Abstract
Natural products and related structures are essential sources of new pharmaceuticals, because of the immense variety of functionally relevant secondary metabolites of microbial and plant species. Furthermore, the development of powerful analytical tools based upon genomics, proteomics, metabolomics, bioinformatics and other 21st century technologies are greatly expediting identification and characterization of these natural products. Here we discuss the synergistic and reciprocal benefits of linking these ‘omics technologies with robust ethnobotanical and ethnomedical studies of traditional medicines, to provide critically needed improved medicines and treatments that are inexpensive, accessible, safe and reliable. However, careless application of modern technologies can challenge traditional knowledge and biodiversity that are the foundation of traditional medicines. To address such challenges while fulfilling the need for improved (and new) medicines - we encourage development of Regional Centres of ‘omics Technologies functionally linked with Regional Centres of Genetic Resources, especially in regions of the world where use of traditional medicines is prevalent and essential for health.
Introduction and Background
Despite the great synthetic diversity derived from the development of combinatorial chemistries and high-throughput screening methods over the past fifty years, natural products and related structures continue to be extremely important elements of pharmacopoeias. Looking forward, natural products and related structures are likely to become even more important for development of improved and new medicines, due to the variety of functionally relevant secondary metabolites of microbial and plant species whose chemical and genetic diversity are being revealed by ultra fast DNA sequencing and related genomics and bioinformatics tools 1-4.
Heretofore, methods for identifying and characterizing the activities of secondary metabolites have been inefficient and often tedious, but recent advances in genomics, informatics, and associated 21st century ‘omics technologies are dramatically accelerating the pace of discovery and analysis. Sophisticated fractionation methods hyphenated to modern spectrometries and spectroscopies as described in this issue (cite reviews by Reynolds5, Hamburger6, Bucar7, and Wurtele8) can define the metabolomes of cells, tissues and even organisms. Multivariate analyses9 and network modeling 10-12 enable comprehensive identification and evaluation of natural product diversity and functionality; and when integrated with systems approaches, it is possible to profile molecular changes caused by mutation and by pathogens and other environmental stressors, and thus to predict the targets and mode(s) of action and toxicities of natural products and derivatives 13, 14.
Considerable synergy and benefit for the development of improved medicines and new drugs can come from linking these powerful scientific tools to robust ethnomedical and ethnobotanical studies of traditional medicines. However, ethical and socio-economic challenges also must be addressed for the development of improved medicines and new drugs to be achieved while also benefiting those that currently rely upon traditional medicines and accompanying natural products for health and well being.
As background, we rely upon these recent contributions: Corson and Crews 15 summarize classical approaches for identifying and studying single active agents of traditional medicines - but these methods are clearly very limited when viewed in context of 21st century ‘omics technologies. Ulrich-Merzenich, et al. 16 and Heinrich 17 describe how genomics, proteomics, metabolomics and systems biologies are contributing to ethnopharmacy, and Prasain and Barnes 18 focus upon the contributions of ‘omics technologies to understanding and validating traditional medicines. These and other similar reviews document the value of ‘omics technologies for advancing analysis of natural products (especially from plants), but in our view the reciprocal and synergistic benefits of linking robust ethnomedical and ethnobotanical studies of traditional medicines with 21st century ‘omics technologies have not been adequately considered.
Here, we focus upon development of improved and new medicines and treatments, especially for ancient diseases such as tuberculosis (Tb) and malaria that continue to plague mankind, and for which traditional medicines have been and will continue to be used.
Tuberculosis and malaria
Throughout recorded history, Mycobacterium tuberculosis has been a leading cause of human morbidity and mortality. With the biomedical advances of the twentieth century, global Tb incidence declined, but the disease has resurfaced with even more vigour in immune-compromised peoples living with HIV/AIDS, and increasingly with diabetes. Presently, about two billion persons are infected or are at risk of M. tuberculosis infection, which is currently responsible for 2.4% of worldwide mortality19,20. Malaria, another ancient and important infectious disease is caused by various Plasmodium species and ranks 5th among causes of death. At particularly high-risk for malaria are children, pregnant women and the many who are immune-compromised; in highly malaria-endemic areas, a child dies every minute from malaria21. In addition to the immediate impact on individuals and families, these ancient diseases have huge economic and social costs that threaten civil societies 22.
Current therapies for Tb require long regimens with significant side effects and toxicities. Consequently, treatments suffers from poor adherence and are compromised by the emergence of multidrug and extreme drug resistant M. tuberculosis 23. For malaria, artemisinin combination therapies (ACT) have reduced disease prevalence and mortality, but the requisite medicines are not accessible to many that need them and their dosing is complex, so monotherapies are still widely used that promote genetic resistance21, 24. New drug scaffolds and treatments that address these shortcomings are critically needed for both Tb and malaria (and other extant and emerging diseases). Importantly, the new treatments should have multiple independent targets and must be accessible and affordable 25, 26. These are often properties of traditional medicines, so better understanding and use of traditional medicines may significantly benefit health and well being.
Significant challenges in developing new treatments for Tb and malaria arise from the complex biology of the pathogens, the expense of research, the lack of financial incentives and inadequate delivery systems 27-29. A “systems biology” approach that integrates epidemiology with ‘omics technologies has been suggested to strengthen such efforts 30. We recommend inclusion in such ‘systems’ approaches, additional elements comprisin robust ethnobotanical and ethnomedical analyses of traditional medicines. Such a ‘hat trick’ of ‘systems studies’ could expand upon the well-established paradigms of important single agent drugs such as aspirin, codeine, quinine and artemisinin31, by adding multicomponent mixtures of treatments derived from traditional medicines that also can be accessible and affordable to those in resource poor regions.
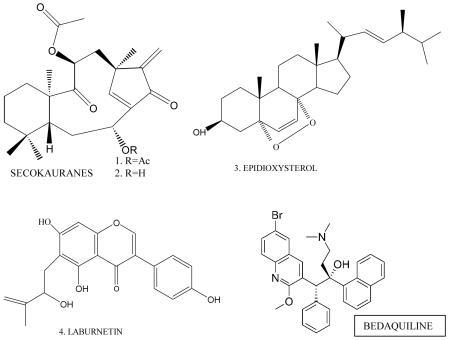
After many years of research, some promising candidates for anti-mycobacterial drugs recently have reached clinical trials 32, 33 and additional candidates will derive from the elucidation of the M. tuberculosis genome 34 and its transcriptome and metabolome and structural and system biology modeling 35, 36 through in silico analyses 37, 38 and from insights into the infection and survival pathways adopted by M. tuberculosis 39-41, as also of host-microbe interactions 42, and informatics resources 43,44-46. Very recently, the U.S Food and Drug Administration (FDA) approved the first new Tb drug in more than 40 years 47. Bedaquilin (shown below) was discovered in 200548 and was assessed by clinical trials of multidrug-resistant Tb patients49. However, treatment requires drug-sensitivity tests which are costly and time-consuming and therefore may not reach many that suffer from Tb in low-income countries.
Numerous natural products in traditional medicines inhibit M. tuberculosis
50 (salient examples are the secokaurenoids 51, epidioxysterol 52, and the isoflavonoid laburnetin 53 shown below), but we are unaware of any current efforts to use these compounds as scaffolds for new drugs. Nor are we aware of concerted efforts to screen other antimicrobial compounds 53 that could become new anti-mycobacterial drug scaffolds.
Similar research advances are being made in the analysis of factors contributing to malaria, including the elucidation of the genomes of Plasmodium species 54-57 , analysis of parasite-host interactions 58, identification of vaccine targets 59-61, transcriptomic, metabolomic and proteomic analyses 62, 63, and new insights into the stages of parasite development 64-66. Moreover, the mechanisms of artemisinin action against Plasmodium spp. are being described and correlated with regions of the Plasmodium genome 67-69 . Genome-scale proteomics and structural biology are helping to elucidate protein structures of potential targets which should facilitate the development of vaccines and new drugs 70. Approaches to further understand the dynamics of artemisinin action and its targets will likely be developed, as has occurred with drugs targeting influenza, bacteria, HIV, and cancer71-74, 75 and should provide deeper insights into the mechanisms for pathogen resistance.
Speeding development of 21st century medicines and treatments
The development of artemisinin and related antimalarial compounds serves as a modern paradigm for the value of traditional medicines in drug discovery, and we assert the potential exists for additional discoveries of similar importance: of the estimated 250,000 – 500,000 extant plant species, only a fraction have been scientifically investigated for biological activity76-79. Unexplored are untold numbers of species that are likely to be included in traditional medicines 80. Plants from widely separated regions of the world that are components of traditional medicines used to treat specific conditions such as malaria are phylogenetically clustered 81; this principle has been recently described for Pterocarpus, which has significant cross-culture patterns that can inform drug development and supports the value of linking robust ethnobotanical and ethnomedical studies with 21st century ‘omics technologies and systems analyses 82-84 to speed identification of functionally relevant bioactivities .
Inclusion of traditional medicines in development of 21st century treatment paradigms can help assure their convenience, acceptability and accessibility 85 . Furthermore, pharmacological synergism, a principle employed by many traditional medicines lessens the likelihood of development of genetic resistance by the pathogen or disease against drug monotherapies. Synergy research inspired by a “reverse pharmacological approach” 86, could lead to a “new generation of phytopharmaceuticals” 87. The use of powerful ‘omics technologies facilitates disentangling such complexity88, 89: metabolomics analyses enable profiling of major and minor metabolites and bioactive components that contribute to synergism; and computational approaches for analysis of multiple-activity networks have become powerful tools for defining the principal components of mixtures with synergistic modes of action, for prediction of drug metabolism and toxicity, and for high-throughput prioritizing of agent combinations 90. Data mining approaches to identify active compounds in mixtures of natural products are being developed 91 and will be essential for the development of effective multiple-agent drugs from traditional medicines.
While U.S. requirements for regulatory approval for health claims made for multicomponent medicines present significant challenges for the development of effective multiple-agent drugs from traditional medicines, this is less so for Europe, and especially not for regions of the world highly impacted by Tb and malaria (and in which there are strong traditions of traditional medicine use). As described below, with the establishment of regional research facilities to confirm the safety and efficacy of traditional medicines through the use of ‘omics tools and robust ethnobotanical and ethnomedical data, significant improvements in development of improved medicines that are accessible and affordable can be expected.
A significant challenge in anti-Tb drug discovery and development is the lack of suitable animal models that can help predict clinical outcomes of infection25. Traditional medicines that have been used for generations can be said to have undergone preliminary preclinical and clinical assessment and thus, with appropriate observational data for safety and efficacy, should offer leads that can supplement or even bypass testing in animal models 31, 92. In turn, application of ‘omics technologies and systems biologies can provide independent evidence for safety and efficacy of traditional medicines 89, 93, 94.
Use of ‘omics technologies for medicinal plant improvement
21st Century ‘omics technologies also can advance the synthesis and production of natural products. An excellent example is artemisinin, traditionally derived from Artemisia annua 95, 96 and very recently, due to the application of genetic technologies, from microbial synthesis 97-100. Traditional medicines containing A. annua have been used for thousands of years and as a consequence, the plant is important in healthcare, agriculture and commerce 101-103. However, artemisinin yields from most outbred varieties is very low, so ‘omics technologies also are being employed to increase yield 96: the synthesis and regulation of artemisinin metabolic pathways have been described 104, 105, and significant attempts have been made to clone biosynthetic cDNAs and ESTs 106, characterize transcription 107, identify key genes 108, 109, and profile expression and metabolite levels 110, 111,112,113.
Plant natural products are intertwined in societies’ cultures, healthcare systems, and economies 114, 115, whose indigenous knowledge 116 can inform natural product research and further development 117, 118. Attention to the conservation of these cultural and biological resources has been assumed by governments and NGOs 119-121 as the livelihoods of people who rely on medicinal plants, especially farmers, marketers and traditional healers, are dependent upon improvement of natural products and related resources 114. Switching to an improved variety of A. annua potentially benefits growers, provided that market demand and prices are assured 122, 123. The cost of artemisinin production in developing countries is usually low, creating competitive markets for the products; for example, in Vietnam, the cultivation of A. annua produced approximately 13 tons of artemisinin at $1000-2000 per ton 124. Other plants with promising medicinal properties also can become potential sources of income125, 126.
Technological development affects the livelihoods of people who participate in the analysis, production and distribution of natural products 114, 118, 127 and can be positive or negative 128, 129. For example, the significant advances in synthetic and microbial artemisinin manufacturing and in A. annua crop yield 113, 130 can contribute to increasing artemisinin supply 131 and may result in decreased cost of artemisinin production for artemisin combination treatments (ACT) 132 . However, while significant improvement in malaria treatment in the early 1990s was associated with increased production of artemisinin in Vietnam 133, 134, artemisinin supply from A. annua cultivation is predicted to meet ACT demand in 2012, hence further increases in artemisinin production are not likely to provide equivalent improvements in malaria treatment 128, 129, 135.
Consequently, key recommendations from the RBM/UNITAID/WHO have been to improve communication and flexibility in the supply chain for artemisinin, rather than to increase yield and production 135. If microbial production of artemisinin becomes significant, plant-derived artemisinin will have to compete with the microbial product, leading to a significant impact on the livelihoods of people who rely on the crop for income.
Preservation of Traditional Knowledge, Biodiversity, and Access and Benefit Sharing of Genetic Resources
The Convention for Biological Diversity (CBD) articulates principles intended to protect traditional knowledge and biodiversity and to facilitate access to genetic resources and the fair and equitable sharing of benefits arising from their utilization. However, these principles have not been translated into reality. Disappointingly, many of the Access and Benefit Sharing (ABS) regimes that developed from the CBD are proving to be significant barriers to the development and utilization of genetic resources, and also poorly define the traditional knowledge systems and the owners that might benefit from ABS regimes136-138.
Recommendations regarding responsibilities and procedures for implementation of the CBD principles and for assuring compliance are articulated by the Nagoya Protocol, which clarifies the scope of the CBD to include research and development of gene sequences and natural products and the traditional knowledge associated with these genetic resources 139. However, much remains to fulfill the vision of the CBD. Parties must develop implementing rules for ratification of the Nagoya Protocol, and there is need for constructive dialogue between parties that view themselves as the ‘providers’ or ‘users’ of genetic resources and traditional knowledge. Such dialogues will be stimulated by development of cooperative research agreements and exchanges of best practices 139 and concomitant commercial successes guided by the CBD principles.
The ‘fair and equitable sharing of benefits’ principles articulated by the CBD are not solely concerned with the sharing of results, products of commercialization and other outcomes of the use of Genetic Resources, but also are meant to include participation in research and development and transfer of technologies 138, 140. Consequently, those who rely primarily upon traditional medicines for their healthcare and livelihoods also should directly benefit from their more detailed analysis and development141. Returning to some of the literature cited earlier, Ulrich-Merzenich, et al.16 recommend use of ‘omics technologies for development of local resources that improve primary care; and similarly, Prasain and Barnes 18 suggest the development of integrative global healthcare systems combining traditional and modern medicines. This latter goal has been espoused by the World Health Organization but not without controversy. Hollenberg, et al. 142 suggest that traditional medical practices can fruitfully coexist with public health and other medical science innovations, but also caution such relationships are especially vulnerable. Parties should appropriately gauge the outcomes of development and encourage only those applications that can be realized without harmful consequences.
Regional Scientific Research and Development Centres
Parties that wish to apply ‘omics technologies to natural product research and drug development are faced with significant start-up and ongoing costs for infrastructure and for trained personnel. These costs can exceed tens of millions of dollars for space and for equipment, and millions of dollars yearly for maintenance of the space and equipment and for training staff. Costs for ‘omics technologies experimentation can exceed tens of thousands of dollars per protocol. While many ‘omics-related informatics resources are readily accessible, the technology and computing infrastructure and properly trained staff required to ensure that the informatics resources reach the potential users can be costly. Many low- and middle-income countries are unlikely to be able to provide such resources without external investments 143, 144. However, those parties that lead the development of public-private partnerships and achieve initial successes are likely to secure long-lasting temporal and market advantages.
To benefit from ‘omics technologies, countries must develop scientific training programs that strengthen know how. This is occurring in Viet Nam, where the Vietnam Academy of Science and Technology (VAST) has built strong networks in research and education with 28 international institutions of 16 countries 145, and the Oxford University Clinical Research Unit in Viet Nam (OUCRU) is providing opportunities for scientists to conduct research programs focusing on malaria, tuberculosis, HIV/AIDS and other major diseases 146. Together with such efforts, building research capacity to study traditional knowledge and traditional medicines is vital.
Analogously, the network of Botanic Gardens in South East Asia, established in 2004 by the Botanic Gardens Conservation International (BGCI) emphasizes conservation of biological diversity and related research and education147. BGCI has built similar strong networks to secure biodiversity, enable training and influence decision making and policy regarding CBD148. We suggest that these centres might link to regional research centres with ‘omics capacities to expedite natural product discovery and drug development. Arguments in favour of regional centres as means of drug development and commercialization have been made by others 138, 149, but we expand upon these suggestions by combining development of ‘omics technologies and biodiversity and indigenous knowledge conservation, access benefit and sharing of resources.
For Africa the BecA-ILRI Hub located in Nairobi, Kenya and managed by ILRI as one of four biosciences centres of excellence that are part of the African Union-New Partnership for Africa’s Development (AU-NEPAD) African Biosciences Initiative is developing ‘omics infrastructure. BecA-ILRI has been created under the Comprehensive African Agricultural Productivity Programme (CAADP) to service the needs of countries in eastern and central Africa. Analogous investment and partnerships in Ghana could provide similar resources to western Africa150. In southern Africa, outstanding ‘omics infrastructure occurs in South Africa’s major research universities and government research agencies, and draft government policies on African Traditional Medicine provide a framework for the registration and regulation of genetic resources, indigenous knowledge and African traditional medicines, and their study through ‘omics technologies. Rigorous clinical evaluation of the more widely used traditional medicines of southern Africa are occurring through partnerships between traditional medical practitioners and biomedical and social scientists151. These partnerships create environments that can guide the choice of traditional medicines to be studied by ‘omics techniques.
Altogether, these efforts should greatly improve treatment programs for HIV/AIDS, Tb, malaria and the chronic diseases for which traditional medicines have been employed for generations, and have much potential to contribute solutions.
Acknowledgements
We thank Dr. Michael Balick, Dr. Paul Hippenmeyer, Professor C. McManis, Dr. Jim Miller, Dr. Hoan Nguyen, Dr. Thach Nguyen, Dr. Phip Ninh, and Mr. Ricki Orford for comments and suggestions. We apologize to many colleagues whose contributions are not cited here for lack of space.
This publication was made possible by NIH Grants P50AT006273 and 3U19AT003264 from the National Center for Complementary and Alternative Medicines (NCCAM), the Office of Dietary Supplements (ODS), the Fogarty International Institute (FIC) and the National Cancer Institute (NCI), the Vietnam Education Foundation (VEF) and the University of Missouri. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, ODS, FIC, NCI, NIH, the VEF or the University of Missouri.
BIBLIOGRAPHY
- 1.Buell R. Natural product reports. 2013 e. al. (Insert citation) [Google Scholar]
- 2.Newman DJ, Cragg GM. Journal of natural products. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh CT, Fischbach MA. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JW, Vederas JC. Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds W. Natural product reports. 2013 e. al. (Insert Citation) [Google Scholar]
- 6.Hamburger M. Natural product reports. 2013 doi: 10.1039/c3np20094a. e. al. (Insert Citation) [DOI] [PubMed] [Google Scholar]
- 7.Bucar F. Natural product reports. 2013 doi: 10.1039/c3np20106f. e. al. (Insert Citation) [DOI] [PubMed] [Google Scholar]
- 8.Wurtele ES. Natural product reports. 2013 e. al. (Insert Citation) [Google Scholar]
- 9.Okada T, Mochamad Afendi F, Altaf-Ul-Amin M, Takahashi H, Nakamura K, Kanaya S. Current Computer - Aided Drug Design. 2010;6:179–196. doi: 10.2174/157340910791760055. [DOI] [PubMed] [Google Scholar]
- 10.Arrell DK, Terzic A. Clinical pharmacology and therapeutics. 2010;88:120–125. doi: 10.1038/clpt.2010.91. [DOI] [PubMed] [Google Scholar]
- 11.Leung EL, Cao Z-W, Jiang Z-H, Zhou H, Liu L. Briefings in Bioinformatics. 2012 doi: 10.1093/bib/bbs043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugrim A, Nikolskaya T, Nikolsky Y. Drug discovery today. 2004;9:127–135. doi: 10.1016/S1359-6446(03)02971-4. [DOI] [PubMed] [Google Scholar]
- 13.Kell DB. Drug discovery today. 2006;11:1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Wishart DS. Drugs in R&D. 2008;9:307–322. doi: 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
- 15.Corson TW, Crews CM. Cell. 2007;130:769–774. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulrich-Merzenich G, Zeitler H, Jobst D, Panek D, Vetter H, Wagner H. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2007;14:70–82. doi: 10.1016/j.phymed.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Heinrich M. Phytochemistry Letters. 2008;1:1–5. [Google Scholar]
- 18.Prasain J, Barnes S. In: Osbourn AE, Lanzotti V, editors. Springer; US: 2009. pp. 533–546. [Google Scholar]
- 19.World Health Organization [Accessed 10/2012, 2012];The top 10 causes of death. http://www.who.int/mediacentre/factsheets/fs310/en/index.html.
- 20.World Health Organization . Global tuberculosis report 2012. World Health Organization; Geneva: 2012. [Google Scholar]
- 21.World Health Organization . World malaria report 2012. World Health Organization; Geneva: 2012. [Google Scholar]
- 22.Sachs J, Malaney P. Nature. 2002;415:680–685. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]
- 23.Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J. Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 24.Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L. Nature reviews. Microbiology. 2010;8:272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- 25.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 26.Kar S. Nature reviews. Drug discovery. 2010;9:511–512. doi: 10.1038/nrd3207. [DOI] [PubMed] [Google Scholar]
- 27.Schmid EF, Smith DA. Drug discovery today. 2005;10:1031–1039. doi: 10.1016/S1359-6446(05)03524-5. [DOI] [PubMed] [Google Scholar]
- 28.Ma Z, Lienhardt C, McIlleron H, Nunn AJ, Wang X. Lancet. 2010;375:2100–2109. doi: 10.1016/S0140-6736(10)60359-9. [DOI] [PubMed] [Google Scholar]
- 29.Bate R, Jensen P, Hess K, Mooney L, Milligan J. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2013 doi: 10.5588/ijtld.12.0355. [DOI] [PubMed] [Google Scholar]
- 30.Kirschner DE, Young D, Flynn JL. Current opinion in biotechnology. 2010;21:524–531. doi: 10.1016/j.copbio.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox PA, Balick MJ. Scientific American. 1994;270:82–87. [PubMed] [Google Scholar]
- 32.Singh R, Manjunatha U, Boshoff HIM, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manjunatha U, Boshoff HI, Barry CE. Communicative & integrative biology. 2009;2:215–218. doi: 10.4161/cib.2.3.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 35.Beste DJ, Hooper T, Stewart G, Bonde B, Avignone-Rossa C, Bushell ME, Wheeler P, Klamt S, Kierzek AM, McFadden J. Genome biology. 2007;8:R89. doi: 10.1186/gb-2007-8-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chindelevitch L, Stanley S, Hung D, Regev A, Berger B. Genome biology. 2012;13:r6. doi: 10.1186/gb-2012-13-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh VK, Ghosh I. Theoretical biology & medical modelling. 2006;3:27. doi: 10.1186/1742-4682-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milne CB, Kim PJ, Eddy JA, Price ND. Biotechnology journal. 2009;4:1653–1670. doi: 10.1002/biot.200900234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boshoff HI, Lun DS. Drug discovery today. Disease mechanisms. 2010;7:e75–e82. doi: 10.1016/j.ddmec.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boshoff HI, Tahlan K. Drug discovery today. Disease mechanisms. 2010;7:e1–e3. doi: 10.1016/j.ddmec.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young D, Stark J, Kirschner D. Nature reviews. Microbiology. 2008;6:520–528. doi: 10.1038/nrmicro1919. [DOI] [PubMed] [Google Scholar]
- 42.Sturdevant DE, Virtaneva K, Martens C, Bozinov D, Ogundare O, Castro N, Kanakabandi K, Beare PA, Omsland A, Carlson JH, Kennedy AD, Heinzen RA, Celli J, Greenberg DE, DeLeo FR, Porcella SF. Future microbiology. 2010;5:205–219. doi: 10.2217/fmb.09.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundaramurthi JC, Brindha S, Reddy TBK, Hanna LE. Tuberculosis. 2012;92:133–138. doi: 10.1016/j.tube.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. PLoS Med. 2009;6:e1000002. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brudey K, Driscoll J, Rigouts L, Prodinger W, Gori A, Al-Hajoj S, Allix C, Aristimuno L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans J, Fauville-Dufaux M, Ferdinand S, de Viedma D, Garzelli C, Gazzola L, Gomes H, Guttierez MC, Hawkey P, van Helden P, Kadival G, Kreiswirth B, Kremer K, Kubin M. BMC Microbiology. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang YT, Marshall GR. Methods Mol Biol. 2011;716:1–22. doi: 10.1007/978-1-61779-012-6_1. [DOI] [PubMed] [Google Scholar]
- 47.Cohen J. Science. 2013;339:130. doi: 10.1126/science.339.6116.130. [DOI] [PubMed] [Google Scholar]
- 48.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 49.Diacon AH, Donald PR, Pym A, Grobusch M, Patientia RF, Mahanyele R, Bantubani N, Narasimooloo R, De Marez T, van Heeswijk R, Lounis N, Meyvisch P, Andries K, McNeeley DF. Antimicrobial agents and chemotherapy. 2012;56:3271–3276. doi: 10.1128/AAC.06126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okunade AL, Elvin-Lewis MP, Lewis WH. Phytochemistry. 2004;65:1017–1032. doi: 10.1016/j.phytochem.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Thongtan J, Kittakoop P, Ruangrungsi N, Saenboonrueng J, Thebtaranonth Y. Journal of natural products. 2003;66:868–870. doi: 10.1021/np030067a. [DOI] [PubMed] [Google Scholar]
- 52.Saludes JP, Garson MJ, Franzblau SG, Aguinaldo AM. Phytotherapy research : PTR. 2002;16:683–685. doi: 10.1002/ptr.1003. [DOI] [PubMed] [Google Scholar]
- 53.Saleem M, Nazir M, Ali MS, Hussain H, Lee YS, Riaz N, Jabbar A. Natural product reports. 2010;27:238–254. doi: 10.1039/b916096e. [DOI] [PubMed] [Google Scholar]
- 54.Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 55.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang’a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 58.Kwiatkowski DP. American journal of human genetics. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GAT, Su X.-z. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 60.Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr., Daily JP, Sarr O, Ndiaye D, Ndir O, Mboup S, Duraisingh MT, Lukens A, Derr A, Stange-Thomann N, Waggoner S, Onofrio R, Ziaugra L, Mauceli E, Gnerre S, Jaffe DB, Zainoun J, Wiegand RC, Birren BW, Hartl DL, Galagan JE, Lander ES, Wirth DF. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 61.Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Nucleic acids research. 2006;34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr., Treatman C, Wang H. Nucleic acids research. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, James K, Rutherford K, Harris B, Harris D, Churcher C, Quail MA, Ormond D, Doggett J, Trueman HE, Mendoza J, Bidwell SL, Rajandream M-A, Carucci DJ, Yates JR, Kafatos FC, Janse CJ, Barrell B, Turner CMR, Waters AP, Sinden RE. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 65.Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, Lemieux J, Barrell B, Pain A, Berriman M, Newbold C, Llinas M. Molecular microbiology. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eckstein-Ludwig U, Webb RJ, Van Goethem ID, East JM, Lee AG, Kimura M, O’Neill PM, Bray PG, Ward SA, Krishna S. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 68.Meshnick SR. Int J Parasitol. 2002;32:1655–1660. doi: 10.1016/s0020-7519(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 69.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vedadi M, Lew J, Artz J, Amani M, Zhao Y, Dong A, Wasney GA, Gao M, Hills T, Brokx S, Qiu W, Sharma S, Diassiti A, Alam Z, Melone M, Mulichak A, Wernimont A, Bray J, Loppnau P, Plotnikova O, Newberry K, Sundararajan E, Houston S, Walker J, Tempel W, Bochkarev A, Kozieradzki I, Edwards A, Arrowsmith C, Roos D, Kain K, Hui R. Mol Biochem Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 71.Ripoll DR, Khavrutskii IV, Chaudhury S, Liu J, Kuschner RA, Wallqvist A, Reifman J. PLoS Comput Biol. 2012;8:e1002665. doi: 10.1371/journal.pcbi.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baldwin ET, Bhat TN, Liu B, Pattabiraman N, Erickson JW. Nature structural biology. 1995;2:244–249. doi: 10.1038/nsb0395-244. [DOI] [PubMed] [Google Scholar]
- 73.Wu CP, Ohnuma S, Ambudkar SV. Current pharmaceutical biotechnology. 2011;12:609–620. doi: 10.2174/138920111795163887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mao Y. BMC structural biology. 2011;11:31. doi: 10.1186/1472-6807-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dančik V, Seiler KP, Young DW, Schreiber SL, Clemons PA. Journal of the American Chemical Society. 2010;132:9259–9261. doi: 10.1021/ja102798t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hostettmann K, Marston A. Phytochemistry Reviews. 2002;1:275–285. [Google Scholar]
- 77.Bernhoft A. S. Proceedings from a symposium held at The Norwegian Academy of and Letters. Bioactive Compounds in Plants: Benefits and Risks for Man and Animals : Proceedings from a Symposium Held in Norwegian Academy of Science and Letters; Oslo. 13 - 14 November 2008 00; Novus Forlag; 2010. [Google Scholar]
- 78.Briskin DP. Plant Physiol. 2000;124:507–514. doi: 10.1104/pp.124.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis WH, Elvin-Lewis MPF. Medical botany: plants affecting human health. J. Wiley; 2003. [Google Scholar]
- 80.Miller J. Econ Bot. 2011;65:396–407. [Google Scholar]
- 81.Saslis-Lagoudakis CH, Klitgaard BB, Forest F, Francis L, Savolainen V, Williamson EM, Hawkins JA. PLoS ONE. 2011;6:e22275. doi: 10.1371/journal.pone.0022275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shyur LF, Yang NS. Current opinion in chemical biology. 2008;12:66–71. doi: 10.1016/j.cbpa.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 83.Ehlting J, Hamberger B, Ginglinger J-F, Werck-Reichhart D. In: Osbourn AE, Lanzotti V, editors. Springer; US: 2009. pp. 475–503. [Google Scholar]
- 84.Clermont G, Auffray C, Moreau Y, Rocke DM, Dalevi D, Dubhashi D, Marshall DR, Raasch P, Dehne F, Provero P, Tegner J, Aronow BJ, Langston MA, Benson M. Genome medicine. 2009;1:88. doi: 10.1186/gm88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cox PA. Ciba Foundation symposium. 1994;185:25–36. discussion 36-41. [PubMed] [Google Scholar]
- 86.Patwardhan B, Mashelkar RA. Drug discovery today. 2009;14:804–811. doi: 10.1016/j.drudis.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 87.Wagner H, Ulrich-Merzenich G. Phytomedicine : international journal of phytotherapy and phytopharmacology. 2009;16:97–110. doi: 10.1016/j.phymed.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 88.Auffray C, Chen Z, Hood L. Genome medicine. 2009;1:2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang M, Lamers RJ, Korthout HA, van Nesselrooij JH, Witkamp RF, van der Heijden R, Voshol PJ, Havekes LM, Verpoorte R, van der Greef J. Phytotherapy research : PTR. 2005;19:173–182. doi: 10.1002/ptr.1624. [DOI] [PubMed] [Google Scholar]
- 90.Li S, Zhang B, Zhang N. BMC systems biology. 2011;5(Suppl 1):S10. doi: 10.1186/1752-0509-5-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, Jin Y, Zhou C, Qu H, Cheng Y. Medical & biological engineering & computing. 2008;46:605–611. doi: 10.1007/s11517-008-0323-1. [DOI] [PubMed] [Google Scholar]
- 92.Fabricant DS, Farnsworth NR. Environmental health perspectives. 2001;109(Suppl 1):69–75. doi: 10.1289/ehp.01109s169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ouedraogo M, Baudoux T, Stevigny C, Nortier J, Colet JM, Efferth T, Qu F, Zhou J, Chan K, Shaw D, Pelkonen O, Duez P. Journal of ethnopharmacology. 2012;140:492–512. doi: 10.1016/j.jep.2012.01.059. [DOI] [PubMed] [Google Scholar]
- 94.Pant AB. Toxicology international. 2011;18:S1–2. doi: 10.4103/0971-6580.85880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merlin W, Gerard B, Genevi_ve B, Vikas D, Jacques F, Jorge F, Bertrand G, Hans-Martin H, Elisabeth H, Pedro Melillo de M. e., Damien P, Colin W. Traditional Medicinal Plants and Malaria. CRC Press; 2004. vol. null. [Google Scholar]
- 96.Liu C, Zhao Y, Wang Y. Applied Microbiology and Biotechnology. 2006;72:11–20. doi: 10.1007/s00253-006-0452-0. [DOI] [PubMed] [Google Scholar]
- 97.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Nature biotechnology. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 98.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 99.Chang MC, Eachus RA, Trieu W, Ro DK, Keasling JD. Nature chemical biology. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 100.Dietrich JA, Fortman JL, Juminaga D, Keasling JD. Biocatalysis for Green Chemistry and Chemical Process Development. John Wiley & Sons, Inc.; 2011. pp. 173–196. [Google Scholar]
- 101.Noorden RV. ch. 3. Nature. 2010 Aug;:672–673. 2010. [Google Scholar]
- 102.Wells TN. Malaria journal. 2011;10(Suppl 1):S3. doi: 10.1186/1475-2875-10-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laughlin JC. Trans R Soc Trop Med Hyg. 1994;88(Suppl 1):S21–22. doi: 10.1016/0035-9203(94)90465-0. [DOI] [PubMed] [Google Scholar]
- 104.Covello PS. Phytochemistry. 2008;69:2881–2885. doi: 10.1016/j.phytochem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 105.Nguyen KT, Arsenault PR, Weathers PJ. vitro cellular & developmental biology. Plant : journal of the Tissue Culture Association. 2011;47:329–338. doi: 10.1007/s11627-011-9343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zeng Q, Zhao C, Yin L, Yang R, Zeng X, Huang Y, Feng L, Yang X. Science in China. Series C, Life sciences / Chinese Academy of Sciences. 2008;51:232–244. doi: 10.1007/s11427-008-0032-x. [DOI] [PubMed] [Google Scholar]
- 107.Wang W, Wang Y, Zhang Q, Qi Y, Guo D. BMC genomics. 2009;10:465. doi: 10.1186/1471-2164-10-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJ, Ross AR, Covello PS. The Journal of biological chemistry. 2008;283:21501–21508. doi: 10.1074/jbc.M803090200. [DOI] [PubMed] [Google Scholar]
- 109.Ma D, Pu G, Lei C, Ma L, Wang H, Guo Y, Chen J, Du Z, Li G, Ye H, Liu B. Plant & cell physiology. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 110.Arsenault PR, Vail D, Wobbe KK, Erickson K, Weathers PJ. Plant Physiology. 2010;154:958–968. doi: 10.1104/pp.110.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma C, Wang H, Lu X, Xu G, Liu B. Journal of chromatography. A. 2008;1186:412–419. doi: 10.1016/j.chroma.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 112.Sangwan RS, Sangwan NS, Jain DC, Kumar S, Ranade SA. Biochemistry and molecular biology international. 1999;47:935–944. doi: 10.1080/15216549900202053. [DOI] [PubMed] [Google Scholar]
- 113.Graham IA, Besser K, Blumer S, Branigan CA, Czechowski T, Elias L, Guterman I, Harvey D, Isaac PG, Khan AM, Larson TR, Li Y, Pawson T, Penfield T, Rae AM, Rathbone DA, Reid S, Ross J, Smallwood MF, Segura V, Townsend T, Vyas D, Winzer T, Bowles D. Science. 2010;327:328–331. doi: 10.1126/science.1182612. [DOI] [PubMed] [Google Scholar]
- 114.Hamilton AC. Biodiversity and Conservation. 2004;13:1477–1517. [Google Scholar]
- 115.Raven PH, Holland D, Howell CH. Flora Mirabilis: How Plants Have Shaped World Knowledge, Health, Wealth, and Beauty. National Geographic Society; 2009. [Google Scholar]
- 116.Trivedi PC. Medicinal Plants: Traditional Knowledge. I.K. International Publishing House; 2006. [Google Scholar]
- 117.Balick MJ, Cox PA. Plants, People, and Culture: The Science of Ethnobotany. Scientific American Library; 1997. [Google Scholar]
- 118.Simmonds MSJ. In: Osbourn AE, Lanzotti V, editors. Springer; US: 2009. pp. 127–140. [Google Scholar]
- 119.Motaleb MA. Approaches to Conservation of Medicinal Plants and Traditional Knowledge: A Focus on the Chittagong Hill Tracts. International Union for Conservation of Nature; Dhaka, Bangladesh: 2010. [Google Scholar]
- 120.Uwe Schippmann DJL. Ninth Regular Session of the Commission on Genetic Resources for Food and Agriculture. Rome, Italy: Oct 12-13, 2002. A. B. Cunningham presented in part at the Biodiversity and the Ecosystem Approach in Agriculture, Forestry and Fisheries. 2002. [Google Scholar]
- 121.World Health Organization . Guidelines on the conservation of medicinal plants. World Health Organization; Geneva: 1993. International Union for Conservation of Nature and Natural Resources. and World Wide Fund for Nature. [Google Scholar]
- 122.Simonnet X. presented in part at the RBM/UNITAID/WHO Artemisinin Conference 2011; Hanoi, Vietnam. November 2-3, 2011.2011. [Google Scholar]
- 123.Bowles D. presented in part at the RBM/UNITAID/WHO Artemisinin Conference 2011; Hanoi, Vietnam. 2011. [Google Scholar]
- 124.Bui UM. presented in part at the RBM/UNITAID/WHO Artemisinin Conference 2011; Hanoi, Vietnam. November, 2011.2011. [Google Scholar]
- 125.Farnsworth NR, Soejarto DD, Olayiwola Akerele VH, Synge H. Global Importance of Medicinal Plants Conservation of Medicinal Plants. Cambridge University Press; 1991. [Google Scholar]
- 126.Canter PH, Thomas H, Ernst E. Trends in biotechnology. 2005;23:180–185. doi: 10.1016/j.tibtech.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 127.Nyigo VA, Malebo HM. Tanzania health research bulletin. 2005;7:154–158. doi: 10.4314/thrb.v7i3.14253. [DOI] [PubMed] [Google Scholar]
- 128.Maxmen A. Nature. 2012;490:13–14. doi: 10.1038/490013a. [DOI] [PubMed] [Google Scholar]
- 129.Maxmen A. Nature medicine. 2012;18:634–635. doi: 10.1038/nm0512-634. [DOI] [PubMed] [Google Scholar]
- 130.Lévesque F, Seeberger PH. Angewandte Chemie International Edition. 2012;51:1706–1709. doi: 10.1002/anie.201107446. [DOI] [PubMed] [Google Scholar]
- 131.White NJ. Science. 2008;320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 132.Hale V, Keasling JD, Renninger N, Diagana TT. The American Journal of Tropical Medicine and Hygiene. 2007;77:198–202. [PubMed] [Google Scholar]
- 133.Claudio S. WHO WPRO and the global Roll Back Malaria Program. 2000 [Google Scholar]
- 134.Barat LM. Am J Trop Med Hyg. 2006;74:12–16. [PubMed] [Google Scholar]
- 135.Cutler M. RBM/UNITAID/WHO Artemisinin Conference 2011; Hanoi, Vietnam. 2011. [Google Scholar]
- 136.Cordell GA. Natural Product Chemistry for Drug Discovery. The Royal Society of Chemistry; 2009. pp. 81–139. [Google Scholar]
- 137.McManis CR. EcoHealth. 2011;8:129–131. [Google Scholar]
- 138.Winter G. ch. 2. In: Kamau EC, Winter G, editors. Genetic resources, traditional knowledge and the law : solutions for access and benefit sharing. Earthscan; London: 2009. pp. 19–36. [Google Scholar]
- 139.Buck M, Hamilton C. Review of European Community & International Environmental Law. 2011;20:47–61. [Google Scholar]
- 140.Stoll P-T. ch. 1. In: Kamau EC, Winter G, editors. Genetic resources, traditional knowledge and the law : solutions for access and benefit sharing. Earthscan; London: 2009. pp. 3–18. [Google Scholar]
- 141.Soejarto DD, Fong HH, Tan GT, Zhang HJ, Ma CY, Franzblau SG, Gyllenhaal C, Riley MC, Kadushin MR, Pezzuto JM, Xuan LT, Hiep NT, Hung NV, Vu BM, Loc PK, Dac LX, Binh LT, Chien NQ, Hai NV, Bich TQ, Cuong NM, Southavong B, Sydara K, Bouamanivong S, Ly HM, Thuy TV, Rose WC, Dietzman GR. Journal of ethnopharmacology. 2005;100:15–22. doi: 10.1016/j.jep.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 142.Hollenberg D, Zakus D, Cook T, Xu XW. World Health Popul. 2008;10:62–75. [PubMed] [Google Scholar]
- 143.Abuduxike G, Aljunid SM. Biotechnology advances. 2012 doi: 10.1016/j.biotechadv.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 144.Nguyen H, Ninh P, Nguyen T. personal communication.
- 145.Vietnam Academy of Science and Technology (VAST) [Accessed August 25, 2012];Overview of International Cooperative Activies. 2012 http://www.vast.ac.vn/index.php?option=com_content&view=article&id=528&Itemid=4 8&lang=en.
- 146.Oxford University Clinical Research Unit Viet Nam Oxford University Clinical Research Unit Viet Nam. 2012 http://www.oucru.org/index.php.
- 147.Botanic Gardens Conservation International South East Asia Botanic Gardens Statement. 2012 http://www.bgci.org/sea/0157/
- 148.Botanic Gardens Conservation International . Botanic Gardens Conservation International: 5 year plan 2007-2012. Botanic Gardens Conservation International; Richmond, Surrey, U.K.: 2006. [Google Scholar]
- 149.Masum H, Daar AS, Al-Bader S, Shah R, Singer PA. Innovations: Technology, Governance, Globalization. 2007;2:129–149. [Google Scholar]
- 150.Winter G. ch. 9. In: Kamau EC, Winter G, editors. Genetic resources, traditional knowledge and the law: solutions for access and benefit sharing. Earthscan; London: 2009. pp. 19–36. [Google Scholar]
- 151.Soejarto DD, Fong HH, Tan GT, Zhang HJ, Ma CY, Franzblau SG, Gyllenhaal C, Riley MC, Kadushin MR, Pezzuto JM, Xuan LT, Hiep NT, Hung NV, Vu BM, Loc PK, Dac LX, Binh LT, Chien NQ, Hai NV, Bich TQ, Cuong NM, Southavong B, Sydara K, Bouamanivong S, Ly HM, Thuy TV, Rose WC, Dietzman GR. J. Ethnopharmacol. 2005;100:15–22. doi: 10.1016/j.jep.2005.05.031. [DOI] [PubMed] [Google Scholar]


