Abstract
We have previously identified tagSNPs at 8q24.21 influencing glioma risk. We have sought to fine-map the location of the functional basis of this association using data from four genome-wide association studies, comprising a total of 4147 glioma cases and 7435 controls. To improve marker density across the 700 kb region, we imputed genotypes using 1000 Genomes Project data and high-coverage sequencing data generated on 253 individuals. Analysis revealed an imputed low-frequency SNP rs55705857 (P = 2.24 × 10−38) which was sufficient to fully capture the 8q24.21 association. Analysis by glioma subtype showed the association with rs55705857 confined to non-glioblastoma multiforme (non-GBM) tumours (P = 1.07 × 10−67). Validation of the non-GBM association was shown in three additional datasets (625 non-GBM cases, 2412 controls; P = 1.41 × 10−28). In the pooled analysis, the odds ratio for low-grade glioma associated with rs55705857 was 4.3 (P = 2.31 × 10−94). rs55705857 maps to a highly evolutionarily conserved sequence within the long non-coding RNA CCDC26 raising the possibility of direct functionality. These data provide additional insights into the aetiological basis of glioma development.
INTRODUCTION
Glioma comprises ∼40% of all primary brain tumours and most are associated with a poor prognosis irrespective of clinical care, with the most common type of glioma, glioblastoma multiforme (GBM), having a median overall survival of only ∼15 months (1).
Little is known about glioma aetiology, and specifically, with the exception of ionizing radiation which accounts for very few cases, and no lifestyle exposure has consistently been linked to glioma risk (1). Evidence for an inherited susceptibility to glioma is provided by the 2-fold increased risk of glioma shown in relatives of patients, but very little of the familial risk can be accounted for by known genetic syndromes (2). Recently genome-wide association studies (GWASs) have identified single-nucleotide polymorphisms (SNPs) mapping to 5p15.33, 7p11.2, 8q24.21, 9p21.3, 11q23.3 and 20q13.33 associated with glioma risk providing evidence for polygenic susceptibility to glioma (3,4) (Supplementary Material, Table S1).
A number of glioma subtypes defined in part by cell subtype (astrocytic, oligodendroglial or mixed) and by malignancy grade (e.g. pilocytic astrocytomas WHO grade 1, diffuse ‘low-grade’ gliomas WHO grade 2, anaplastic gliomas WHO grade 3 and GBM WHO grade 4) can be distinguished. Accumulating data indicate that these subtypes have different molecular genetic profiles possibly resulting from different etiologic pathways, which might be shared by different tumour subtypes or be type specific. Support for this assertion is shown in the strength of SNPs for specific glioma subtypes at the six GWAS loci (5).
The tagSNPs genotyped in GWAS are generally not themselves candidates for causality, but simply act as markers for functional variants. While the functional variant may also be common, it is increasingly being recognized that some SNP associations can be a consequence of correlations with low-frequency risk variants which have greater impact on disease risk. Furthermore, some associations can arise owing to independent correlation of a tagSNP with more than one functional SNP. The discovery of functional variants may be aided by deep examination of genetic variation in the linkage disequilibrium (LD) blocks in which the tagSNPs reside. Such discovery is likely to benefit from recent efforts such as the 1000 Genomes Project, where a comprehensive discovery of novel variants has been carried out in several populations.
In our original GWAS of glioma, we found some evidence for the existence of two independent disease loci at 8q24.21 defined by rs4295627 and rs891835. At that time, sequence cataloguing of variation at 8q24.21 in reference panels was insufficient to fully disentangle the association signals. Recent advances have now allowed us to revisit the 8q24.21 association with two primary aims. First, to refine the most likely location of the ‘disease-causing/functional’ variant based on association testing at genotyped and imputed SNPs and their relationship with risk by subtype. Secondly, to determine the SNP(s) most likely to be functional within the fine-mapped regions based on effect sizes and strengths of association and identify potentially functional variants by annotating SNPs.
To achieve these goals, we used data from four different GWAS of glioma to fine-map genetic variation at 8q24.21. In addition to using data from the 1000 Genomes Project, we used high-coverage sequencing data from 253 individuals to generate in silico genotypes for a large number of additional SNPs. This combination of high-density genotyping and imputation allowed a wide and deep examination of SNPs with minor allele frequency (MAF) > 0.01 in this region. Coupled with these studies we performed bioinformatic analysis of the most strongly associated variant to provide insight into the functional basis for the association.
RESULTS
We studied four non-overlapping glioma case–control series of Northern European ancestry providing data on 4147 cases and 7435 controls (Supplementary Material, Table S1). We used Haploview to define the haplotype blocks and recombination hotspots containing the tagSNPs rs4295627 and rs891835, previously found to be associated with glioma risk at 8q24.21. We determined the haplotype block in which this tagSNP resided and identified from dbSNP (build 136) all SNPs with MAF > 0.01 between the recombination hotspots flanking the haplotype block, irrespective of their relationship to the original tagSNP. On the basis of these metrics and to include the possibility of long-range synthetic associations, we considered the 8q24.21 region of association to be defined by a 700 kb interval (rs147853379 at 130 200 197 bp to rs298617 at 130 899 961 bp).
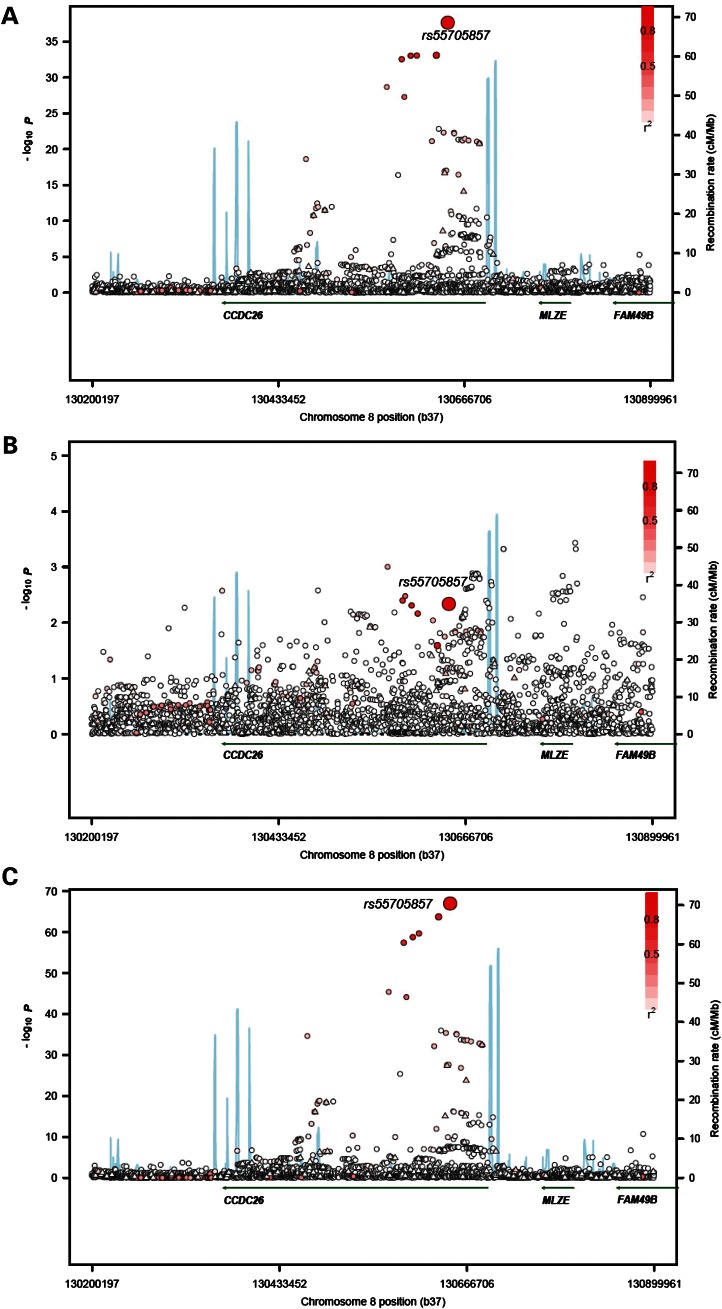
Genotypes from 130 directly typed SNPs which mapped to this 700 kb region were available from each of the four GWASs. In a meta-analysis of these data, the strongest association was provided by rs4295627 which maps at 130 685 457 bp (P = 4.60 × 10−21; Fig. 1, Table 1). The possible second loci at rs891835 showed a weaker association of P = 1.54 × 10−12.
Figure 1.
Case–control association plot for glioma and genomic structure of the chromosome 8q24.21 region: (A) All glioma, (B) GBM and (C) non-GBM tumours. Overview of single-point SNP association results of both genotyped (triangles) and imputed (circles) SNPs are shown with recombination rates within the region. The region shown is chr8: 130 200 197–130 899 961 bp (HG19 Build 37). −log10 P values (y-axis) of the SNPs are shown according to their chromosomal positions (x axis). The genotyped and imputed SNPs are labelled by diamonds and circles, respectively. rs55705857 in each analysis is labelled by a large circle . The colour intensity of each symbol reflects the extent of LD with rs55705857, from white (r2 = 0) to dark red (r2 = 1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Also shown are the relative positions of genes and transcripts mapping to the region of association.
Table 1.
Summary statistics from top imputed SNPs and two SNPs previously observed in GWAS (rs4295627 and rs891835), showing single SNP P-values and P-values conditioned on rs55705857 as well as conservation, allele frequency and LD metrics
| SNP | Positiona | Allelesb | Phast_consc | GERPc | TFBSd | Europeane | Africane | Asiane | fr2 | fD′ | P | Pconditional |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs891835 (tagSNP) | 130 491 752 | T/G | 0.001 | 0.47 | None | 0.255 | 0.020 | 0.138 | 0.16 | 0.92 | 3.40 × 10−12 | 0.38 |
| rs72714295 | 130 569 398 | C/A | 0.00 | −0.12 | None | 0.095 | 0.008 | 0.005 | 0.44 | 0.82 | 2.11 × 10−29 | 0.07 |
| rs72714302 | 130 588 045 | G/C | 0.00 | −0.72 | None | 0.065 | 0.002 | 0.00 | 0.79 | 0.92 | 2.78 × 10−33 | 0.55 |
| rs149644757 | 130 591 380 | A/G | 0.384 | 0.12 | None | 0.094 | 0.006 | 0.005 | 0.43 | 0.82 | 5.10 × 10−28 | 0.17 |
| rs72716319 | 130 599 332 | A/G | 0.00 | −2.78 | None | 0.065 | 0.002 | 0.00 | 0.79 | 0.92 | 9.08 × 10−34 | 0.49 |
| rs72716328 | 130 606 932 | C/T | 0.040 | 1.68 | None | 0.059 | 0.002 | 0.00 | 0.83 | 0.98 | 8.60 × 10−34 | 0.54 |
| rs147958197 | 130 631 395 | T/C | 0.002 | 0.66 | None | 0.062 | 0.00 | 0.00 | 0.90 | 1.00 | 7.86 × 10−34 | 0.24 |
| rs55705857 | 130 645 692 | A/G | 1.00 | 5.98 | OCT1 | 0.069 | 0.002 | 0.00 | 1(ref) | 1(ref) | 2.24 × 10−38 | 1.00 |
| rs4295627 (tagSNP) | 130 685 457 | T/G | 0.00 | −6.72 | None | 0.193 | 0.138 | 0.260 | 0.21 | 0.98 | 1.71 × 10−21 | 0.37 |
aPosition from NCBI b37.
bReference/non-reference alleles.
cPhast_cons and GERP conservation scores, with maxima of 1.00 and 6.00, respectively.
dTranscription factor binding site information as obtained from UCSC web portal.
eNon-reference allele frequency.
fLD metrics obtained from 1000 Genomes.
To improve marker density across the 700 kb region, we imputed genotypes in cases and controls using data from the 1000 Genomes Project dataset as well as deep sequencing (>30×) data generated on 253 individuals. In total, an additional 5897 non-monomorphic SNPs mapping to the interval were successfully imputed, of which 1959 were not catalogued by 1000 Genomes. A cluster of seven SNPs provided greatly superior evidence for an association with glioma risk (Fig. 1; Table 1). The strongest signal was shown by rs55705857 which maps within the long non-coding RNA sequence CCDC26 at 130 645 692 bp (P = 2.24 × 10−38). Pairwise LD metrics, r2 and D′ with rs4295627 and rs891835 are 0.15, 1.0 and 0.11, 1.0, respectively. The OR for glioma associated with rs55705857 was 2.40 with minimal evidence of between-study heterogeneity (Phet = 0.77 and I2 = 0%; Table 2). Logistic regression analysis of SNPs with evidence of association in the meta-analysis at P< 5.0 × 10−4 did not identify any additional independent risk alleles in the region. Conditional analysis on rs55705857 showed that rs55705857 genotype was sufficient to capture the allelic variation impacting on disease risk within the region (Table 1). To check for any biases incorporated by using a reference panel enriched for colorectal cancer cases, imputation was also performed using only the 1000 Genomes Project data, rs55705857 was again the top SNP (Supplementary Material, Table S2).
Table 2.
Association between glioma risk and rs55705857 genotype for all glioma, GBM only and non-GBM
| Stage | Study | All glioma |
GBM |
Non-GBM |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases/controls | OR | P-value | Cases/controls | OR | P-value | Cases/controls | OR | P-value | ||
| GWAS | French | 1423/1190 | 2.7 | 1.87 × 10−14 | 430/1190 | 1.3 | 0.11 | 993/1190 | 3.9 | 3.38 × 10−21 |
| German | 846/1310 | 2.3 | 1.91 × 10−8 | 431/1310 | 1.6 | 0.02 | 415/1310 | 4.3 | 4.83 × 10−13 | |
| USA | 1247/2236 | 2.3 | 1.35 × 10−12 | 652/2236 | 1.3 | 0.11 | 595/2236 | 5.1 | 8.27 × 10−25 | |
| UK | 631/2699 | 2.3 | 9.76 × 10−8 | 270/2699 | 1.1 | 0.74 | 361/2699 | 4.8 | 4.60 × 10−14 | |
| Combined | 4147/7435 | 2.4 | 2.24 × 10−38 | 1783/7435 | 1.3 | 0.005 | 2364/7435 | 4.4 | 1.07 × 10−67 | |
| Phet (I2%) | 0.77 (0%) | 0.63 (0%) | 0.57 (0%) | |||||||
| Replication | French | 273/1425 | 3.1 | 1.66 × 10−7 | 72/1425 | 0.6 | 0.19 | 201/1425 | 5.4 | 7.75 × 10−12 |
| German | 290/428 | 2.5 | 0.0001 | 172/428 | 2.3 | 0.006 | 118/428 | 3.8 | 5.70 × 10−5 | |
| UK | 306/559 | 3.5 | 3.50 × 10−15 | |||||||
| Combined Phet (I2%) | 563/1853 | 2.8 | 1 × 10−10 | 244/1853 | 1.4 | 0.14 | 625/2412 | 4.0 | 1.41 × 10−28 | |
| 0.45 (0%) | 0.008 (85.6%) | 0.37 (0%) | ||||||||
| Combined | 4710/9288 | 2.5 | 2.5 × 10−47 | 2027/9288 | 1.3 | 0.001 | 2989/9847 | 4.3 | 2.31 × 10−94 | |
| 0.78 (0%) | 0.12 (43%) | 0.61 (0%) | ||||||||
Using data from all studies, we examined risk for GBM and non-GBM tumours associated with rs55705857 (Table 2). Respective ORs for GBM and non-GBM tumours were 1.33 (P = 0.005) and 4.43 (P = 1.07 × 10−67). This is consistent with our previous observation based on the original tagSNP that the 8q24.21 association is strongly driven by the association with low-grade glioma (5). To validate this association, we genotyped rs55705857 in three additional series totalling 869 cases and 2412 controls (Table 2). This analysis confirmed that the association is non-GBM specific (P = 1.41 × 10−28) with non-significant findings for GBM cases (P = 0.14). In the combined analysis of all data, the OR for non-GBM glioma associated with rs55705857 was 4.3 (P = 2.31 × 10−94; Table 2).
We then examined the association by subtype in more detail using histology data of WHO grade. This analysis showed that the association was seen in both grade II and III gliomas (Table 3). Subsequent to this we examined the association between glioma risk and rs55705857 stratified by molecular characteristics of WHO II and III tumours. We restricted this analysis to the French dataset since this provided the largest collection of centrally reviewed pathology and for which molecular data were available for tumours (Tables 4 and 5). This analysis indicated that the association was not driven by a relationship with specific cell lineage, with strong associations being shown for low-grade tumours of oligo- and astrocyte origin (Table 4). Within non-GBM tumours, the strongest associations were shown for IDH mutation, EGFR non-amplification, P16 non-deleted, 10q non-deleted and 1p/19q co-deleted. None of these associations extended to GBM tumours (Table 5).
Table 3.
Glioma risk and rs55705857 stratified by morphology grade
| Grade | French |
Germany |
USA |
UK |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | OR | P-value | Cases | OR | P-value | Cases | OR | P-value | Cases | OR | P-value | |
| II | 406 | 11.8 | 5.79 × 10−30 | 135 | 4.8 | 8.76 × 10−6 | 209 | 21.7 | 3.83 × 10−26 | 130 | 4.1 | 1.06 × 10−4 |
| III | 335 | 5.3 | 1.07 × 10−9 | 191 | 9.6 | 8.73 × 10−14 | 295 | 5.0 | 5.32 × 10−12 | 141 | 11.9 | 1.48 × 10−12 |
| IV | 430 | 1.4 | 0.15 | 431 | 1.6 | 0.024 | 652 | 1.3 | 0.11 | 270 | 1.1 | 0.74 |
Table 4.
Glioma risk and rs55705875 by morphology in the French case–control series
| Grade | Histology | Cases | OR | P-value |
|---|---|---|---|---|
| II | Low-grade astrocytoma | 85 | >100 | 3.04 × 10−24 |
| Oligoastrocytoma | 98 | 21.4 | 1.53 × 10−10 | |
| Oligodendroglioma | 223 | 38.5 | 1.00 × 10−28 | |
| III | Anaplastic astrocytoma | 49 | 4.7 | 0.02 |
| Anaplastic oligoastrocytoma | 101 | 19.7 | 4.94 × 10−8 | |
| Anaplastic oligodendroglioma | 185 | 6.9 | 1.26 × 10−6 | |
| IV | GBM | 430 | 1.3 | 0.11 |
Table 5.
Glioma risk and rs55705875 stratified by morphology type: IDH1 mutation, EGFR amplification, CDKN2A-p16-INKa homozygous deletion, 9p and 10q loss, 1p-19q co-deletion status in the French case-control series
| Tumour alteration | Non-GBM |
GBM |
||||
|---|---|---|---|---|---|---|
| Cases | OR | P-value | Cases | OR | P-value | |
| IDH1+ | 281 | 27.5 | 3.64 × 10−29 | 24 | 3.0 | 0.24 |
| IDH1− | 210 | 13.8 | 8.79 × 10−13 | 239 | 1.2 | 0.51 |
| EGFR amplified | 37 | 1.9 | 0.47 | 82 | 2.5 | 0.08 |
| EGFR non-amplified | 452 | 11.0 | 2.65 × 10−28 | 186 | 1.1 | 0.77 |
| p16 deleted | 75 | 7.8 | 2.69 × 10−4 | 98 | 1.8 | 0.20 |
| p16 non-deleted | 406 | 12.8 | 2.30 × 10−27 | 164 | 1.2 | 0.66 |
| 9p deleted | 148 | 17.0 | 1.99 × 10−10 | 128 | 2.0 | 0.07 |
| 9p non-deleted | 325 | 15.9 | 1.23 × 10−25 | 137 | 0.9 | 0.89 |
| 10q deleted | 142 | 23.7 | 1.46 × 10−11 | 203 | 1.4 | 0.28 |
| 10q non-deleted | 328 | 13.8 | 9.02 × 10−24 | 62 | 1.0 | 0.99 |
| 1p-19q co-deleted | 152 | >100 | 1.88 × 10−42 | 28 | 1.7 | 0.48 |
| 1p-19q non-co-deleted | 323 | 8.7 | 6.95 × 10−16 | 237 | 1.4 | 0.29 |
Differences in the overall sample numbers are due to some samples not being tested or failing for some of the tumour alterations.
Sequence conservation in non-coding regions has been shown to be a good predictor of cis-regulatory sequences. Moreover, it has been proposed that variation with evolutionary-conserved regions is likely to be associated with phenotypic differences that may contribute to expression of traits. Cross-species sequence comparison between vertebrates of the genomic region encompassing rs55705857 revealed strong evidence of evolutionary conservation notably at rs55705857 (Table 1) with Phast_cons and GERP scores of 1.00 and 5.98, respectively. This contrasts significantly to the other associated SNPs which did not show evidence of conservation.
Using RegulomeDB (6) rs55705857 was shown to overlap a DNase hypersensitivity site. Assessment of regulatory features using UCSC GenomeBrowser revealed through sequencing of brain cell lines (7) that rs55705857 resides within an unmethylated CpG cluster.
To gain insight into the biological basis of the 8q24.21 association, we interrogated Tumor Cancer Genome Atlas (TCGA) RNA-seq expression and Affymetrix 6.0 SNP data (dbGaP accession number: phs000178.v7.p6) on 110 low-grade glioma and 171 GBM cases using rs16893247 as the best proxy for rs55705857 (r2 = 0.19, D′ = 1.0; LD metrics r2 and D′ with rs4295627 and rs891835 are 0.8, 1.0 and 0.28, 0.70, respectively). In a Gene Set Enrichment Analysis (GSEA) of association P-values after thresholding by false discovery rate, 25 gene-sets enriched for inflammation, leucocyte function and apoptosis remained that were significantly enriched for association with rs16893247 in low-grade glioma (Supplementary Material, Table S3). In contrast, GSEA of GBM revealed only two significantly enriched gene sets which were not related to inflammation (Supplementary Material, Table S3). These observations are consistent with the 8q24.21 association being highly specific to low-grade disease.
To further explore the role of rs55705857 in low-grade development, GSEA was applied to expression data from individuals homozygous for rs16893247 rare allele (low-grade glioma n = 8, GBM = 4). All 25 of the significantly enriched gene sets in low-grade glioma were significantly down-regulated compared with GBM (data not shown). Although speculative, this observation is compatible with rs55705857-mediated down-regulation of apoptosis/inflammation/leucocyte activation playing a role in non-GBM development.
DISCUSSION
We have carried out a comprehensive evaluation of the 8q24.21 glioma risk locus discovered by GWAS. Using data from the 1000 Genomes Project together with additional high-coverage sequencing data has led to a large increase in informative markers, thereby providing more accurate and comprehensive information about local patterns of LD within the region. Through this analysis we were able to demonstrate that the 8q24.21 signal for glioma can be captured by a low frequency SNP with the risk allele having a population frequency of ∼5% in the European population. Glioma incidence varies between countries and is much lower in individuals of African/Asian descent than European (8). It is possible that these differences reflect differences in genetic predisposition. In this regard, it is intriguing that the rs55705857 is monomorphic in Asians, and has an MAF 30-fold lower in Africans compared with Europeans (Table 1).
The development of the various histological forms of glioma is increasing being shown to have a different aetiological basis. In this respect, the 8q24.21 association is informative in that the association is confined to risk of low-grade gliomas. Moreover, our findings show that the association did not extend to IDH GBM tumours indicative of secondary GBM.
While it is not necessarily the case that SNPs with the strongest evidence of association with disease are those that are truly functional, the strength of the association provided by rs55705857 is several orders of magnitude greater than other SNPs mapping to the region. It is possible that rs55705857 is highly correlated with much lower frequency variants which we have not been able to harvest through imputation. However, there is strong a priori evidence supporting a direct role in glioma aetiology, notably the base changed by the SNP is highly conserved across divergent species.
rs55705857 is located within CCDC26 a presumptive long intergenic non-coding RNA (lincRNA) and resides within a conserved unmethylated CpG cluster. Such motifs are sufficient for recruitment of repressive polycomb complex components which are implicated in control of cell fate, development and cancer through gene silencing (9–11). Up-regulation of the CCDC26 RNA was associated with the reduction in CD14, a differentiation marker for monocytes (12). CD-14 immunoreactive microglia cells are detectable in astrocytic brain tumours (13). Intriguingly, in the TCGA data, homozyosity for the risk allele at rs16893247 was seen to be significantly associated with reduced CD14 expression in low-grade glioma (Supplementary Material, Fig. S1). Since CCDC26 has a role in leukaemia differentiation (12), an attractive hypothesis for the functional basis of rs55705857 on low-grade glioma risk is via alteration of polycomb complex recruitment to CCDC26, affecting glial stem cell fate determination.
Since the allele frequency of rs55705857 is ∼5% in the European population, the associated genotype risks mean that the SNP impacts significantly on the heritable risk for low-grade glioma accounting for ∼40% of the familial risk (95% CI: 35–46%). This observation and increasing evidence for different aetiological basis of the various glioma subtypes indicates that future searches for novel genetic variants influencing risk should be directed to specific histological subtypes. In contrast to most SNPs identified through GWASs rs55705857 in combination with family history information has the potential value in the clinic setting for risk prediction.
In conclusion, we have conducted a large fine-mapping and annotational study of the 8q24.21 glioma risk locus. We have refined the size of the disease-associated region and provided evidence for a candidate SNP underscoring the association and its association with a specific subtype. Although in part speculative, the data suggest that the functional consequence of rs55705857 may be mediated through lymphocyte–tumour interaction rather than simply through autonomous effects on the oligodendrocyte/astrocytes. Finally, while much of the current search for rarer disease-causing variants is being directed towards analysis of exomes, rs55705857 serves to illustrate that moderate penetrance susceptibility to cancer can embodied by non-coding sequence and that some of these variants may be harvestable by exploitation of existing GWAS datasets.
MATERIALS AND METHODS
Subjects
We used GWAS data previously generated on four non-overlapping case–control series of Northern European ancestry which have been the subject of previous studies (3,4); summarized in Supplementary Material, Table S4. Briefly, the UK GWAS was based on 636 cases (401 males; mean age 46 years) ascertained through the INTERPHONE study (3,14). Individuals from the 1958 Birth Cohort (n = 2930) served as a source of controls. The US GWAS was based on 1281 cases (786 males; mean age 47 years) ascertained through the MD Anderson Cancer Center, Texas, between 1990 and 2008. Individuals from the Cancer Genetic Markers of Susceptibility (CGEMS, n = 2245) studies served as controls (3,15,16). The French GWAS study comprised 1495 patients with glioma ascertained through the Service de Neurologie Mazarin, Groupe Hospitalier Pitié-Salpêtrière Paris (4). The controls (n = 1213) were ascertained from the SU.VI.MAX (SUpplementation en VItamines et MinerauxAntioXydants) study of 12 735 healthy subjects (women aged 35–60 years; men aged 45–60 years) (17). The German GWAS comprised 880 patients who underwent surgery for a glioma at the Department of Neurosurgery, University of Bonn Medical Center, between 1996 and 2008 (18). Control subjects were taken from three population studies: KORA (Co-operative Health Research in the Region of Augsburg; n = 488) (19), POPGEN (Population Genetic Cohort; n= 678) (20) and from the Heinz Nixdorf Recall study (n= 380) (21).
For replication we made use of three additional case–control series: (i) 625 non-GBM cases (187 males, mean age 44 years) ascertained through the National Study of Brain Tumour Genetics and 559 healthy individuals with no personal history of malignancy from the National Study of Colorectal Cancer (199 males, mean age 57 years); both cases and controls were UK residents and had self-reported European ancestry; (ii) 291 patients (173 males, mean age 54 years) who underwent surgery for glioma at the Department of Neurosurgery, University of Bonn Medical Center and were not included in the German GWAS. Controls (n = 428) were healthy blood donors from a location close to the case collection matched by age (mean age 49, SD 12) and sex at the group level recruited through the Institute of Transfusion Medicine, Mannheim (Germany); (iii) 279 patients (171 males) with glioma ascertained through the Service de Neurologie Mazarin, Groupe Hospitalier Pitié-Salpêtrière Paris, which had been ascertained subsequently to those included in the French GWAS. The 1425 controls were from the SU.VI.MAX (SUpplementation en VItamines et MinerauxAntioXydants) study of 12 735 healthy subjects (women aged 35–60 years; men aged 45–60 years) which were not part of the French GWAS (17).
Collection of blood samples and clinical information from subjects was undertaken with informed consent and relevant ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Genotyping
Full details of the genotyping of cases and quality control using Illumina Infinium HD Human610-Quad BeadChips (Illumina, San Diego, CA, USA) are detailed in previously published work (3,18). Briefly, duplicate samples were used to check genotyping quality. SNPs and samples with <95% SNPs genotyped were eliminated from the analyses. Genotype frequencies at each SNP were tested for deviation from the Hardy–Weinberg equilibrium and rejected at P< 10−4. We have previously confirmed an absence of systematic genetic differences between cases and controls and shown no significant evidence of population stratification in these sample sets. Replication genotyping was conducted using either Kaspar allele-specific PCR or Taqman implemented on ABI 7900HT platforms (Applied Biosystems, Foster City, USA; assay details are available on request). Genotyping quality control in all assays was evaluated through inclusion of duplicate DNA samples in SNP assays, together with direct sequencing of specific SNP in a subset of samples to confirm imputation accuracy (PCR details are available on request).
Statistical analysis
Analyses were primarily undertaken using R (v2.6), STATA v.10 (State College) and PLINK (v1.07) software. The association between each SNP and risk of glioma was assessed by the Cochran-Armitage trend test. Odds ratios and associated 95% confidence intervals were calculated by unconditional logistic regression. Prediction of the ungenotyped SNPs across all cases and controls in the four studies was carried out using IMPUTEv2 based on data from the 1000 Genomes Project (Phase 1, February 2012 release). In addition, we made use of an in-house European reference panel generated by high-coverage next-generation sequencing of 199 colorectal cancer cases and 54 healthy individuals. Sequencing of these individuals was carried out using unchained combinatorial probe anchor ligation chemistry on arrays of self-assembling DNA nanoballs (22) and paired-end reads aligned to the Human Genome NCBI (National Center for Biotechnology Information) Build 37. Average coverage across the region 8q24.21 (130 200 000–130 900 000 bp) was 52.7. Phasing of these data was performed using the enhanced hidden Markov model chain program SHAPEIT. Imputed data were analysed using SNPTEST v2 to account for uncertainties in SNP prediction. Association meta-analyses only included markers with proper_info scores >0.5, imputed call rates/SNP > 0.9 and MAFs > 0.01.
Meta-analyses were carried out with META using the genotype probabilities from Imputev2, where an SNP was not directly typed. To test for the presence of additional independent risk alleles in each region, we carried out logistic regression analysis that included all SNPs with evidence of association in the meta-analysis at P < 5.0 × 10−4. We calculated Cochran's Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation that was caused by heterogeneity. I2 values ≥75% were considered to indicate substantial heterogeneity.
The familial relative risk of glioma attributable to a variant was calculated using the formula:
where p is the population frequency of the minor allele, q = 1 – p, and r1 and r2 are the relative risks (approximated by odds ratios) for heterozygotes and the rarer homozygotes relative to the more common homozygotes respectively. From λ*, it is possible to quantify the influence of the locus on the overall familial risk of glioma in first-degree relatives of glioma patients. Assuming a multiplicative interaction between risk alleles, the proportion of the overall familial risk attributable to the locus is given by log (λ*)/log (λ0), where λ0, the overall familial risk of glioma, shown in epidemiological studies is 1.8 (2).
Measurement of LD between SNPs were based on Data Release 27, phase 3 (February 2009 on NCBI B36 assembly, dbSNP26) and additional data from 1000 Genomes analysed using Plink. Association plot were drawn using SNP annotation and proxy search (SNAP).
Sequence conservation metrics GERP and Phast_cons as well as conserved transcription factor binding sites were obtained (http://snp.gs.washington.edu/SeattleSeqAnnotation134/ and http://genome.ucsc.edu/cgi-bin/hgGateway). Genomic evolutionary rate profiling (GERP) is an estimate of evolutionary constraint whose score reflects the proportion of substitutions at that site rejected by selection compared with observed substitutions expected under a neutral evolutionary model, using a sequence alignment of 34 mammalian species; the score ranges from −12 to 6, with 6 being indicative of complete conservation (23). Phast_cons is a measure of conservation whose score reflects the probability that a given nucleotide is conserved, based on sequence alignment of 17 vertebrate species; the score ranges from 0 to 1, where 1 is most conserved (24). Conserved transcription factor bindings sites were predicted by the UCSC track ‘tfbsCons’, which searches for conserved elements in TransFac Matrix Database v4 after alignment in Human/Rat/Mouse. A z-score threshold of 1.64 was applied.
Tumour genotyping
Tumour samples were available from a subset of the patients ascertained through the Service de Neurologie Mazarin, Groupe Hospitalier Pitié-Salpêtrière Paris. Tumours were snap-frozen in liquid nitrogen and DNA was extracted using the QIAmp DNA minikit, according to the manufacturer's instructions (Qiagen, Venlo, LN, USA). DNA was analysed for large-scale copy number variation by CGH array as previously described (5). In the cases not analysed by CGH array, 9p, 10q, 1p and 19q status was assigned using PCR microsatellites, and EGFR-amplification and CDKN2A-p16-INK4a homozygous deletion by quantitative PCR. IDH1 codon R132 status was determined by sequencing.
Relationship between genotype and gene expression
To examine the relationship between SNP genotype and gene expression, we made use of RNA seq V2 expression data (20 531 gene probes) and Affymetrix 6.0 SNP Array data for 110 low-grade and 171 GBM tumours from the TGCA Pilot Project. The association between SNP and gene expression was quantified using the Kruskal–Wallis trend test. To explore inter-relationships between P-values in low-grade glioma and GBM datasets, we conducted a ‘Preranked’ gene-set enrichment analysis (GSEA) using the MSigDB database v3.1 of gene-sets (updated 27 September 2012) and GSEA software (25,26) using default options (min gene set 15 genes, max 500 genes) and applying thresholds of P < 0.05, FDR q < 0.05 and FWER < 0.05.
URL
The R suite can be found at http://www.r-project.org/.
Detailed information on the tagSNP panel can be found at http://www.illumina.com/.
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/.
HAPMAP: http://www.hapmap.org/.
CGEMS: http://cgems.cancer.gov/.
SNPTEST: http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html.
KORA: http://epi.helmholtz-muenchen.de/kora-gen/index_e.php.
POPGEN: http://www.popgen.de/.
SHAPEIT: http://www.shapeit.fr/.
SNAP Plots: http://www.broadinstitute.org/mpg/snap.
Gene-set enrichment analysis: http://www.broadinstitute.org/gsea/index.jsp.
TCGA: http://cancergenome.nih.gov/.
SUPPLEMENTARY MATERIAL
FUNDING
In the UK, funding was provided by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), the Wellcome Trust and the DJ Fielding Medical Research Trust. B.K. is supported by a PhD studentship funded by the Sir John Fisher Foundation. The UK INTERPHONE study was supported by the European Union Fifth Framework Program ‘Quality of life and Management of Living Resources’ (QLK4-CT-1999-01563) and the International Union against Cancer (UICC). The UICC received funds from the Mobile Manufacturers' Forum and GSM Association. Provision of funds via the UICC was governed by agreements that guaranteed INTERPHONE's scientific independence (http://www.iarc.fr/ENG/Units/RCAd.html) and the views expressed in the paper are not necessarily those of the funders. The UK centres were also supported by the Mobile Telecommunications and Health Research (MTHR) Programme and the Northern UK Centre was supported by the Health and Safety Executive, Department of Health and Safety Executive and the UK Network Operators. In the USA, funding was provided by NIH grants 5R01 (CA119215&5R01 CA070917). Support was also obtained from the American Brain Tumor Association and the National Brain Tumor Society. In France, funding was provided by the Délégation à la Recherche Clinique (MUL03012), the Association pour la Recherche sur les Tumeurs Cérébrales (ARTC), the Institut National du Cancer (INCa; PL046) and the French Ministry of Higher Education and Research. In Germany, funding was provided to M.S. and J.S. by the Deutsche Forschungsgemeinschaft (Si552, Schr285), the Deutsche Krebshilfe (70-2385-Wi2, 70-3163-Wi3, 10-6262) and BONFOR. Funding for the WTCCC was provided by the Wellcome Trust (076113&085475). The KORA Augsburg studies are supported by grants from the German Federal Ministry of Education and Research (BMBF) and were mainly financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg. This work was financed by the German National Genome Research Network (NGFN) and supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ.
NOTE ADDED IN PROOF
The assertion this SNP is directly functional is supported by a contemporaneous sequencing effort of the 8q24.21 region showing that rs55705857 genotype is sufficient to capture the glioma association (27). However, in contrast to our study although the association was strongest for IDH-mutated oligodendroglial tumors and astrocytoma an association was also shown for IDH-mutated GBM which was not seen in our study.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all the patients and individuals for their participation and we would also like to thank the clinicians and other hospital staff, cancer registries and study staff in respective centres who contributed to the blood sample and data collection. For the UK GWAS, we acknowledge the participation of the clinicians and other hospital staff, cancer registries, study staff and funders who contributed to the blood sample and data collection for this study as listed in Hepworth et al. (BMJ 2006, 332, 883). MD Anderson acknowledges the work on the USA GWA study of Phyllis Adatto, Fabian Morice, Hui Zhang, Victor Levin, Alfred W.K. Yung, Mark Gilbert, Raymond Sawaya, Vinay Puduvalli, Charles Conrad, Fredrick Lang and Jeffrey Weinberg from the Brain and Spine Center. For the German GWA study, we are indebted to B. Harzheim (Bonn) and Dr A. Müller-Erkwoh (Bonn) for help with the acquisition of clinical data and R. Mahlberg (Bonn) who provided technical support. The UK GWA study made use of control genotyping data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. The US GWA study made use of control genotypes from the CGEMS prostate and breast cancer studies. A full list of the investigators who contributed to the generation of the data is available from http://cgems.cancer.gov/. French controls were taken from the SU.VI.MAX study. The German GWA study made use of genotyping data from three population control sources: KORA-gen, The Heinz-Nixdorf RECALL study and POPGEN. We are extremely grateful to all investigators who contributed to the generation of these datasets. The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI (dbGaP accession number: phs000178.v7.p6). Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bondy M.L., Scheurer M.E., Malmer B., Barnholtz-Sloan J.S., Davis F.G., Il'yasova D., Kruchko C., McCarthy B.J., Rajaraman P., Schwartzbaum J.A., et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113:1953–1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemminki K., Tretli S., Sundquist J., Johannesen T.B., Granstrom C. Familial risks in nervous-system tumours: a histology-specific analysis from Sweden and Norway. Lancet Oncol. 2009;10:481–488. doi: 10.1016/S1470-2045(09)70076-2. [DOI] [PubMed] [Google Scholar]
- 3.Shete S., Hosking F.J., Robertson L.B., Dobbins S.E., Sanson M., Malmer B., Simon M., Marie Y., Boisselier B., Delattre J.Y., et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanson M., Hosking F.J., Shete S., Zelenika D., Dobbins S.E., Ma Y., Enciso-Mora V., Idbaih A., Delattre J.Y., Hoang-Xuan K., et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum. Mol. Genet. 2010;20:2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon M., Hosking F.J., Marie Y., Gousias K., Boisselier B., Carpentier C., Schramm J., Mokhtari K., Hoang-Xuan K., Idbaih A., et al. Genetic risk profiles identify different molecular etiologies for glioma. Clin. Cancer Res. 2010;16:5252–5259. doi: 10.1158/1078-0432.CCR-10-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrow R., Darefsky A.S. Demographic variation in incidence of adult glioma by subtype, United States, 1992–2007. BMC Cancer. 2011;11:325. doi: 10.1186/1471-2407-11-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparmann A., van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 10.Lynch M.D., Smith A.J., De Gobbi M., Flenley M., Hughes J.R., Vernimmen D., Ayyub H., Sharpe J.A., Sloane-Stanley J.A., Sutherland L., et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin W., Rossin A., Clifford J.L., Gronemeyer H. Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. Oncogene. 2006;25:3735–3744. doi: 10.1038/sj.onc.1209410. [DOI] [PubMed] [Google Scholar]
- 13.Deininger M.H., Meyermann R., Schluesener H.J. Expression and release of CD14 in astrocytic brain tumors. Acta Neuropathol. 2003;106:271–277. doi: 10.1007/s00401-003-0727-9. [DOI] [PubMed] [Google Scholar]
- 14.Cardis E., Richardson L., Deltour I., Armstrong B., Feychting M., Johansen C., Kilkenny M., McKinney P., Modan B., Sadetzki S., et al. The INTERPHONE study: design, epidemiological methods, and description of the study population. Eur. J. Epidemiol. 2007;22:647–664. doi: 10.1007/s10654-007-9152-z. [DOI] [PubMed] [Google Scholar]
- 15.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 17.Hercberg S., Galan P., Preziosi P., Bertrais S., Mennen L., Malvy D., Roussel A.M., Favier A., Briancon S. The SU.VI.MAX Study: a randomized, placebo-controlled trial of the health effects of antioxidant vitamins and minerals. Arch. Intern. Med. 2004;164:2335–2342. doi: 10.1001/archinte.164.21.2335. [DOI] [PubMed] [Google Scholar]
- 18.Sanson M., Hosking F.J., Shete S., Zelenika D., Dobbins S.E., Ma Y., Enciso-Mora V., Idbaih A., Delattre J.Y., Hoang-Xuan K., et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum. Mol. Genet. 2011;20:2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holle R., Happich M., Lowel H., Wichmann H.E. KORA—a research platform for population based health research. Gesundheitswesen. 2005;67(Suppl. 1):S19–S25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 20.Wichmann H.E., Gieger C., Illig T. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 21.Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Commun. Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 22.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 23.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins R.B., Xiao Y., Sicotte H., Decker P.A., Kollmeyer T.M., Hansen H.M., Kosel M.L., Zheng S., Walsh K.M., Rice T., et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat. Genet. 2012;44:1122–1125. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.