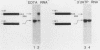Abstract
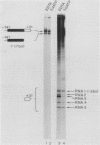
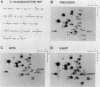
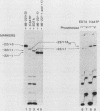
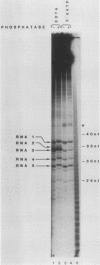
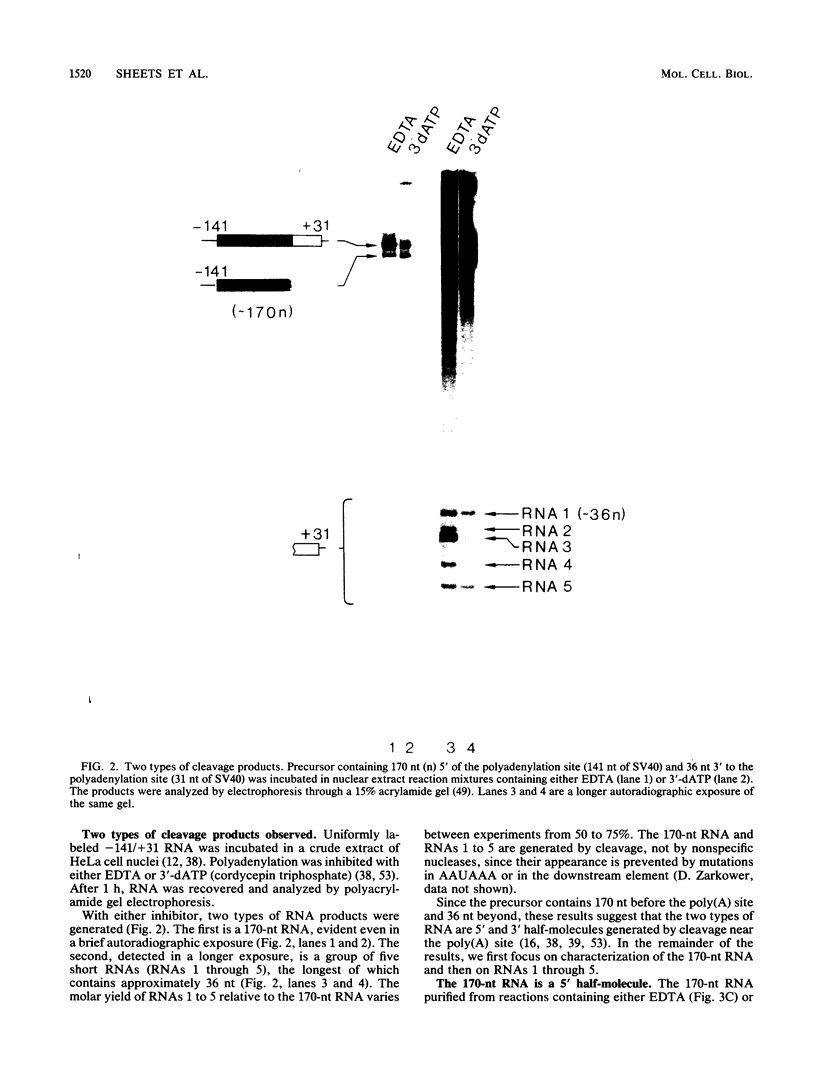
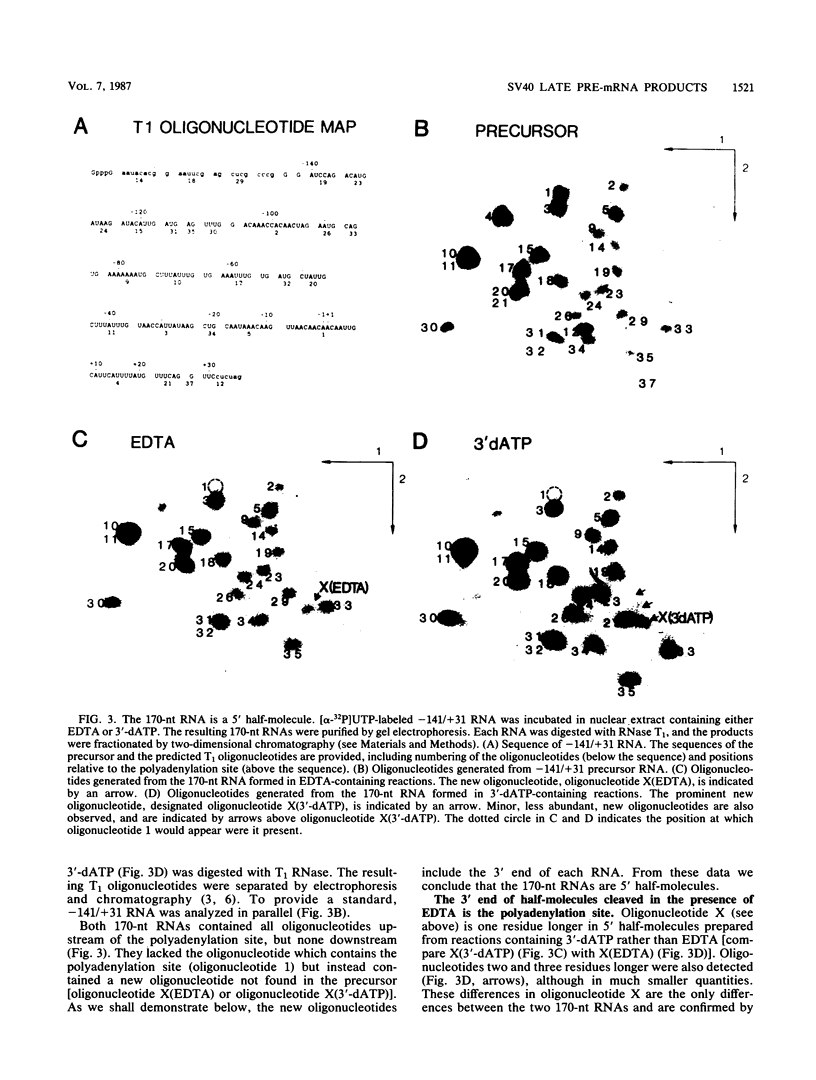
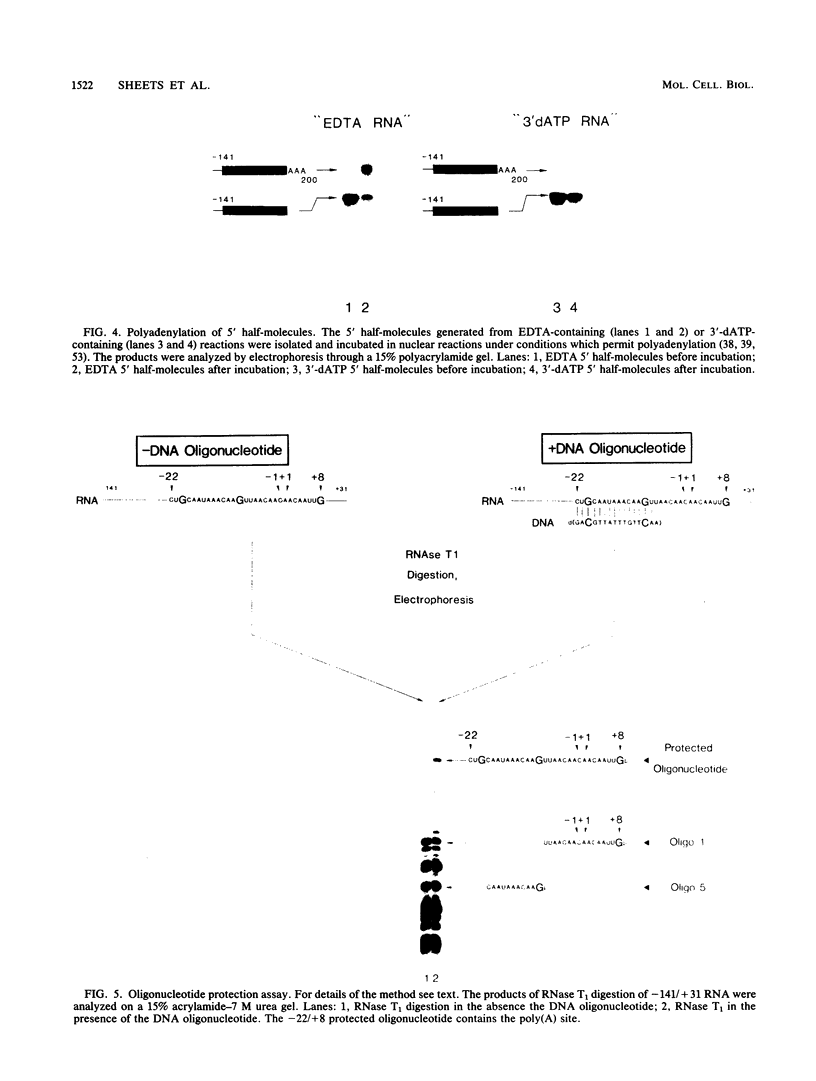
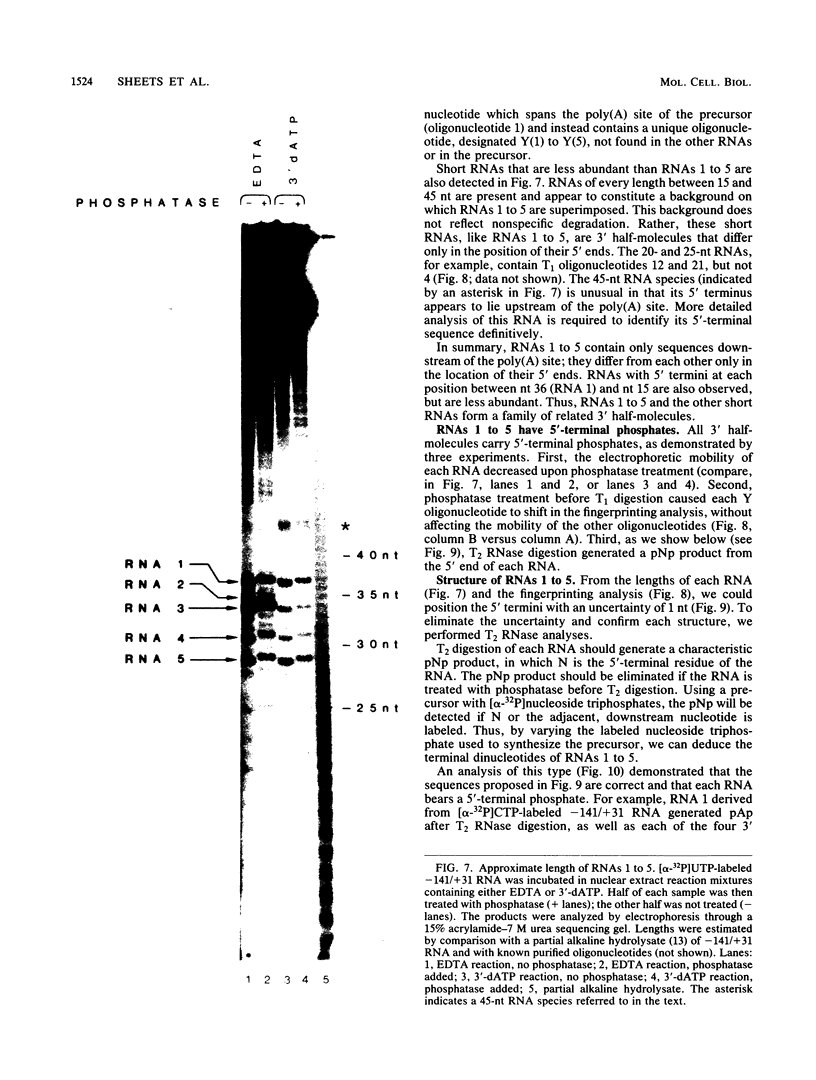
Formation of mRNA 3' termini involves cleavage of an mRNA precursor and polyadenylation of the newly formed end. Cleavage of simian virus 40 late pre-mRNA in a crude nuclear extract generated two RNAs, 5' and 3' half-molecules. These RNAs were unmodified and linear. The 5' half-molecule contained sequences upstream but not downstream of the poly(A) site and ended in a 3'-terminal hydroxyl. The 3' half-molecules comprised a family of RNAs, each of which contains only sequences downstream of the poly(A) site, and ends in a 5'-terminal phosphate. These RNAs differed only in the locations of their 5' terminus. The 3' terminus of the 5' half-molecule was the adenosine 10 nucleotides downstream of AAUAAA, at the +1 position. The 5' terminus of the longest 3' half-molecule was at +2. Thus, these two RNAs contain every nucleoside and phosphate of the precursor. The existence of these half-molecules demonstrates that endonucleolytic cleavage occurs near the poly(A) site. 5' half-molecules generated in the presence of EDTA (which blocks polyadenylation, but not cleavage) ended at the adenosine at position +1 of the precursor. When incubated in the extract under suitable conditions, they became polyadenylated. 5' half-molecules formed in 3'-dATP-containing reactions contained a single 3'-deoxyadenosine (cordycepin) residue added onto the +1 adenosine and were poor polyadenylation substrates. We infer that the +1 adenosine of the precursor becomes the first A of the poly(A) tract and provides a 3' hydroxyl group to which poly(A) is added posttranscriptionally.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Rosbash M. Behavior of individual maternal pA+ RNAs during embryogenesis of Xenopus laevis. Dev Biol. 1982 Nov;94(1):79–86. doi: 10.1016/0012-1606(82)90070-7. [DOI] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudny H., Deutscher M. P. Apparent involvement of ribonuclease D in the 3' processing of tRNA precursors. Proc Natl Acad Sci U S A. 1980 Feb;77(2):837–841. doi: 10.1073/pnas.77.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Philipson L., Wall R., Adesnik M. Polyadenylic acid sequences: role in conversion of nuclear RNA into messenger RNA. Science. 1971 Oct 29;174(4008):507–510. doi: 10.1126/science.174.4008.507. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri G. L. Formation of low molecular weight RNA species in HeLa cells. J Cell Physiol. 1980 Feb;102(2):199–207. doi: 10.1002/jcp.1041020211. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. A small nuclear ribonucleoprotein associates with the AAUAAA polyadenylation signal in vitro. Cell. 1986 May 23;45(4):581–591. doi: 10.1016/0092-8674(86)90290-4. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984 Dec;39(3 Pt 2):423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- Knapp G., Ogden R. C., Peebles C. L., Abelson J. Splicing of yeast tRNA precursors: structure of the reaction intermediates. Cell. 1979 Sep;18(1):37–45. doi: 10.1016/0092-8674(79)90351-9. [DOI] [PubMed] [Google Scholar]
- Koch S., Niessing J. Mode of inhibition of nuclear poly(A)polymerase by 2'-dATP and 3'-dATP. FEBS Lett. 1978 Dec 15;96(2):354–356. doi: 10.1016/0014-5793(78)80435-9. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Konarska M., Filipowicz W., Domdey H., Gross H. J. Formation of a 2'-phosphomonoester, 3',5'-phosphodiester linkage by a novel RNA ligase in wheat germ. Nature. 1981 Sep 10;293(5828):112–116. doi: 10.1038/293112a0. [DOI] [PubMed] [Google Scholar]
- Konarska M., Filipowicz W., Gross H. J. RNA ligation via 2'-phosphomonoester, 3'5'-phosphodiester linkage: requirement of 2',3'-cyclic phosphate termini and involvement of a 5'-hydroxyl polynucleotide kinase. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1474–1478. doi: 10.1073/pnas.79.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Maale G., Stein G., Mans R. Effects of cordycepin and cordycepintriphosphate on polyadenylic and ribonucleic acid-synthesising enzymes from eukaryotes. Nature. 1975 May 1;255(5503):80–82. doi: 10.1038/255080a0. [DOI] [PubMed] [Google Scholar]
- Madore S. J., Wieben E. D., Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol. 1984 Jan;98(1):188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendecki J., Lee S. Y., Brawerman G. Characteristics of the polyadenylic acid segment associated with messenger ribonucleic acid in mouse sarcoma 180 ascites cells. Biochemistry. 1972 Feb 29;11(5):792–798. doi: 10.1021/bi00755a018. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984 Mar;36(3):581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Skolnik-David H., Sharp P. A. Analysis of RNA cleavage at the adenovirus-2 L3 polyadenylation site. EMBO J. 1986 Aug;5(8):1929–1938. doi: 10.1002/j.1460-2075.1986.tb04446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rose K. M., Bell L. E., Jacob S. T. Specific inhibition of chromatin-associated poly(A) synthesis in vitro by cordycepin 5'-triphosphate. Nature. 1977 May 12;267(5607):178–180. doi: 10.1038/267178a0. [DOI] [PubMed] [Google Scholar]
- Rosenthal E. T., Tansey T. R., Ruderman J. V. Sequence-specific adenylations and deadenylations accompany changes in the translation of maternal messenger RNA after fertilization of Spisula oocytes. J Mol Biol. 1983 May 25;166(3):309–327. doi: 10.1016/s0022-2836(83)80087-4. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Alwine J. C. Sequences on the 3' side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984 Aug;4(8):1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., Alwine J. C. Identification of a sequence element on the 3' side of AAUAAA which is necessary for simian virus 40 late mRNA 3'-end processing. Mol Cell Biol. 1985 Oct;5(10):2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- Ziff E. B. Transcription and RNA processing by the DNA tumour viruses. Nature. 1980 Oct 9;287(5782):491–499. doi: 10.1038/287491a0. [DOI] [PubMed] [Google Scholar]