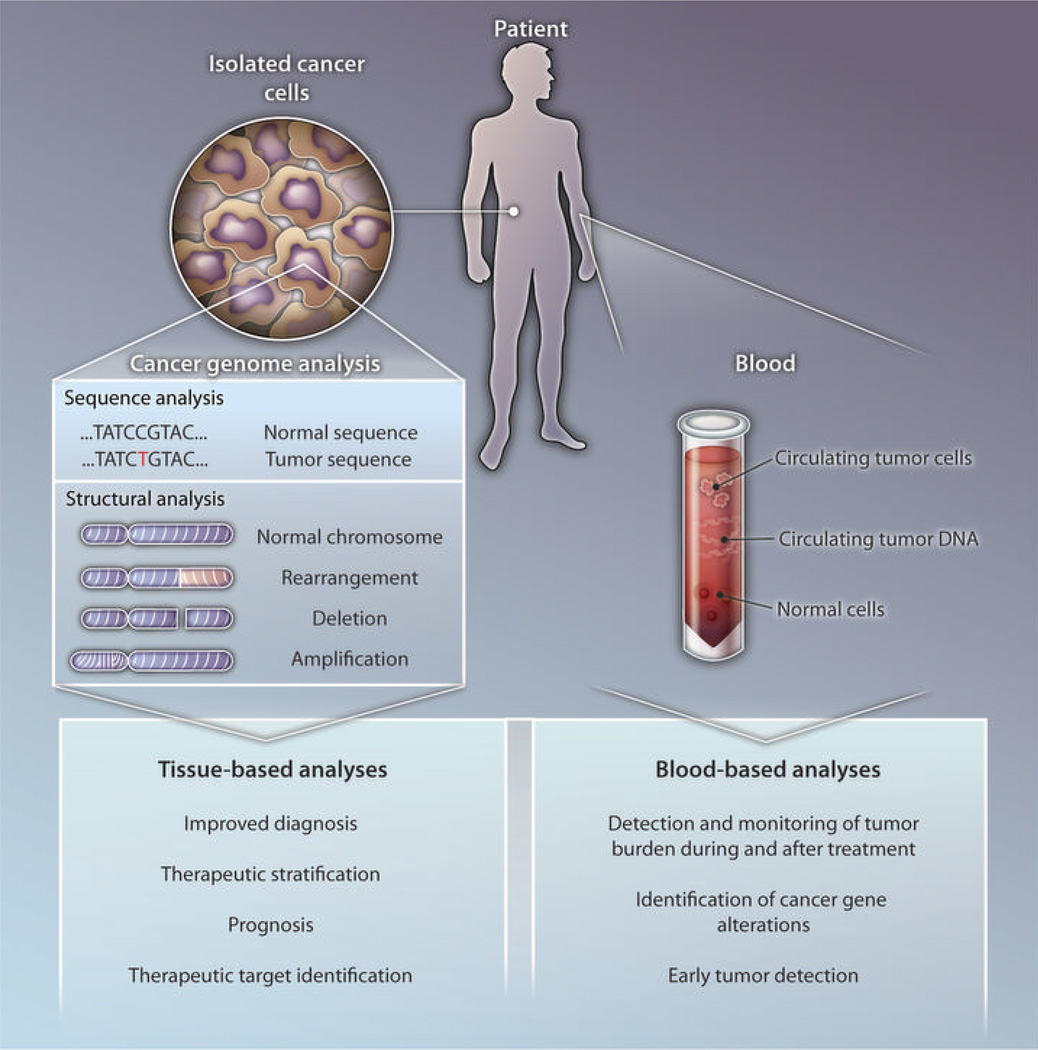
The specificity of these gene alterations has provided new opportunities for cancer diagnosis and prognosis (Fig. 1). For example, the discovery of alterations in IDH genes has provided a specific marker to distinguish between primary and secondary glioblastomas (5). The precise genetic alterations can be detected by direct sequencing of tumor tissue DNA at several regions of the IDH gene sequence that constitute mutational hotspots. Sequence analysis of such selectively mutated genes is also useful in classifying visibly indistinguishable tumor subtypes that have different clinical outcomes. For example, IDH mutations have been shown to represent an independent prognostic marker for improved survival in patients with gliomas (3, 5). Similarly, mutations in ATRX or DAXX identify a subset of pancreatic neuroendocrine patients with improved clinical outcomes (9). Surprisingly, the same gene may have different effects in different tumor types, as mutations in IDH and in the DNA methyltransferase–encoding DNMT3A gene have been reported to be indicators of worse prognosis in patients with AML (14, 15).
Fig. 1. Combating cancer.
Approaches for translating cancer-genome analyses to patient care.
In addition to identifying tumor subtypes with different courses of natural progression, tumor-specific alterations may be useful in stratifying patients for specific therapies. In some cases, the proteins encoded by the altered genes provide direct therapeutic targets. the classic example of such rational therapeutic targeting is the protein kinase inhibitor imatinib (Gleevec) and its use in CML patients with the BCR/ABL translocation. More recently, lung cancers and neuroblastomas that contain alterations in the ALK tyrosine kinase provide an exciting use for new protein kinase inhibitors such as crizotinib (16–22). Mutations in IDH genes have spurred efforts to identify compounds that inhibit its newly acquired metabolic function (23, 24). Alterations in BRAF genes have led to the development of various targeted compounds that block the aberrant protein kinase activity of the encoded mutated signaling protein; the most recent of these drugs, vemurafenib, has shown dramatic clinical effects in patients with melanoma—the tumor type in which this gene is most frequently mutated (25, 26). Importantly, the effect of such targeted inhibition is efficacious only in tumor cells that express the genetically altered version of the target gene. Given the high expression of the WWTR1/CAMTA1 fusion gene in EHEs, it is conceivable that this gene could serve as a useful therapeutic target if the fusion gene is required for continued proliferation of EHEs and the targeting compounds are specific for the altered cells.
Because they are not present in normal cells, somatic alterations should be highly specific for tumor cells and, thus, have the potential to serve as biomarkers for tumor detection and monitoring. Mutation-associated biomarkers also could be useful for monitoring tumor response to specific therapies, detection of residual disease after surgery, and long-term clinical management. For example, quantitative reverse transcription–polymerase chain reaction (qRT-PCR) measurement of BCR/ABL mRNA transcripts has provided a sensitive method for determining the response, at the molecular level, to imatinib therapy in CML patients (27). More broadly generalizable approaches have been developed recently with the use of cancer-specific gene-sequence alterations (28) and genomic rearrangements (29, 30). Tumor-specific recurrent rearrangements, such as the WWTR1/CAMTA1 translocation in EHEs, may be particularly useful as biomarkers because these genetic disruptions can be detected with a direct PCR-based test that analyzes the DNA sequence across the rearranged fusion gene junction.
Although understanding the complexity of cancer provides new avenues for clinical intervention, a number of challenges must be met before such approaches can be made clinically practicable. First is the transfer of high-throughput genomic analyses from the research laboratory to clinical evaluation of individual patient samples. Although such mutational analyses can be performed on a gene-by-gene basis in clinical labs (including the newly described FISH analysis of the WWTR1/CAMTA1 translocation), it is now possible to routinely analyze the entire genomic landscape of each patient’s cancer (31). Although this proposition may seem daunting, technology has reached the point at which a large number of individual gene tests could be performed more efficiently through the simultaneous analysis of all protein-coding genes. Such assays take full advantage of new DNA sequencing technologies; however, substantial effort is required to make such large-scale tests affordable, timely, and integrated in routine care.
Perhaps more important, the immense amount of data obtained by such tumor sequencing needs to be carefully analyzed to extract bona fide somatic alterations and to interpret the results of these molecular events from a biological and a clinical perspective. Using genomic sequencing information for tumor diagnosis, therapeutic decision making, and clinical monitoring will require a new generation of appropriately trained scientists and physicians who can incorporate the ever-changing connections between genetic alterations and novel therapies into clinical practice. Much remains to be done, but with such molecular tools, we are on the verge of understanding the complexity of human tumors in individual patients. Once the enemy is known, writes Sun Tzu, “a thousand battles, a thousand victories” (1).
Acknowledgments
Funding: This work was supported by grants CA121113 and CA129825 from the U.S. National Institutes of Health, the Virginia and D. K. Ludwig Fund for Cancer Research, the AACR Stand Up To Cancer-Dream Team Translational Cancer Research Grant, and Swim Across America.
Footnotes
Competing interests: V.E.V. and L.D. are cofounders of Personal Genome Diagnostics and Inostics, are members of their Scientific Advisory Boards, and own Personal Genome Diagnostics and Inostics stock, which is subject to certain restrictions under Johns Hopkins University policy.
REFERENCES AND NOTES
- 1.Tzu S. The Art of War (written in 6th century B.C.), English translation by Samuel B. Griffith. Oxford Univ. Press; Oxford: 1971. [Google Scholar]
- 2.Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci. Transl. Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Jones S, Wang TL, Shih I-M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1Ain ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih I-M, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTORpathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, Jackman S, Krzywinski M, Scott DW, Trinh DL, Tamura-Wells J, Li S, Firme MR, Rogic S, Griffith M, Chan S, Yakovenko O, Meyer IM, Zhao EY, Smailus D, Moksa M, Chittaranjan S, Rimsza L, Brooks-Wilson A, Spinelli JJ, Ben-Neriah S, Meissner B, Woolcock B, Boyle M, McDonald H, Tam A, Zhao Y, Delaney A, Zeng T, Tse K, Butterfield Y, Birol I, Holt R, Schein J, Horsman DE, Moore R, Jones SJ, Connors JM, Hirst M, Gascoyne RD, Marra MA. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, Späth D, Kayser S, Zucknick M, Götze K, Horst HA, Germing U, Döhner H, Döhner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 16.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 18.George RE, Sanda T, Hanna M, Fröhling S, Luther W, 2nd, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, Xue L, Zozulya S, Gregor VE, Webb TR, Gray NS, Gilliland DG, Diller L, Greulich H, Morris SW, Meyerson M, Look AT. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, Nakagawara A, Hayashi Y, Mano H, Ogawa S. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 20.Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, Valteau-Couanet D, Frebourg T, Michon J, Lyonnet S, Amiel J, Delattre O. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 21.Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, Laureys G, Speleman F, Kim C, Hou C, Hakonarson H, Torkamani A, Schork NJ, Brodeur GM, Tonini GP, Rappaport E, Devoto M, Maris JM. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non�small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: Biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 25.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D’Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur AG BRIM-3 Study Group. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, Baccarani M, Cortes J, Cross NC, Druker BJ, Gabert J, Grimwade D, Hehlmann R, Kamel-Reid S, Lipton JH, Longtine J, Martinelli G, Saglio G, Soverini S, Stock W, Goldman JM. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: Review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci. Transl. Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, Hämäläinen E, Stebbings LA, Andersson LC, Flanagan AM, Durbecq V, Ignatiadis M, Kallioniemi O, Heckman CA, Alitalo K, Edgren H, Futreal PA, Stratton MR, Campbell PJ. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49:1062–1069. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingsmore SF, Saunders CJ. Deep sequencing of patient genomes for disease diagnosis: When will it become routine? Sci. Transl. Med. 2011;3:87ps23. doi: 10.1126/scitranslmed.3002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah SP, Köbel M, Senz J, Morin RD, Clarke BA, Wiegand KC, Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E, Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y, Zeng T, Glover JN, Vanderhyden B, Zhao C, Parkinson CA, Jimenez-Linan M, Bowtell DD, Mes-Masson AM, Brenton JD, Aparicio SA, Boyd N, Hirst M, Gilks CB, Marra M, Huntsman DG. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 33.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie T-X, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Treviño L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño JC, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, Bassaganyas L, Baumann T, Juan M, López-Guerra M, Colomer D, Tubío JM, López C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernández JM, Puente DA, Freije JM, Velasco G, Gutiérrez-Fernández A, Costa D, Carrió A, Guijarro S, Enjuanes A, Hernández L, Yagüe J, Nicolás P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjosé S, Piris MA, de Alava E, San Miguel J, Royo R, Gelpí JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigó R, Bayés M, Heath S, Gut M, Klatt P, Marshall J, Raine K, Stebbings LA, Futreal PA, Stratton MR, Campbell PJ, Gut I, López-Guillermo A, Estivill X, Montserrat E, López-Otín C, Campo E. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McConechy MK, Anglesio MS, Kalloger SE, Yang W, Senz J, Chow C, Heravi-Moussavi A, Morin GB, Mes-Masson AM, Carey MS, McAlpine JN, Kwon JS, Prentice LM, Boyd N, Shah SP, Gilks CB, Huntsman DG. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J. Pathol. 2011;223:567–573. doi: 10.1002/path.2848. [DOI] [PubMed] [Google Scholar]