Abstract
Development of axons and dendrites constitutes a critical event in neuronal maturation and seems to require signaling through the planar cell polarity (PCP) pathway. Mutations in components of the PCP pathway lead to a spectrum of neurological phenotypes and disorders. For example, a missense mutation in Prickle 1 (Pk1) is associated with progressive myoclonus epilepsy (PME) in humans, and its reduced gene dosage increases sensitivity to induced seizure in mice. In an effort to unravel the role of the PCP pathway in mammalian neuronal development, we examined the expression of Pk1 in the central nervous system (CNS) using in situ hybridization (ISH) in combination with a genetic knock-in approach. We show that Pk1 transcripts are detected in the postmitotic cells of the subplate and cortical plate during mid- and late stages of cortical neurogenesis. In adult brain, Pk1 is expressed in distinct neuronal and glial cell populations, with dynamic formation of dendrites and glial processes during development. Of all the cell types in the mature retina, the highest expression of Pk1 is detected in cholinergic amacrine neurons. Knockdown of Pk1 by shRNA or dominant-negative constructs causes reduced axonal and dendritic extension in hippocampal neurons. Similarly, Pk1 knockdown in neonatal retina leads to defects in inner and outer segments and axon terminals of photoreceptors. Our studies implicate Pk1 function in axonal-dendritic development associated with the maturation of CNS neurons.
INTRODUCTION
Neuronal morphogenesis in the central nervous system (CNS) involves active cytoskeletal rearrangements and membrane compartmentalization, which seem to require signaling through the planar cell polarity (PCP) pathway. The PCP pathway is highly conserved in the animal kingdom from arthropods to vertebrates; in mammals, PCP governs developmental events including convergent extension during gastrulation, tissue closures and hair orientation (1). The three integral membrane proteins (Frizzled, Flamingo/Stan/Celsr and Vangl/Strabismus) and three membrane-associated proteins (Dishevelled, Diego/Inversin and Prickle) constitute the core PCP components (1,2). Other molecules, such as fat and dachsous cadherins, also play significant roles in PCP in parallel to the core pathway. Frizzleds, Celsrs, Dishevelled and Vangl contribute to neurogenesis (3,4), neuronal survival (5), axon guidance, target innervation and dendritic development (6–11). Atypical cadherin Fat3 is reportedly essential for correct dendritic configuration of a subset of retinal amacrine cells (ACs) (12). However, little is known about the role of Prickle proteins during CNS development.
In Drosophila, Prickle (Pk) coordinates hair orientation and ommatidia polarity (13). The ommatidium orientation is coupled to photoreceptor R3 and R4 fate determination, which is controlled by PCP (14,15). Total four prickle homologs have been identified in mammalian genomes with Pk1 and Pk2 are more similar to one another than Pk3 and Pk4. Along the line with classic PCP function, Pk1 and Pk2 have been implicated in inner ear hair cell development (16,17) and in the establishment of cell polarity during early embryogenesis as well (18,19). Interestingly, mouse Pk1 and Pk2 are differentially regulated by NRL (20), a transcription factor necessary for rod photoreceptor development (21). The data bring out an interesting question of whether Prickle genes function in mammalian photoreceptor development as well, and an even bigger question of whether there is a PCP in mammalian retina. In separate lines, a recessive missense mutation in the PET domain of human Pk1 gene is recently shown to be associated with progressive myoclonus epilepsy (PME) (22,23). Both Pk1 and Pk2 genes share three LIM (Lin 11, Isl-1 and Mec-3) and one PET domain (Prickle, Espinas and Testin) at the N-terminus of the protein, and a CAAX prenylation motif at the C-terminus, functions of which have not been addressed in a biochemical context. Mice with a reduced gene dosage of either Pk1 or Pk2 are prone to drug-induced seizures (23). Additionally, overexpression of Pk1 or Pk2 promotes neurite outgrowth in neuroblastoma cells (24). These studies suggest that Prickle genes have broader roles in the CNS.
Albeit all these findings, cell-type specific expression and function of Prickle genes as general PCP players have not been fully addressed. Here, we have investigated the expression of Pk1 in mouse brain and retina and identified Pk1-expressing cells using a genetic approach. We have also explored Pk1 function in neuronal morphogenesis using cultured hippocampal neurons and by in vivo electroporation in the retina. We found that Pk1 is expressed in the cortical subplate, an important site for early neuronal maturation. Our findings provide a framework for elucidating the broader function of Pk1 in the CNS and may assist in defining the mechanism of corresponding human disease(s).
RESULTS
Expression of Pk1 in the developing mouse brain and retina
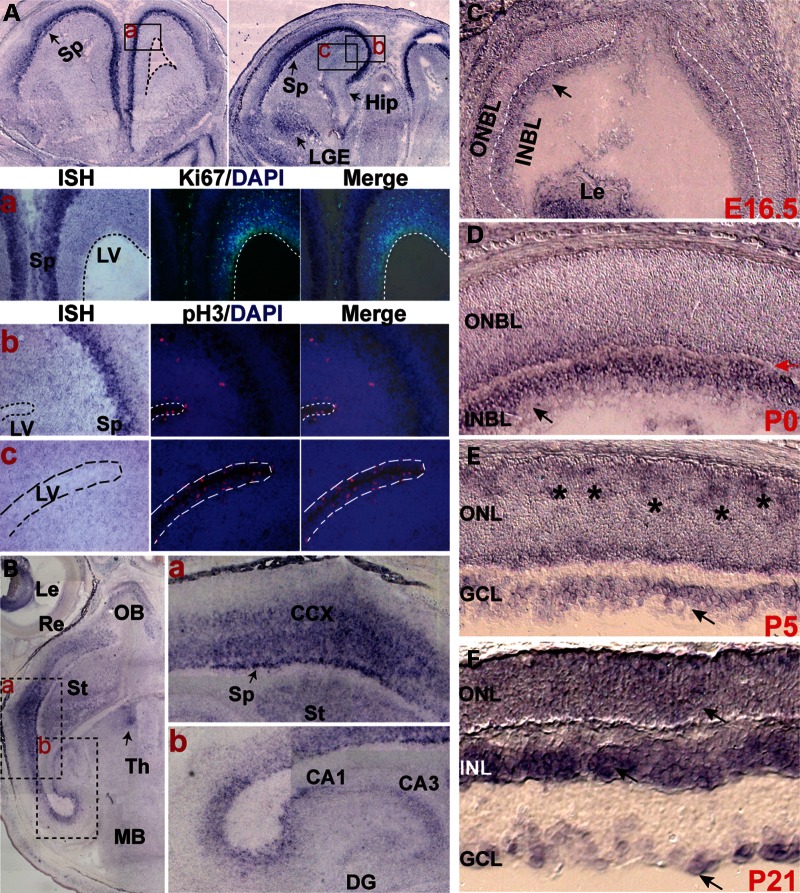
We first investigated Pk1 expression in the CNS by in situ hybridization (ISH). Pk1 expression is first detected before gastrulation (19) and remained through all stages of development (25). At E13.5, Pk1 is weakly expressed in developing neocortex without a distinct pattern (Supplementary Material, Fig. S1A). At E15.5, Pk1 expression is largely in cortical intermediate and subventricular zones and lateral ganglion eminence (LGE) (Supplementary Material, Fig. S1E–G). Intriguingly, Pk1 expression becomes more restricted to the subplate and early maturing cortical plate at E16.5 (Fig. 1A and B), and mostly free of ventricular, subventricular and intermediate zones (Fig. 1A and B, dashed lines demarcate the right lateral ventricle, LV). The dynamic Pk1 expression seems to follow the progressive neurogenesis from early to late stages during cortical neuronal maturation. Consistent with this idea, Pk1-expressing cells are postmitotic at stage E16.5 demonstrated by ISH in combination with immunohistochemistry (IHC) using Ki67 and pH3 antibodies (Fig. 1Aa–c). Similarly, Pk1 transcripts are also detected in the subplate and cortical plate at E17.5, when majority of cortical neurons are undergoing maturation (Fig. 1Ba). Pk1 is also expressed in other brain areas such as hippocampus (Fig. 1Bb) and non-neural tissues as well such as lens (Fig. 1B, left panel; Supplementary Material, Fig. S1J). A weak expression of Pk1 hippocampus anlage is detected from E13.5 (Supplementary Material, Fig. S1A, domain between yellow lines); however, its discrete expression in hippocampus started to be seen at E17.5, since a barely detectable ISH signal is found in the hippocampal area at E16.5 (Fig. 1A, right panel). Hippocampal Pk1 expression continues to be seen at P1 and P5 (Supplementary Material, Fig. S1G and H). Together, Pk1 is expressed in postmitotic cells during mid- and late stages of cortical neurogenesis.
Figure 1.
Expression of Pk1 in the developing brain and retina detected by ISH. (A) ISH shows Pk1 expression in the anterior forebrain at E16.5. Top panels are coronal sections from anterior levels of E16.5 forebrain with the left panel more anterior to the right. Pk1 transcripts are restricted to the subplate area (Sp), but not in the ventricular germinal zone (dashed lines demarcate a lateral right ventricle). A weak hybridization signal is also present in LGE. A barely detectable ISH signal is observed in developing hippocampus (Hip, black arrow). Boxed areas of (a, b and c) roughly correspond to the three rows of panel sets below (a, b and c) (a) Sequential detection of Pk1 mRNA and Ki67 protein by ISH and IHC, respectively. Pk1 mRNA (purple, left panel) is in the subplate (Sp) of the developing cortex; Ki67 (green, middle panel) is expressed in proliferating and dividing neuronal progenitors of the LV and intermediate zone; superimposed image of Pk1 mRNA and Ki67 indicates non-overlapping Pk1 and Ki67. (b, c) Sequential detection of Pk1 mRNA and phospho-histone H3 (pH3) protein by ISH and IHC, respectively. Pk1 mRNA, purple, left panels; pH3, red, middle panels; superimposed images, right panels. (b) shows the Sp area where Pk1 is expressed, and the partial LV zone shows pH3 expression. (c) The ventricular area labeled with pH3, but not Pk1. (B) ISH on horizontal sections of E17.5 brain showing Pk1 mRNA in lens (Le), olfactory bulb (OB), retina (Re), cerebral cortex (CCX), striatum (St) (Fig. 1B), subplate (Sp) (Fig. 1Ba), hippocampal DG and Cornu Ammonis (CA) (Fig. 1Bb). (C), Pk1 mRNA is detected in E16.5 (arrow) retinal INBL, which is primarily populated by early-born ganglion and ACs. Le, lens; ONBL, outer neuroblast layer. (D) A similar Pk1 expression is observed in postnatal day 0 (P0) (black arrow) INBL, separated from the ONBL by a thin newly formed inner plexiform layer (red arrow). (E) Pk1 transcripts are broadly expressed through retinal areas at P5, with a patchy staining pattern in the prospective ONL (asterisks). (F) At postnatal day 21 (P21), Pk1 expression is upregulated in the INL, with only a weak expression in the ONL and GCL (arrows).
We extended our investigations to the retina, a well-defined model of neural development. Pk1 is only weakly expressed in early developing retina at E13.5 (Supplementary Material, Fig. S1B). A weak expression is then observed in the inner neuroblast layer (INBL) at E16.5 (Fig. 1C). Pk1 expression is enhanced in P0 retina, primarily in the differentiating inner neuroblastic layer (INBL) (Fig. 1D). As more retinal progenitors become postmitotic by P5, Pk1 is detected throughout the retina (Fig. 1E). Patches of higher expression are visible in newly postmitotic cells that are being organized in the apical outer NBL (ONBL) (asterisks in Fig. 1E). By P21, Pk1 is largely restricted to the inner nuclear layer (INL) (Fig. 1F), with enhanced patchy staining along the inner portion of the INL, likely representing ACs (Fig. 1F). In addition to the ganglion cell layer (GCL) and the INL, Pk1 is also weakly expressed in the outer nuclear layer (ONL) of photoreceptors (Fig. 1F). We did not detect significant signals using a Pk1 sense probe in different stages of retina and brain sections (Supplementary Material, Fig. S1K–O, data not shown).
Pk1 expression revealed by a knock-in reporter gene
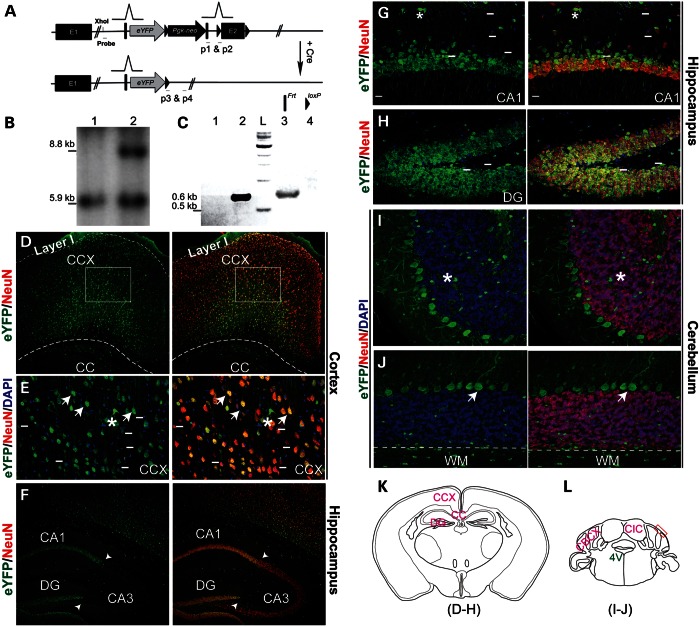
For detailed analysis of Pk1 expression and function, we created a Pk1 gene-trap allele and genetically labeled Pk1-expressing neurons using an eYFP reporter, inserted into the endogenous Pk1 locus via homologous recombination (Fig. 2A). Southern analysis and PCR genotyping confirmed correct integration of the engineered construct (Fig. 2B and C). We explored Pk1 expression by following the eYFP reporter in combination with a pan-neuronal marker, NeuN. Extensive colabeling of eYFP and NeuN was detected in the cerebral cortex, hippocampus, thalamus, hypothalamus and cerebellum (Fig. 2D–L, Supplementary Material, Fig. S2). Notably, a small population of neurons in the CNS does not express Pk1 (cells above short lines in Fig. 2E, G and H, data not shown). In hippocampus, Pk1 exhibits much lower expression in CA3 in contrast to CA1 and dentate gyrus (DG) (Fig. 2F–H). In the cerebellum, Pk1 is expressed in NeuN-negative Purkinje cells, but not in NeuN-positive granular cells (Fig. 2I and J). Pk1 is also present in glia throughout the brain (asterisks in Fig. 2E, G and I), as clearly distinguishable in neuron-free areas such as corpus callosum (CC, Fig. 2D) and white matter (WM) of the cerebellum (Fig. 2J). The Pk1 expression pattern revealed by the knock-in reporter is largely concordant with the data given by ISH.
Figure 2.
Expression of Pk1 in adult brain, revealed by an eYFP reporter driven by the endogenous promoter. (A) An eYFP reporter gene, PGK-neo cassette, Frt and loxP sites are targeted into the Pk1 exon 2 region in the order shown here. An En1 gene splicing acceptor is placed before eYFP start codon and several polyadenylation signals are after eYFP stop codon. This gene-trap allele can be excised by Cre to generate a straight knockout allele. The Sox2-Cre transgenic line is used to cross with Pk1 gene-trap mice in this study. (B) Southern analysis of XhoI-digested genomic DNA using a 5′ probe reveals an 8.8 kb band, showing correct integration of the knock-in gene-trap construct into the Pk1 locus (lane 2) and a 5.9 kb fragment for the WT allele (lane 1). (C) PCR genotyping of gene-trap and straight knockout alleles after Cre excision. Primers 1 and 2 will amplify a 0.6 kb fragment from the straight KO allele (lane 2). Primers 3 and 4 will amplify a 0.65 kb fragment from the gene-trap allele (lane 3). L, DNA ladder. Lanes 1 and 4 are wild-type controls with no PCR products. (D–J) Images shown in these panels are from adult brain at 4-week-old. (D) Pk1 is broadly expressed in the cerebral cortex, as identified by eYFP-positive cells, except for layer I. A majority of the eYFP-positive cells (green) are NeuN positive (red). Boxed areas are shown in (E). (E) High magnification shows expression of eYFP in both neurons (NeuN positive, arrows) and glia (asterisks). Short lines indicate a fraction of eYFP-negative neurons. Note that most eYFP-positive neuronal somata are triangular, a typical shape for pyramidal neurons. (F) Pk1 expression in hippocampus, with strong labeling in CA1 and DG and weaker expression in CA3 (arrowheads demarcate the boundary). (G and H) A high magnification image shows overlapping of Pk1 expression (green) with NeuN in CA1 and DG. Note that some NeuN-positive neurons in the molecular layer are eYFP negative (above the short lines). (I and J) Cerebellar Pk1 expression in Purkinje, but not in granular neurons. Purkinje neurons are NeuN negative; granular neurons are NeuN positive (red). Asterisks indicate glia; arrows indicate Purkinje cells. Below the dashed lines, we observe WM showing sparsely distributed glia (green dots). (K and L) Schematic illustrations of sections of imaged areas for the relevant panels, as indicated below the drawings. 4V, 4th ventricle; red box is the photo area in F and G; CIC, central nucleus of inferior colliculus; CBCX, cerebellar cortex.
Characterization of Pk1-expressing cells in the adult brain
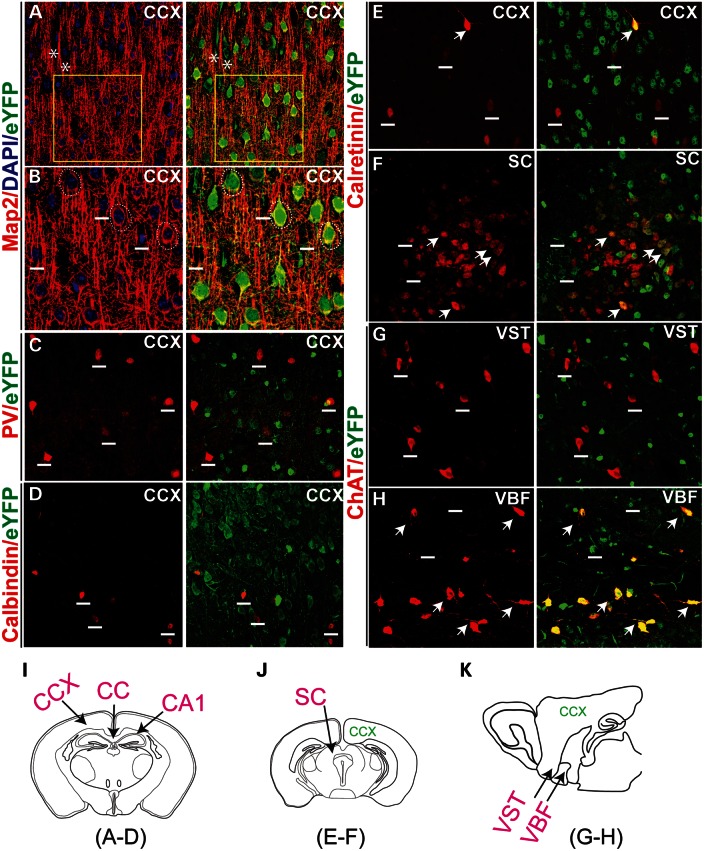
Over 75% of cortical neurons are pyramidal neurons, which spread to all layers, except layer I. The others are inhibitory GABAergic interneurons, which are distributed sparsely among the pyramidal neurons and glial cells (26). A majority of Pk1-expressing cortical neurons are typical pyramidal neurons, judging by their soma shapes (triangular shape, GFP stained soma in Fig. 2D), distributions (mostly excluded from layer I, Fig. 2D) and density (overlapping with most of the NeuN staining) (27). The pyramidal expression of Pk1 (eYFP) could also be identified by MAP2 staining, which envelops the neuronal somata (Fig. 3A and B, dashed circles) and dendrites. Pk1-expressing glial cells (eYFP) (Fig. 3A, asterisks) did not show somata MAP2 staining, and MAP2-negative neurons do not show Pk1 in any instance (Fig. 3B, white short lines). To further classify Pk1-expressing neurons, we investigated whether Pk1 was present in the inhibitory interneurons of the cortex by examining several calcium-binding proteins. We observed that neurons expressing parvalbumin (PV) (Fig. 3C), calbindin (Fig. 3D), calretinin (Fig. 3E and F) and ChAT (Fig. 3G) largely excluded Pk1 expression. A few non-bipolar calretinin neurons co-localize with Pk1 in the cortex (Fig. 3E) and other areas, such as superior collicullus (SC) (Fig. 3F). Pk1 is completely absent in ChAT neurons in most brain areas (Fig. 3G) including the cortex (not shown), except for a subset of ChAT neurons in the ventral basal forebrain (VBF, Fig. 3H) adjacent to the ventral striatum (VST, Fig. 3G). Pk1 expression is, therefore, absent in most interneurons in the brain.
Figure 3.
Description of Pk1-expressing neurons in the brain. (A) Pk1 expression overlaps with Map2, a neuronal maker that labels all pyramidal neurons in the cortex (red). Asterisks indicate two Map2 negative glia cells expressing Pk1 gene (eYFP, green). Boxed areas are zoomed in (B). (B) Pk1 colocalizes with soma staining of Map2 enclosing the neurons’ nuclei indicated by dashed circles. Neurons negative for Map2 do not express Pk1 (above the short lines). (C–E) A majority of PV (C), calbindin (D) and calretinin (E)-stained interneurons in the cortex do not express Pk1 (eYFP). Short lines indicate interneurons expressing the calcium-binding proteins, but not eYFP. (F) In the SC, a few more calretinin neurons express a low level of Pk1 as indicated by arrows. (G) Cholinergic neurons that are labeled with ChAT (red) in the VST do not express Pk1 (short lines in G). (H) ChAT neurons in VBF coexpress Pk1 (arrows in H). (I–K) Schematic illustrations of sections of imaged areas for the relevant panels that are indicated below the diagram.
We next examined Pk1 expression in neural glia. The three major types of glia in the brain are microglia, oligodendrocytes and astrocytes. eYFP expression is correlated with two populations of glia—CD11b-positive microglia (Fig. 4A) and CNPase-positive oligodendrocytes (Fig. 4B and C). Interestingly, eYFP expression is not detected in GFAP-positive astrocytes in any instance (Fig. 4D). Therefore, Pk1 is expressed in two types of neuroglia that are known for their dynamic cytoskeletal and membrane modifications.
Figure 4.
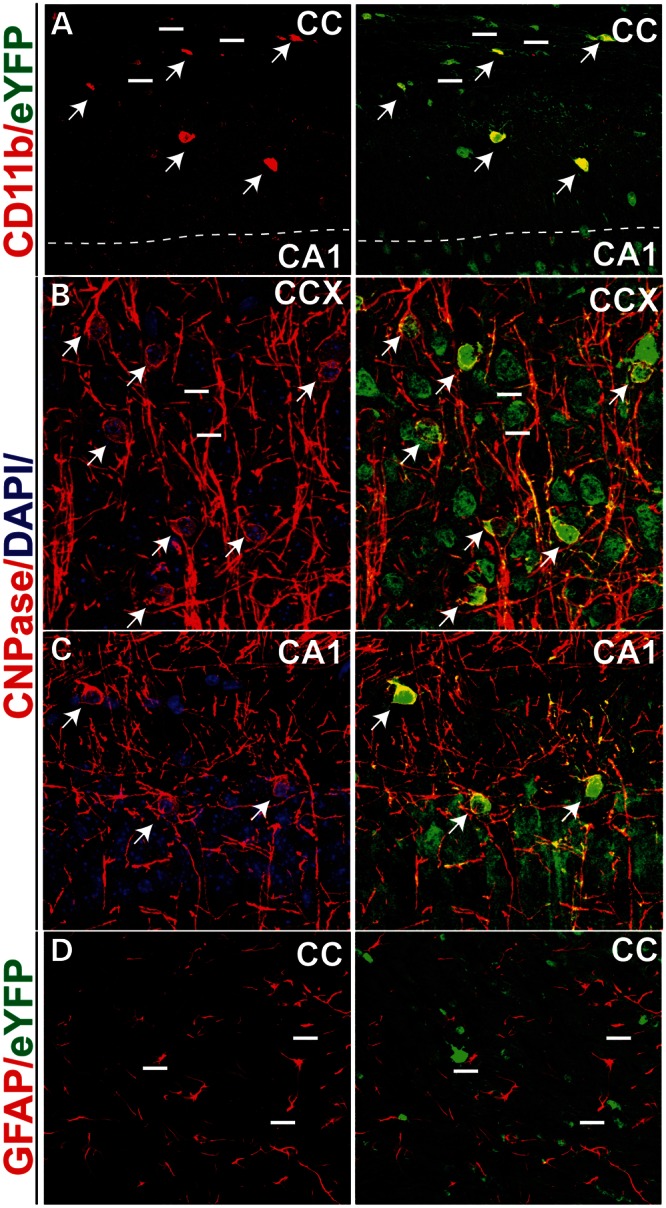
Identification of Pk1-expressing glia in the brain. (A) Colabeling of CD11b and Pk1 (eYFP, green) in microglia (red) of the CC area (arrows). Not all eYFP cells are CD11b positive (short lines), but all CD11b cells are eYFP positive. (B and C) CNPase, a myelin protein staining oligodendrocytes (red), colocalizes with Pk1 expression in the brain (arrows). All CNPase-positive cells are eYFP positive. (B and C) The CCX and CA1 areas, respectively. (D). Pk1 is not expressed in astrocytes marked by GFAP (red) in either CC or other brain areas (not shown). Imaged sections for panels refer to Figure 3I.
Pk1 expression in retinal neurons
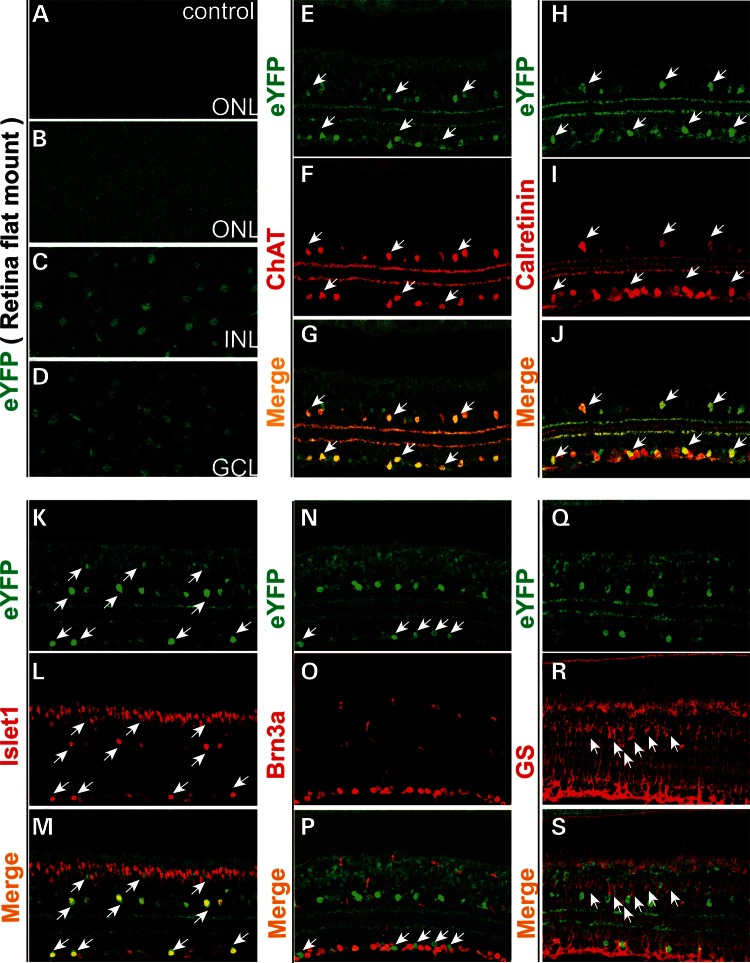
To identify Pk1-expressing cells in the retina, we first confirmed that eYFP expression in the adult retina recapitulates the expression detected by ISH. Pk1 is weakly expressed throughout the ONL in a flat mount preparation (Fig. 5B, compared with the control in 5A). Moderate eYFP signals can be readily detected in the INL (Fig. 5C) and GCL (Fig. 5D) in agreement with the Pk1 pattern detected by ISH. Pk1 is highly expressed in and fully overlaps with cells expressing ChAT, a specific marker of cholinergic starburst ACs (Fig. 5E–G). Pk1-positive cells also express calretinin and Islet-1, the two markers of inner retina neurons including amacrine, ganglion and bipolar cells (arrows, Fig. 5H–M). Lower expression of Pk1 is observed in the remainder of INL, partially overlapping with Islet1 at the outer part of the INL (the top row of arrows, Fig. 5K–M). Pk1 expression is undetectable in Brn3a-positive retinal ganglion cells (RGCs) (Fig. 5N–P) or in Muller glia marked by glutamine synthetase (GS) (Fig. 5Q–S). Therefore, Pk1 expression in the retina is specific to neurons and highly enriched in Starburst amarcine cells.
Figure 5.
Characterization of Pk1-expressing neurons in the adult retina. (A–D) Endogenous eYFP from flat mount retina of 4-week-old Pk1-eYFP knock-in and wild-type mice. (A) ONL of the wild-type mouse (control) does not reveal an EYFP signal. (B) ONL of the heterozygous Pk1-eYFP knock-in mouse shows a faint eYFP signal, consistent with weaker Pk1 expression in the ONL detected by ISH at the same age. (C) eYFP expression in a fraction of neurons in the INL. (D) Expression of eYFP in the retinal GCL. (E–S) Immunohistochemical identification of Pk1 (eYFP)-expressing neurons using an anti-GFP/eYFP antibody and additional markers. (E–G) Pk1 is strongly expressed in cholinergic starburst ACs, as marked by ChAT (in red). Arrows indicate examples of starburst ACs that are coexpressing Pk1 (eYFP green) and ChAT. (H–J) Pk1-expressing amacine cells are calretinin positive. Arrows indicate colocalization of calretinin and Pk1 gene. (K–M) Pk1 is weakly expressed in cells of the upper half of the INL, some of which are Islet1 positive. Arrows indicate colabeled neurons. (N–P) Pk1 expression is excluded from Brn3a-labeled retinal ganglion cells (RGCs) (red). Arrows indicate Pk1 expressing eYFP-positive cells (green). (Q–S) Pk1 is not detectable in Muller glia marked by glutamine synthetase (GS, red) in the INL. Arrows point to Muller glia somata (red).
Aberrant axon–dendrite growth in cultured hippocampal neurons by Pk1 knockdown
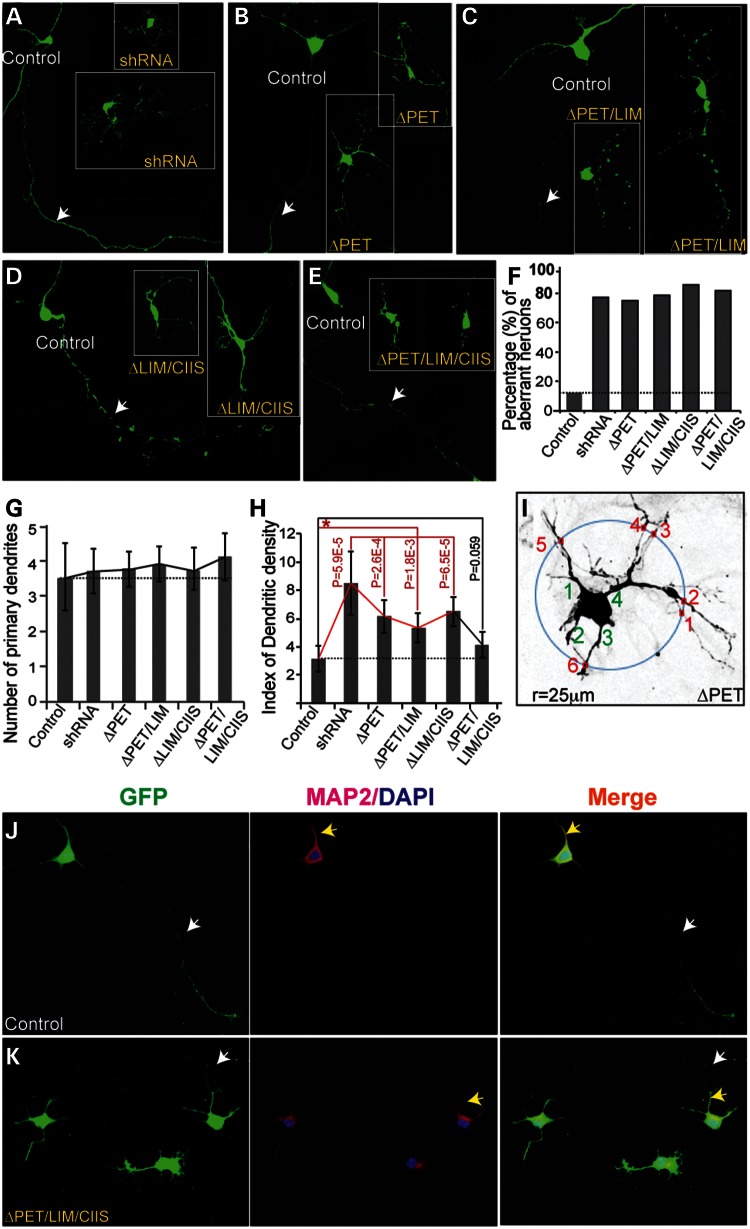
The general expression of Pk1 seems to coincide with specific neurons and glia that exhibit complex and/or dynamic morphological presentations. We, therefore, explored Pk1 function in axonal and dendritic development using two approaches designed to disrupt Pk1 function in cultured hippocampal neurons one by delivering shRNA and the other by delivering Pk1 deletion constructs lacking selected domains and predicted to act as dominant-negative mutants. Pk1 shRNA was validated in transfected HEK 293 cells (Supplementary Material, Fig. S3A). Cultured neurons grown on an astrocyte feeder layer (28) appear healthy and are positive for MAP2 (Supplementary Material, Fig S3B–D). Introduction of Pk1 shRNA into hippocampal neurons causes a pronounced change in neurite outgrowth, with neurons extending short, stubby and punctate dendrites and/or axons (Fig. 6A, 9 out of 12 cells). As PET and LIM domains in Pk1 mediate protein interactions and the CAAX prenylation motif confers the ability to associate with membranes, we examined whether the removal of these domains would impact axonal and dendritic growth. Myc-tagged versions of Pk1 mutant constructs (Supplementary Material, Fig. S4A) were transfected into HEK 293 cells for validation of their successful expression (Supplementary Material, Fig. S4B and C). Interestingly, we also found that differential localization of Pk1 mutant proteins in HEK293 cells (Supplementary Material, Fig. S4B–E). Transfection of ΔPET, ΔPET/LIM, ΔLIM/CIIS or ΔPET/LIM/CIIS construct into hippocampal neurons results in significant axon–dendrite phenotypes. The severity of phenotype induced by ΔPET/LIM/CIIS (Fig. 6E; 9 out of 11 cells) is similar to that of shRNA knockdown, whereas milder defects are observed in ΔPET (8 out of 11), ΔPET/LIM (11 out of 14) and ΔLIM/CIIS-transfected neurons (12 out of 14) (Fig. 6B–D, F and Supplementary Material, Table S1). Therefore, the deletion mutations of Pk1 likely serve as dominant-negative proteins, competing with the normal Pk1 function in transfected neurons. We further quantified the dendritic growth by looking at primary dendrites and dendritic branches of shRNA and deletion constructs transfected neurons (see Materials and Methods, Fig. 6I). While the primary dendritic number retains little affected (Fig. 6G), the branching index significantly increased in shRNA and the mutant construct-transfected neurons (Fig. 6H), except for that of ΔPET/LIM/CIIS transfections, where the severity of the phenotype could have hindered the dendritic outgrowth. In most cases, both control and mutant construct-transfected neurons exhibit Map2-positive and negative regions, implying generally normal specification of dendrite and axon (Fig. 6J and K). A similar staining pattern is also detected in neurons transfected with other mutant constructs (data not shown). Our results argue in favor of a role of Pk1 in modulating axon and dendrite development.
Figure 6.
Functional analysis of Pk1 in cultured hippocampal neurons. (A) Cultured hippocampal neurons transfected with a GFP construct and Pk1 shRNA. Control GFP-transfected neurons show gross normal neuronal morphologies with certain degree of variations (control, 14 out of 16 neurons, 87.5%). Knocking down Pk1 caused aberrant axon–dendrite growth (9 out of 12 neurons, 75%). The dendrites cannot extend well and became stubby and branchy. Arrow indicates an axon of a GFP (control)-transfected neuron. (B–E) Transfection of constructs with deletions of Pk1 key domains and prenylation motif into cultured hippocampal neurons. (B) ΔPET, 8 out of 11 neurons (72%); (C) ΔPET/LIM, 11 out of 14 neurons (78%). (D) ΔLIM/CIIS, 12 out of 14 (85.7%). (E) ΔPET/LIM/CIIS, 9 out of 11 (81%). (F) Graphic presentation of affected neurons with transfection of different mutant Pk1 DNA constructs. (G) Graphic presentation of primary dendrites (green numbers in I) of different mutant Pk1 DNA constructs. There exists a trend of increased primary dendrites in average of mutant construct-transfected neurons but with a big error range. (H) The dendritic branching in mutant construct-transfected neurons is significantly increased compared with the controls (red lines), except for the ΔPET/LIM/CIIS Pk1 construct. The index of dendritic branching is derived from counting the crossing points that a 25 µm radius circle maximally meets with dendrites when placed outside of primary dendrites of a neuron (small red squares and numbers in I). (I) An example of counting primary dendrites (green numbers) and dendritic branching of a ΔPET Pk1-construct transfected neuron. Note: some crossing points with the fine dendritic arborizations in neurons transfected with Pk1 mutant constructs are ignored for the sake of reliability in counting. (J) GFP and Map2 (red) labeled a control neuron showing dendrites (yellow arrows) and axon(s) (white arrows). (K) GFP and Map2 labeled neurons transfected with ΔPET/LIM/CIIS.
Rod photoreceptor defects by in vivo knockdown of Pk1 in mouse retina
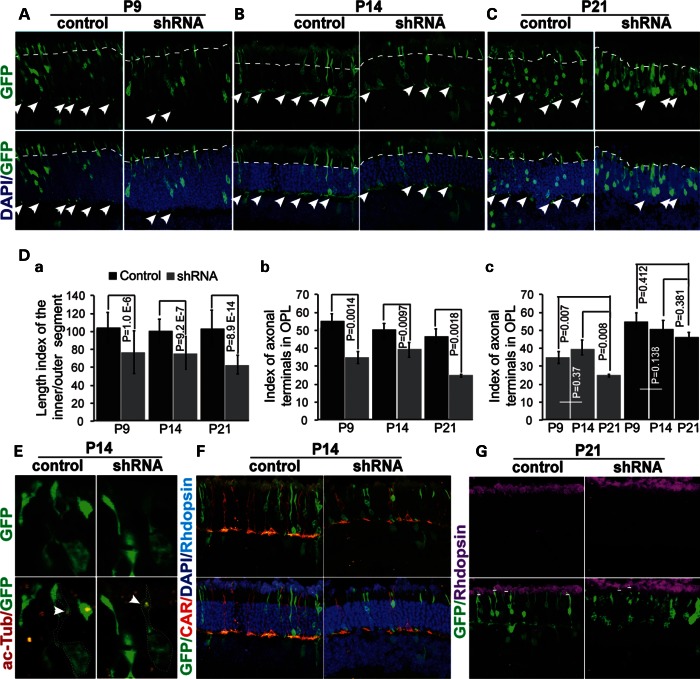
As Pk1 expression is significantly reduced in the cone-only Nrl−/− retina (20,29), we hypothesized that Pk1 might play a role in rod photoreceptor morphogenesis. We first examined dynamic Pk1 expression during retinal development by qPCR (Supplementary Material, Fig. S5A) and confirmed the down-regulation of Pk1 protein in Nrl−/− retina by immunoblotting (Supplementary Material, Fig. S5B). Then, we performed in vivo electroporation to deliver Pk1 shRNA into P0 mouse retina with ubiquitin promoter-driven GFP as a transfection and morphological indicator. shRNA-transfected rods show significantly smaller inner/outer segments from P9 to P21 (above dashed lines of the ONL, Fig. 7A–C). Interestingly, the number of rod terminals in the outer plexiform layer (OPL) is also significantly reduced in shRNA-transfected photoreceptors (arrows, Fig. 7A–C). Quantification analysis corroborates the above observations (Fig. 7Da and b). The defects in axon terminals and IS and OS of photoreceptors become more pronounced at P21 (Fig. 7Dc). However, the defective photoreceptors do possess cilia as marked by acetylated alpha tubulin (Fig. 7E), indicating that loss or shortening of inner/outer segments probably reflects both impaired photoreceptor biogenesis and maintenance. Photoreceptors maintain their rod phenotype even after Pk1-knockdown, showing rhodopsin but not cone arrestin expression (Fig. 7F and G). Additionally, we did not observe any altered protein trafficking at P21or P8 based on the localization of rhodopsin and rds proteins (Fig. 7G, Supplementary Material, Fig. S6A–E). Pk1 was expressed in all the cells of the ONL (Supplementary Material, Fig. S5C), including cones (Supplementary Material, Fig. S5D–F, above short lines). Hence, these results indicate a broader role of Pk1 in photoreceptors and during neuronal morphogenesis.
Figure 7.
Rod photoreceptor defects in Pk1 shRNA electroporated retina in vivo. (A). Shortened inner/outer segments (above dashed lines) and a reduced number of axon terminals at P9 caused by Pk1 shRNA (arrows in the inner plexiform layer, IPL). (B) Similar defects are observed at P14 compared with P9. (C). Severe defects in both inner/outer segments and synaptic terminals at P21 (dashed lines and arrows). (D) (a) A significant decrease of inner/outer segments’ length in shRNA electroporated retina at different ages. A length index is generated by measuring the inner/outer segments of GFP-positive photoreceptors using ImageJ. (b) A significant decrease of synaptic terminals in shRNA-electroporated retina at different ages. The axon/synaptic terminal index is the ratio of GFP dots in the OPL to the total GFP-positive cells on the section, multiplied by a factor of “100”. (c) Progressed reduction of axonal terminals in shRNA electroporated retinas occurs between P14 and P21, but not P9 and P14. The original data used to plot the graph are the same as in (b). (E) Acetylated alpha tubulin (red) stains cilia in both control and Pk1-shRNA photoreceptors (green). (F) Pk1-shRNA rods (green) do not change to cones (red) identified by cone arrestin. (G) Pk1-shRNA photoreceptors still express rhodopsin (above the short lines).
DISCUSSION
During mammalian development, complex interactions among multiple signaling pathways produce millions of distinct cell types, determine their unique organizational patterns and generate billions of specific connections. While established in Drosophila and other model organisms, the role of PCP signaling in mammals, and particularly in the CNS, is only recently being delineated. Prickle is a core PCP component, and two mammalian Prickle proteins—Pk1 and Pk2—appear to have significant function in the CNS, as revealed by human and mouse mutations (19), cell culture studies (24) and expression data (30,31). Here, we have performed a comprehensive analysis of Pk1 expression in developing and adult brain and retina and loss-of-function studies in vitro and in vivo. Together with the published data, our results suggest a significant role of Pk1 in neuronal morphogenesis and maturation, specifically in axonal and dendritic configuration.
Our data reveal an unambiguous postmitotic expression of Pk1 during the mid–late stage of cortical neurogenesis at E16.5, consistent with another report (31). Immediately after exiting cell cycle at the ventricular zone, neurons start migrating through the intermediate zone to reach the subplate and cortical plate. Predominant Pk1 expression at the subplate indicates that it is important for early onset neuronal maturation since subplate harbors the earliest born and mature neurons that make the earliest connections (32). The axon–dendrite development and maturation of subplate neurons precede neurons in other layers and undergoes more dynamic cytoskeleton rearrangement, which may require more polarity-related molecules, such as Pk1. Similarly in the retina, Pk1 is expressed in the early differentiating cells (see Fig. 1, E16.5). Pk1 expression in postnatal retina may reflect active shaping and polarization of retinal neurons during maturation as well.
Genetic labeling of Pk1-expressing neurons, using an eYFP knock-in reporter, allowed us to identify their characteristics by co-localization with specific markers. In most adult brain areas, eYFP co-localized with pan-neuronal markers, NeuN and MAP2, at variable intensities. Cells in the cortex and other brain areas showing no Pk1 expression consist primarily of inhibitory interneurons, as revealed by expression of distinct calcium-binding proteins. In the cerebellum, eYFP is completely absent in granular cells but is strongly expressed in Purkinje neurons. A wide expression of Pk1 in the brain suggests a broad generic function in neuronal maturation; yet in the retina, Pk1 is highly expressed in Starburst ACs and virtually excluded from ganglion cells and Muller glia. A low level of Pk1 is detected in a subpopulation of bipolar cells and in photoreceptors. The selectivity of Pk1 expression in distinct cell types of the adult brain likely reflects its functional relevance. Pk1-expressing pyramidal neurons, Purkinje cells and retinal cholinergic ACs have a large dendritic field and extensive arborization. The morphogenesis, maturation and synaptic plasticity of these neurons conceivably place a greater demand on polarity components. This hypothesis is also validated by strong Pk1 expression in microglia and oligodendrocytes. Microglia are immune cells with multiple processes constantly monitoring the neuronal environment by virtue of its mobility, and requiring dynamic cytoskeletal and membrane rearrangements. Similarly, oligodendrocytes have active membrane rearrangements for wrapping axons. Taken together, Pk1 is expressed in distinct cell populations with dynamic morphological configurations in the CNS.
In accordance with its predicted role in neuronal morphogenesis, Pk1 knockdown by shRNA or deletion constructs compromised axon and dendrite extension in cultured hippocampal neurons. One interesting observation is that although the shRNA-transfected neurons are stubby and less extended in general, the dendritic branching index significantly increased (by calculation of 25 µm surrounding area from the soma). In vivo evidence has shown that knocking out Fat3, a PCP component in Drosophila, causes the growth of ectopic dendrites (12), which is consistent with shRNA knockdown in hippocampal neurons here. As Pk1 has several well-characterized protein domains and motif, we investigated whether these domains are critical for Pk1 function in neurons by constructing and introducing domain/motif deletion constructs into cultured neurons. We observed a similar dendritic branching phenotype to shRNA knockdown of Pk1, which strongly suggests that these domain/motif-deleted proteins function in a dominant-negative manner in competition with wild-type Pk1. However, a direct biochemical assay needs to be established to examine the functional properties of each Pk1 domain.
Although Pk1 has been shown to be regulated by NRL previously, knockdown of Pk1 in rod photoreceptors led to morphological defects in inner/outer segments and axonal terminals, but not the cell fate. The less-extended axon terminals and stubby inner segments in Pk1 knockdown photoreceptors may also reflect the same aspects of neuronal morphogenesis in hippocampal neurons. Growth of outer segments distal to the connecting cilia is a key step in photoreceptor differentiation and requires precise control of microtubule and actin filament dynamics at the ciliary stalk, where new outer segment morphogenesis takes place (33,34). A well-characterized PCP target is the cytoskeletal system. Since knockdown of Pk1 does not seem to disrupt the initiation of ciliogenesis, which highly relies on microtubule functioning, it is plausible that the actin assembling at the ciliary stalk is compromised, which in turn causes abnormal shape of the outer segment. The more pronounced reductions of axonal terminals at the late stage of retinal development could imply two functional aspects of Pk1: one is to promote axonal/dendritic extension at early stages, for instance, from P9 to P14; and second is to maintain/stabilize the synaptic connections at later stages, for instance, from P14 to P21. The data further imply that Pk1 might not be important for synaptogenesis since synaptic connection occurs actively before the mouse eye opens around P12–P14 (35). We did not observe any significant rhodopsin trafficking and programmed cell death. One interpretation could be that the general metabolic state of shRNA-transfected photoreceptor has been slowing down such that little rhodopsin is accumulated. Another possibility is that Pk1 might affect rhodopsin expression, as it is a potential target of NRL. NRL is a key transcription factor regulating the expression of a large cohort of genes required for terminal differentiation of rods including the rhodopsin gene. Consistent with downregulation of Pk1 in NRL retina, NRL ChIP-Seq data identified two binding peaks within the first intron of the Pk1 gene (36). Additionally, we noticed that Pk1 is also expressed in cones, which probably functions in a similar way as in rods; however, this might be regulated by different mechanisms from that of rods. It will be of interest to further investigate the differential regulations of Pk1 gene in rods versus cones in future. As complete loss of Pk1 appears to result in early embryonic lethality, future studies employing tissue-specific and conditional gene targeting will be needed to obtain in vivo functional insights into the role of Pk1 in the CNS.
MATERIALS AND METHODS
Histology, in situ hybridization, immunolocalization and imaging
Procedures for preparation of flat mount retina, light microscopy, ISH and IHC have been described previously (4,37). To generate probes for ISH, C-termini of Pk1 and Pk2 complementary DNAs (cDNAs) were cloned into a pBluescript II SK (+) plasmid. RNA probes were synthesized using a T3 or T7 RNA polymerase. In some experiments, after completion of color development in ISH using X-phos substrates (37), the sections were subjected to blocking with 1X TBST containing 5% goat serum before performing IHC using standard protocols. Frozen, 4% paraformaldehyde fixed retina sections were used for IHC staining, whereas brain tissue IHC was performed on 50–100 µm thick floating vibratome sections.
Antibodies and dilutions used in this study are as follows: mouse anti-Brn3a, 1:1000 (Santa Cruz, sc-8429); mouse anti-GFAP, 1:1000 (Millipore, MAB3402); mouse anti-Islet1, 1:2000 (DSHB, 40.3A4); mouse anti-calretinin 1:1000 (MAB 1568); goat anti-ChAT 1:100 (Millipore AB144P); rat anti-CD11b 1:200 (Millipore, MAB1387Z); rabbit anti-phospho-histone H3, 1:150 (Cell Signaling, 9701); mouse anti-Ki67, 1:200 (Abcam, ab8191); rabbit anti-cone arrestin, 1:500 (Millipore, ab15282); mouse anti-rhodopsin 1D4, 1:200 (ab5417); mouse anti-GS, 1:1000 (laboratories, 610518); rabbit anti-GFP 1:1000 (Torrey Pines Biolabs, Inc, TP401); mouse anti-NeuN, 1:1000 (Millipore, MAB 377), mouse anti-myc, 1:1500 (DSHB, 9E10), mouse anti-CNPase, 1:200 (Millipore, clone 11–5B), rabbit anti-Pk1, 1:1000 (custom-made, and a gift from Dr Bassuk), mouse anti-MAP2, 1:1000 (Millipore, MAB3418), mouse anti-calbindin D28, 1:2000 (Sigma, C9848, CB-955), mouse anti-PV, 1:2000 (Swant, code No. 235), rabbit anti-Arl13B, 1:1000 (proteintech, 17711-1-AP); mouse anti-acetylated alpha tubulin, 1:1000 (Sigma, T6793); chick anti-Rds/peripherin, 1:2000 (Customized). Fluorescent images were collected using a Leica SP5 confocal microscope.
qRT–PCR and immunoblot analysis
Total RNA and protein extracts were prepared from different stages of mouse retina. Quantitative RT–PCR and immunoblot analysis have been described previously (4).
Generation of Pk1 mutant alleles
A Pk1 gene-trap mutant strain was generated by NEI Genetic Engineering Core. All procedures involving the use of mice were approved by National Eye Institute Animal Care and Use Committee. The knockout strain was engineered similar to the ‘knocjou-first’ (38). Briefly, a gene-trap construct was engineered with a Frt site and En2 gene splicing acceptor sequence (SA) preceding the eYFP reporter gene, followed by a polyadenylation signal, loxP, PGK-neo, Frt and loxP. This gene-trap cassette was placed into the upstream of Pk1 exon 2 without disruption of its splicing acceptor (Fig. 3). A third loxP was then placed right after exon 2, which is used for the generation of a conditional knockout allele later. The engineered allele was named Pk1 gene-trap allele, in which eYFP reporter expression is under the control of an endogenous Prickle 1 promoter. Upon a complete excision by Cre, this allele is converted to a straight knockout carrying eYFP reporter. The Sox2-Cre transgenic line (JAX laboratory) was used to cross with the gene-trap allele.
Southern analysis was used to identify the F1 knock-in Pk1 gene-trap allele using a 5′ probe outside of the left arm after XhoI site. XhoI was used for digesting the genomic DNA that was transferred to Amicon nylon membrane for Southern hybridization using α32P dCTP labeled probes as described (5). Labeled probes detected 8.8 and 5.9 kb DNA fragments, which corresponded to the wild-type and mutant alleles, respectively. CL-pk1-7 (p1) and CL-pk1-8 (p2) primer pair was used to genotype the Pk1 straight knockout allele, giving a 610 bp product. Neo (p3) and CL-pk1-6 (p4) pair was used to genotype the Pk1 gene-trap allele, giving a 650 bp product. PCR procedure was as follows: 94°C 1 min; 35 cycles: 94°C 30 s, 60°C, 30 s; 72°C 45 s; 72°C 10 min. Genotyping primers: CL-pk1-6: ACCCATGCATACTCTTCAAGTGCACAG; CL-pk1-7: ATCACTCTCGGCATGGACGAGCTGTACAAG; CL-pk1-8: GTTGTCAAAGGCCATGTTTATCAGCAAC; Neo: ATCGCCTTCTTGACGAGTTCTTCTGA.
Pk1 shRNA and deletion constructs
A DNA primer used for generating Pk1 shRNA is as follows (the targeted sequence is underlined): 5′-GGCAGCACTGGCACGCCACAAAGCTTTGTGGCGTGCCAGTGCTGCCCTTTTTG-3′. The embedded HindIII site (bold in sequence) was used for the hairpin structure. The oligomer DNA was cloned into the pBS/U6 vector with an U6 promoter (39). Pk1 deletion constructs were made using a set of PCR primers to put together the desired Pk1 cDNA fragments, a myc-tag and an optimized Kozak sequence into the pRK5 vector with a CMV promoter. A Pk1 cDNA clone was originally purchased from Open Biosystems.
Tissue culture and DNA transfection
HEK293 cells were cultured in DMEM/F12 (Invitrogen, Cat. 12660-012) medium with supplement of 10% FBS and penicillin/streptomycin (Invitrogen, Cat. 15070-063). Transfections were performed using FuGENE 6, as recommended by the manufacturer (Roche, 11814443001). Five nanograms of shRNA were applied to 2 × 104 cells for evaluating shRNA knockdown efficacy (Fig. 6A); 100 ng DNA was used for expression of Pk1 deletion constructs. Cells were grown on poly-d-lysine-coated coverslips, methanol-fixed after 48 h transfection and stained with anti-myc and/or anti-Pk1 antibody.
The hippocampal neuronal culture primarily followed the published protocol (28), with some modifications. Briefly, astrocytes were prepared separately from the mouse cerebral cortex at P0, cultured for 7 days, split into 24-well dishes at 2 × 104 cells per well, grown for 3 days in glia medium and then changed into neuronal culture medium. Freshly dissociated hippocampal neurons were plated on 12 mm poly-d-lysine coated coverslips in a 24-well dish with 3 to 4 wax posts on the cell growing surface. After 3 to 4h, coverslips with settled hippocampal neurons were transferred into 24-well dishes of feeder glial cells with the wax posts facing the feeder glia. Neurons were cultured on day 3, transfected with DNA constructs using Lipofectamine 2000 (Invitrogen, 11668-019). Coverslips were flipped with neurons facing up during the transfection and flipped back afterwards. Neurons were continually cultured on day 7. Coverslips with cells were fixed with methanol for IHC.
In vivo electroporation in mouse retina
Retinal electroporation was essentially performed as described (36). Eyes of P0 CD1 pups (Charles River Laboratories) were injected with DNA constructs containing Pk1 shRNA into the sub-retinal space, followed by in vivo electroporation with BTX EC M830. pUb-GFP plasmids were coinjected and electroporated into mouse retina for tracking the transfection efficiency and to analyze photoreceptor morphology.
Quantification neuronal processes
For cultured neurons, primary dendrites and dendritic branching were quantified. The dendritic branching index is derived from counting the crossing points that a 25 µm radius circle maximally meets with dendrites when placed outside of primary dendrites of a neuron. The more the crossing points are, the more complex the dendritic branching is. Some crossing points with the fine dendritic arborizations in neurons transfected with Pk1 mutant constructs are subjectively ignored for achieving more reliable data (Fig. 6I). For quantifying the length of inner/outer segments of the photoreceptors, a length index is generated by measuring the inner/outer segments of GFP-positive photoreceptors using ImageJ. The length is, therefore, in pixels, which is not calibrated to the real length.
SUPPLEMENTARY MATERIAL
FUNDING
This work is supported by the intramural research program of the National Eye Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Alexander Bassuk (University of Iowa) for anti-Pk1 rabbit antibody, Robert Fariss (Imaging Core) for assistance with confocal imaging, Tudor Badea and Rivka Rachel for helpful comments, Douglas Kim for shRNA, Hong Hao for Chip-Seq data, Matthew Brooks for qPCR and Helen May-Simera for discussions. We thank Yide Mi, Linn Gieser and Jacob Nellissery for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wang Y., Nathans J. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development. 2007;134:647–658. doi: 10.1242/dev.02772. [DOI] [PubMed] [Google Scholar]
- 2.Seifert J.R., Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat. Rev. Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong A., Ryu Y.K., Chieco D., Kuruvilla R. Frizzled3 is required for neurogenesis and target innervation during sympathetic nervous system development. J. Neurosci. 2011;31:2371–2381. doi: 10.1523/JNEUROSCI.4243-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C., Bakeri H., Li T., Swaroop A. Regulation of retinal progenitor expansion by frizzled receptors: implications for microphthalmia and retinal coloboma. Hum. Mol. Genet. 2012;21:1848–1860. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C., Wang Y., Smallwood P.M., Nathans J. An essential role for frizzled5 in neuronal survival in the parafascicular nucleus of the thalamus. J. Neurosci. 2008;28:5641–5653. doi: 10.1523/JNEUROSCI.1056-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 7.Fenstermaker A.G., Prasad A.A., Bechara A., Adolfs Y., Tissir F., Goffinet A., Zou Y., Pasterkamp R.J. Wnt/planar cell polarity signaling controls the anterior–posterior organization of monoaminergic axons in the brainstem. J. Neurosci. 2010;30:16053–16064. doi: 10.1523/JNEUROSCI.4508-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jessen J.R., Topczewski J., Bingham S., Sepich D.S., Marlow F., Chandrasekhar A., Solnica-Krezel L. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyuksyutova A.I., Lu C.C., Milanesio N., King L.A., Guo N., Wang Y., Nathans J., Tessier-Lavigne M., Zou Y. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 2003;302:1984–1988. doi: 10.1126/science.1089610. [DOI] [PubMed] [Google Scholar]
- 10.Tissir F., Bar I., Jossin Y., De Backer O., Goffinet A.M. Protocadherin Celsr3 is crucial in axonal tract development. Nat. Neurosci. 2005;8:451–457. doi: 10.1038/nn1428. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Thekdi N., Smallwood P.M., Macke J.P., Nathans J. Frizzled-3 is required for the development of major fiber tracts in the rostral CNS. J. Neurosci. 2002;22:8563–8573. doi: 10.1523/JNEUROSCI.22-19-08563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deans M.R., Krol A., Abraira V.E., Copley C.O., Tucker A.F., Goodrich L.V. Control of neuronal morphology by the atypical cadherin Fat3. Neuron. 2011;71:820–832. doi: 10.1016/j.neuron.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenny A., Mlodzik M. Planar cell polarity signaling: a common mechanism for cellular polarization. Mt. Sinai J. Med. 2006;73:738–750. [PubMed] [Google Scholar]
- 14.Jenny A., Darken R.S., Wilson P.A., Mlodzik M. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 2003;22:4409–4420. doi: 10.1093/emboj/cdg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenny A., Reynolds-Kenneally J., Das G., Burnett M., Mlodzik M. Diego and Prickle regulate frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol. 2005;7:691–697. doi: 10.1038/ncb1271. [DOI] [PubMed] [Google Scholar]
- 16.Deans M.R., Antic D., Suyama K., Scott M.P., Axelrod J.D., Goodrich L.V. Asymmetric distribution of prickle-like 2 reveals an early underlying polarization of vestibular sensory epithelia in the inner ear. J. Neurosci. 2007;27:3139–3147. doi: 10.1523/JNEUROSCI.5151-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narimatsu M., Bose R., Pye M., Zhang L., Miller B., Ching P., Sakuma R., Luga V., Roncari L., Attisano L., et al. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell. 2009;137:295–307. doi: 10.1016/j.cell.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Tao H., Inoue K., Kiyonari H., Bassuk A.G., Axelrod J.D., Sasaki H., Aizawa S., Ueno N. Nuclear localization of Prickle2 is required to establish cell polarity during early mouse embryogenesis. Dev. Biol. 2012;364:138–148. doi: 10.1016/j.ydbio.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao H., Suzuki M., Kiyonari H., Abe T., Sasaoka T., Ueno N. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity. Proc. Natl Acad. Sci. USA. 2009;106:14426–14431. doi: 10.1073/pnas.0901332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J., He S., Friedman J.S., Akimoto M., Ghosh D., Mears A.J., Hicks D., Swaroop A. Altered expression of genes of the Bmp/Smad and Wnt/calcium signaling pathways in the cone-only Nrl−/− mouse retina, revealed by gene profiling using custom cDNA microarrays. J. Biol. Chem. 2004;279:42211–42220. doi: 10.1074/jbc.M408223200. [DOI] [PubMed] [Google Scholar]
- 21.Swaroop A., Kim D., Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassuk A.G., Wallace R.H., Buhr A., Buller A.R., Afawi Z., Shimojo M., Miyata S., Chen S., Gonzalez-Alegre P., Griesbach H.L., et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am. J. Hum. Genet. 2008;83:572–581. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao H., Manak J.R., Sowers L., Mei X., Kiyonari H., Abe T., Dahdaleh N.S., Yang T., Wu S., Chen S., et al. Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am. J. Hum. Genet. 2011;88:138–149. doi: 10.1016/j.ajhg.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimura L., Watanabe-Takano H., Sato Y., Tokuhisa T., Hatano M. Prickle promotes neurite outgrowth via the Dishevelled dependent pathway in C1300 cells. Neurosci. Lett. 2009;467:6–10. doi: 10.1016/j.neulet.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 25.Tissir F., Goffinet A.M. Expression of planar cell polarity genes during development of the mouse CNS. Eur. J. Neurosci. 2006;23:597–607. doi: 10.1111/j.1460-9568.2006.04596.x. [DOI] [PubMed] [Google Scholar]
- 26.Chan C.H., Godinho L.N., Thomaidou D., Tan S.S., Gulisano M., Parnavelas J.G. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cerebral Cortex. 2001;11:1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- 27.Parnavelas J.G., Lieberman A.R., Webster K.E. Organization of neurons in the visual cortex, area 17, of the rat. J. Anat. 1977;124:305–322. [PMC free article] [PubMed] [Google Scholar]
- 28.Kaech S., Banker G. Culturing hippocampal neurons. Nat. Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 29.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 30.Crompton L.A., Du Roure C., Rodriguez T.A. Early embryonic expression patterns of the mouse Flamingo and Prickle orthologues. Dev. Dyn. 2007;236:3137–3143. doi: 10.1002/dvdy.21338. [DOI] [PubMed] [Google Scholar]
- 31.Okuda H., Miyata S., Mori Y., Tohyama M. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Lett. 2007;581:4754–4760. doi: 10.1016/j.febslet.2007.08.075. [DOI] [PubMed] [Google Scholar]
- 32.McAllister A.K. Subplate neurons: a missing link among neurotrophins, activity, and ocular dominance plasticity? Proc. Natl Acad. Sci. USA. 1999;96:13600–13602. doi: 10.1073/pnas.96.24.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams D.S., Linberg K.A., Vaughan D.K., Fariss R.N., Fisher S.K. Disruption of microfilament organization and deregulation of disk membrane morphogenesis by cytochalasin D in rod and cone photoreceptors. J. Comp. Neurol. 1988;272:161–176. doi: 10.1002/cne.902720202. [DOI] [PubMed] [Google Scholar]
- 34.Chaitin M.H., Burnside B. Actin filament polarity at the site of rod outer segment disk morphogenesis. Invest. Ophthalmol. Vis. Sci. 1989;30:2461–2469. [PubMed] [Google Scholar]
- 35.Regus-Leidig H., Tom Dieck S., Specht D., Meyer L., Brandstatter J.H. Early steps in the assembly of photoreceptor ribbon synapses in the mouse retina: the involvement of precursor spheres. J. Comp. Neurol. 2009;512:814–824. doi: 10.1002/cne.21915. [DOI] [PubMed] [Google Scholar]
- 36.Hao H., Kim D.S., Klocke B., Johnson K.R., Cui K., Gotoh N., Zang C., Gregorski J., Gieser L., Peng W., et al. Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by In Vivo NRL Targetome Analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C., Nathans J. An essential role for frizzled 5 in mammalian ocular development. Development. 2008;135:3567–3576. doi: 10.1242/dev.028076. [DOI] [PubMed] [Google Scholar]
- 38.Testa G., Schaft J., van der Hoeven F., Glaser S., Anastassiadis K., Zhang Y., Hermann T., Stremmel W., Stewart A.F. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda T., Cepko C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl Acad. Sci. USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.