Abstract
Despite efficient antiretroviral therapy, eradication of HIV-1 infection is challenging and requires novel biological insights and therapeutic strategies. Among other physiological and environmental factors, intracellular iron greatly affects HIV-1 replication. Higher iron stores were shown to be associated with faster progression of HIV-1 infection and to inversely correlate with the survival of HIV-1 infected patients. Iron is required for several steps in the HIV-1 life cycle, including reverse transcription, HIV-1 gene expression and capsid assembly. Here, the authors present a comprehensive review of the molecular mechanisms involved in iron- and oxygen-mediated regulation of HIV-1 replication. We also propose key intracellular pathways that may be involved in regulating HIV-1 replication, via protein kinase complexes, CDK9/cyclin T1 and CDK 2/cyclin E, protein phosphatase-1 and other host factors.
Keywords: CDK2, CDK9, HIV-1, hypoxia, iron chelators, protein phosphatase-1, Tat
Despite efficient antiretroviral therapy, eradication of HIV-1 infection continues to be a daunting challenge and requires new biological insights for the development of effective therapeutic strategies. Eradication of latent HIV-1 provirus still remains ineffective, as integrated HIV-1 is not affected by the existing antiretro-viral drugs until viral transcription is activated [1]. Both environmental and physiological factors can contribute to the progression of HIV-1 disease. For example, higher intracellular iron stores correlate with rapid progression of HIV-1 infection, thus associating with the lower rate of survival of HIV-1-infected patients [2]. Iron is required for several steps in the HIV-1 life cycle, including reverse transcription, HIV-1 gene expression, and capsid assembly [3]. Lower levels of physiological oxygen concentrations also affect HIV-1 transcription and replication [4]. Here, the authors review the molecular mechanisms involved in the iron and hypoxia-mediated HIV-1 transcription regulation and discuss how these pathways could be potentially used to inhibit or activate HIV-1 for the development of future therapeutic interventions for the treatment of HIV-1 infections (Tables 1 & 2).
Table 1.
Iron and oxygen regulatory pathways and their effect on HIV-1.
| Modulation | Mechanism | Mediating pathways | HIV-related consequence | Ref. |
|---|---|---|---|---|
| ↑ Iron | Iron suppl. | Unknown | ↑ HIV-1 replication | [57] |
| ↑ Iron | Hepcidin. | Ferroportin degradation | ↑ HIV-1 replication | [14] |
| ↓ Iron | Iron chelation | ↓ CDK2 activity, ↓CDK9 activity | ↓ HIV-1 transcription | [12,13] |
| ↓ CDK2 | siRNA | ↓ CDK9 Ser-90 phosphorylation | ↓ HIV-1 transcription, ↓ HIV-1 replication | [56] |
| ↓ CDK9 | Inhibition by ARC | ↓ CDK9 activity | ↓ HIV-1 transcription, ↓ HIV-1 replication | [100] |
| ↓ Iron | Ferroportin | Unknown | ↓ HIV-1 replication | [14] |
| ↑ HO-1 | Heme suppl. | Unknown | ↓ HIV-1 replication | [77,78] |
| ↓ Oxygen | 3% O2 culture | ↓ CDK9 activity | ↓ HIV-1 transcription, ↓ HIV-1 replication | [4] |
| ↓ Oxygen | 1% O2 culture | ↓ PP1 activity due to association with NIPP1 | Unknown | [103] |
| ↓ Oxygen | 1% O2 culture | p27 expression | Unknown | [104] |
| ↓ PP1 | Expression of PP1 inhibitors | CDK9 Ser-175 and Thr-186 phosphorylation | ↓ HIV-1 transcription | [53–55] |
Table 2.
Epidemiological evidence for iron and oxygen pathways effect on HIV-1.
| Epidemiological evidence | Hypothesized mechanism | HIV-1 related consequence | Ref. |
|---|---|---|---|
| Thalassemia major, oral iron administration, haptoglobin 2–2 polymorphism | ↑ Iron, ↑ ferritin | ↑ Mortality | [2] |
| Zimbabwe HIV cohort | ↑ Ferritin | ↑ HIV-1 infection | [7] |
| Gambian HIV cohort | ↑ Iron, ↑ ferritin | ↑ Mortality | [8] |
| SLC11A1 (NRAMP1) polymorphisms | ↑ ↓ Ferritin | ↑ ↓ Mortality | [9] |
| Sickle cell disease | ↑ Hemolysis | ↓ HIV-1 infection; ↓ HIV-1 progression | [107,108,111] |
| Sickle cell disease | ↑ HO-1, ↑ p21 | Unknown | [113] |
| Gambian HIV cohort | ↑ Iron, ↑ ferritin | ↑ Mortality | [8] |
| HIV-1 elite controllers | ↑ p21, ↓ CDK9 activity | ↓ HIV-1 progression | [74] |
Iron overload & HIV-1 progression
The development of AIDS is accompanied by an increasing iron accumulation in macrophages, microglia, endothelial cells, myocytes, bone marrow, brain white matter, muscle and liver [5,6]. Increased iron stores correlate with rapid HIV-1 progression in AIDS patients, in iron-loaded thalassemia major patients, in HIV-positive patients administered with oral iron, and in those with the haptoglobin 2–2 polymorphism [2]. This association was supported by a retrospective study demonstrating the lower survival of HIV-1-infected individuals to be inversely correlated to higher iron stores [2]. Nonanemic HIV-1-positive women in Zimbabwe with high serum ferritin concentration are reported to exhibit increased viral load, which provides further evidence for the association of increased iron stores with more severe HIV-1 infection [7]. Elevated iron level has been suggested as a potential predictive marker for higher mortality in HIV-1-infected Gambian adults [8]. Different solute carrier family 11/natural resistance-associated macrophage protein 1 polymorphisms have been shown to be associated with either protection or greater mortality [9]. Therefore, iron metabolism is intrinsically connected to HIV-1 infection and disease progression.
Effect of cellular iron on HIV-1 replication
Iron serves as an important element during various steps in the life cycle of HIV-1 such as reverse-transcription, activation of NF-κB, regulation of HIV-1 transcription, translation of viral mRNA, and viral assembly [3]. During viral entry, HIV-1 reverse transcription is dependent on the activity of host cell ribonucleotide reductase that contains nonheme iron, which is important for enzymatic activity [10]. Transcription of integrated HIV-1 proviral DNA is induced by HIV-1 Tat protein that relieves RNA polymerase II pausing by recruiting positive transcription elongation factor b (P-TEFb), which contains CDK9/cyclin T1 and other transcriptional regulatory factors, to the HIV-1 long terminal repeat [11].
Recent studies from the authors’ laboratory have shown that HIV-1 transcription is inhibited by iron chelators that inhibit the enzymatic activity of CDK2 [12,13], and by the expression of the iron export protein, ferroportin [14]. The HIV-1 promoter contains several binding sites for host transcription factors, including two NF-κB binding sites [15]. NF-κB availability can be increased by iron efflux that activates IκB kinase [16–18]. Cellular iron levels can also be increased by Nef, an HIV-1 accessory protein, that downregulates the expression of HFE, which controls iron uptake by macrophages and is mutated in hereditary hemochromatosis [5]. Deregulation of HFE by Nef increases iron levels, which coincides with increased HIV-1 gag expression, suggesting a beneficial effect of increased iron on the production of HIV-1 virions and viral replication [5].
Export of unspliced HIV-1 mRNA requires HIV-1 Rev protein and host eIF5α. The eIF5α protein contains N-epsilon-4-amino-2-hydroxybutyl-lysine (hypusine) that is produced by deoxyhypusine hydroxylase, an iron-containing enzyme [19]. The topical fungicide, ciclopirox, and the iron chelator, deferiprone, inhibit HIV-1 gene expression interfering with the hydroxylation step in the hypusine modification of eIF5α [20]. Assembly of HIV capsid requires an ATP-binding cassette subfamily E member 1 protein, which contains iron-sulfur clusters [21] and binds to HIV-1 Gag protein [22].
Regulation of HIV-1 transcription
HIV-1 Tat protein recruits CDK9/cyclin T1, a component of P-TEFb, to transactivation response (TAR) element RNA, a hairpin-loop structure located at the 5′-end of all nascent HIV-1 transcripts (Figure 1) [23]. Inefficient transcription activation by Tat or the absence of essential transcription factors may contribute to the establishment of latency and protect HIV-1 provirus from anti-HIV-1 drugs, preventing HIV-1 eradication [1]. Formation of TAR RNA may disrupt the efficient recognition of HIV-1 TATA box by the cellular preinitiation complexes that require CTGC motifs for the accurate formation and also contribute to the establishment of latency [24]. HIV-1 transcription through the nucleosomal structure of integrated HIV-1 provirus requires chromatin-associated Spt6 and PAAF1 that protects Spt6 from proteasomal degradation [25]. Loss of Spt6 or PAAF1 leads to the synthesis of transcripts that are not efficiently translated [25].
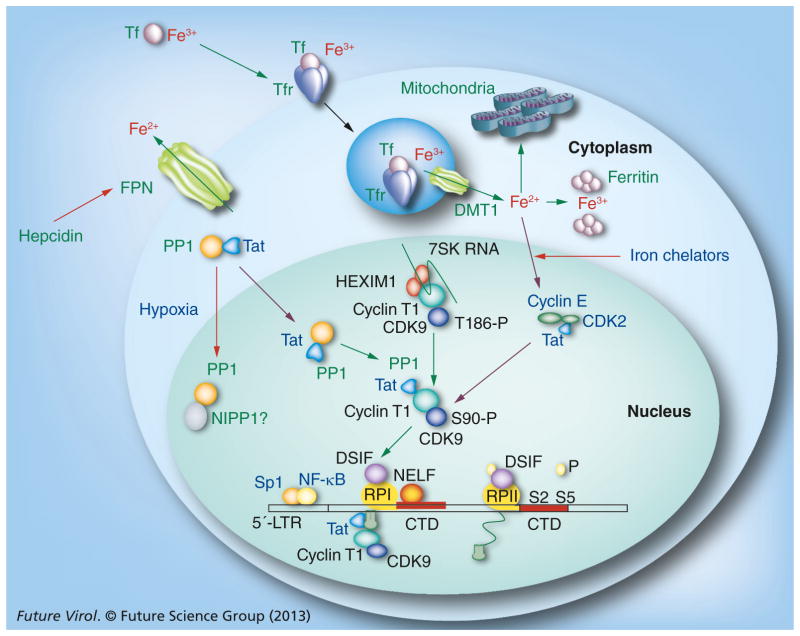
Figure 1. Schematic representation of the effect of iron and hypoxia on Tat-mediated HIV-1 transcription.
Tf-bound iron is uptaken through the Tfr and then exported to cytoplasm by DMT1. Ferritin stores cellular iron. Excess iron is exported by ferroportin, which is negatively regulated by extracellular hepcidin that binds to ferroportin and leads to its internalization and degradation. HIV-1 transcription is activated by Tat protein, which recruits CDK9/cyclin T1 to the HIV-1 5′ LTR promoting the phosphorylation of NELF, DSIF and the CTD of RPII. CDK2/cyclin E activates CDK9 by phosphorylation on Ser-90. Iron chelation inhibits CDK2 and CDK9 activities. Tat also shuttles PP1 to the nucleus, where PP1 dephosphorylates CDK9 and facilitates the dissociation of CDK9/cyclin T1 from the inactive complex containing 7SK RNA and HEXIM1. Hypoxia affects PP1, facilitating its binding to NIPP1. Expression of ferroportin inhibits HIV-1, and inhibition of ferroportin by hepcidin induces HIV-1.
CTD: C-terminal domain; DMT1: Divalent metal transporter 1; DSIF: DRB-sensitivity inducing complex; HEXIM1: Hexamethylene bis-acetamide-inducible protein 1; LTR: Long terminal repeat; NELF: Negative elongation factor; NIPP1: Nuclear inhibitor of PP1; PP1: Protein phosphatase-1; RPII: RNA polymerase; Tf: Transferrin-bound iron; Tfr: Transferrin receptor.
Activation of HIV-1 transcription by Tat requires P-TEFb that exists in two molecular-weight forms. The lower-molecular-weight kinase active form of P-TEFb consists of CDK9 and cyclin T1 [26,27]. The high molecular weight inactive form of P-TEFb contains 7SK RNA, a dimer of CDK9/cyclin T1 and several additional proteins including HEXIM1 dimer, LARP7 protein [28–30] and MePCE [31,32]. HEXIM1 positions its inhibitory PY NT sequence to the active site of CDK9 inhibiting its enzymatic activity [33]. The high molecular weight P-TEFb complex serves as a source of CDK9/cyclin T1 for the recruitment by HIV-1 Tat [34].
HIV-1 Tat protein may also recruit CDK9/cyclin T1 to the HIV-1 promoter in inactive complex with 7SK RNA, and formation of TAR RNA can displace 7SK RNA [35]. Tat has recently been shown to facilitate the formation of a super-elongation complex containing active P-TEFb and additional elongation factors and co-activators [36,37]. P-TEFb triggers elongation of RNA polymerase II transcription by phosphorylating the negative elongation factor and the DRB-sensitivity inducing complex (DSIF/Spt4/Spt5), thus promoting the release of negative elongation factor [38]. P-TEFb can also phosphorylate Ser-5 residues of the C-terminal domain of the largest subunit of RNA polymerase II, especially when C-terminal domain is prephosphorylated on Ser-7 residues [39].
We have shown that HIV-1 transcription is activated by Tat in the G1 phase, but not in the G2 phase [40,41], suggesting that Tat might function in concert with a host cell factor that is expressed in G1. Association of Tat with CDK2/cyclin E- and CDK2-activated HIV-1 transcription in vitro [42,43], and inhibition of HIV-1 transcription and replication in the CDK2 knock-down cells, further supports a close interaction of CDK2 with the HIV-1 promoter both in vitro and in vivo [44]. Inhibition of CDK2 with small-molecule inhibitor roscovitin [45] inhibits HIV-1 replication and prevents the association of CDK2 with the HIV-1 promoter [44]. Roscovitin and its analog CR8 also inhibit CDK9 activity [46], suggesting a cooperative interaction between CDK2 and CDK9 in regulating HIV-1 replication [47,48].
CDK9 contains several phosphorylation sites, including Thr-186, that are critical for its functional activity [49,50] and the association of CDK9/cyclin T1 with 7SK RNA snRNP [49,50]. Dephosphorylation of CDK9 at Thr-186 by protein phosphatase-1 (PP1) in stress-induced cells dissociates 7SK RNA and HEXIM1 and activates CDK9/cyclin T1 [51]. Additional phosphorylation sites on CDK9 include autophosphorylated Thr-29 [52] and the C-terminal residues Ser-329, Thr-330, Thr-333, Ser-334, Ser-347, Thr-350, Ser-353, and Thr-354 [49]. Dephosphorylation of CDK9’s T-loop Ser-175 residue by PP1 induces CDK9 activity and activates HIV-1 transcription [53]. Accordingly, expression of the central domain of nuclear inhibitor of PP1 (cdNIPP1) in cultured cells, or as part of HIV-1 pNL4–3 in place of nef, efficiently inhibits HIV-1 transcription and replication [54]. Stable expression of cdNIPP1 disrupts the interaction of Tat with PP1, increases CDK9 phosphorylation of Thr-186, and induces association of CDK9 with 7SK RNA [54]. Expression of cdNIPP1 also increases CDK9’s Ser-175 phosphorylation and inhibits the enzymatic activity of CDK9 [53].
The authors have recently developed small molecules that target a noncatalytic site of PP1 and identified inhibitors [55] as well as activators [Nekhai S & Kumari N, Unpublished Data] of HIV-1 transcription. The functional link between CDK2 and CDK9 is further supported by the inhibition of CDK2 by iron chelators (discussed in the next section) that also inhibit CDK9 activity and thus HIV-1 transcription [12,13]. It may be possible that CDK2 directly phos-phorylates CDK9 (or Ser-90 of CDK9) and, therefore, may serve as a potentially key CDK2 phosphorylation site [56]. Phosphorylation of CDK9’s Ser-90 may likely be a requirement for the functional CDK9 activity to activate HIV-1 transcription more efficiently because of the increased association with large P-TEFb complex [56]. Thus, recent studies may direct investigation to potentially novel and intriguing regulatory pathways linking CDK2 and CDK9 to cellular iron metabolism in the regulation of HIV-1 transcription.
Inhibition of HIV-1 transcription by iron chelators
In cultured T-cells, excess of iron is associated with an increased HIV-1 viral replication, whereas iron chelation with desferrioxamine correlates with lower viral replication [57]. Iron chelators deferoxamine and deferiprone inhibit HIV-1 replication in human primary peripheral blood lymphocytes and macrophages; however, the inhibition was largely attributed to decreased cellular proliferation [58]. Recently, the topical fungicide, ciclopirox, and the iron chelator, deferiprone, have been shown to inhibit HIV-1 gene expression at the level of transcription initiation by interfering with the hypusine modification of eIF5α [20]. Iron chelators negatively affect the activity or expression of CDK2 [13,59,60] and thus mediate the deregulation of CDK2 and arrest cell cycle progress [61,62]. The iron chelator, desferriexochelin, inhibits the binding of cyclin A and E to CDK2 in human mammary epithelial cells, whereas in human breast cancer cells the binding is increased [59]. On the other hand, iron chelator 311 inhibits the expression of cyclins D1, D2, D3, A and B1 and also CDK2, but not cyclin E [60]. HIV-1 transcription is inhibited in CEM T-cells by iron chelators 311 and ICL670 (deferasirox or Exjade®, Novartis Oncology, NJ, USA) [13]. These iron chelators inhibit the cellular activity of CDK2 and inhibit both basal and Tat-induced HIV-1 transcription [13].
Previously described di-2-pyridylketone thiosemicarbazone- and 2-benzoylpyridine thiosemicarbazone-based tridentate iron chelators markedly inhibit HIV-1 transcription and virus replication at much lower concentrations than 311 or ILC670 [12]. Whilst the Bp4aT and Bp4eT iron chelators inhibit CDK2 activity, as expected, they also inhibit CDK9 activity [12]. CDK2 is positively regulated by the binding of cyclin E (G1/S transition) or cyclin A (S phase transition) and by CDK7-mediated phosphorylation of Thr-160 [63].
Negative regulation of CDK2 includes its association with the p21 (CIP1/WAF1) and p27 (Kip1) inhibitory proteins and Tyr15 phosphorylation by Wee1 kinase [64]. The mechanism of CDK2 inhibition by iron chelators may be related to the expression of the cyclin-dependent kinase inhibitors p21 or p27 [65,66]. Iron depletion removes iron from prolyl hydroxylase and increases HIF-1α and HIF-2α protein levels, mimicking the effect of hypoxia [67]. Induction of p21 expression by 9-aminoacridine inhibits HIV-1 transcription [68,69]. In a recent study, overexpression of p21 was found in a group of elite HIV-1 controllers [70]. Blockade of p21 in CD4+ T cells from elite controllers markedly increased viral reverse transcripts and mRNA production, leading to higher enzymatic activities of CDK9, indicating p21 as a barrier against HIV-1 infection in CD4+ T cells [70]. Thus, based on the earlier observation that CDK2 and CDK9 associate in a complex with HIV-1 Tat [71], and the fact that CDK9 is directly phosphorylated by CDK2 [56], it is likely that p21 inhibits CDK9 indirectly by interacting with CDK2 and inhibiting its activity.
HIV-1 infection & iron metabolism regulation in macrophages
HIV-1 infection of macrophages has been recognized as an important component of viral pathogenesis [72]. An important function of human macrophages is a recycling of iron to the bone marrow from aged red blood cells, which involves iron export by ferroportin [73]. Macrophages recycle approximately 20–25 mg of iron per day, meeting the bioavailability of iron requirement for erythropoiesis [74]. After degradation of red blood cells in phagolysosomes, heme reaches the cytoplasm, where it activates the transcription of several cellular genes, including HO-1 and ferroportin [74]. HO-1 degrades heme to release CO, biliverdin and iron, which is stored in ferritin barrels or exported back to the plasma by ferroportin [74].
Activation of HO-1 and production of CO by HO-1 has recently been shown to protect against cerebral malaria in A/S sickle cell trait individuals [70], and may explain the survival advantage of sickle cell trait against malaria [75,76]. Hemin-induced HO-1 induction also efficiently inhibits HIV-1 infection in vitro and in vivo [77,78]. Hemoglobin-stimulated macrophages undergo a selective differentiation program and demonstrate increased ferroportin expression and reduced iron levels [79].
Ferroportin is the only iron exporter found in mammals [80] and is present in cells and tissues where major iron flows are regulated, such as red blood cells, placenta, hepatocytes and macrophages [81–83]. Ferroportin is also expressed on the basolateral side of duodenal enterocytes and helps to release iron into plasma, thus controlling both systemic iron absorption and iron recycling [84]. Mutations in the ferroportin gene, A77D, V162del and G490D, result in reduced iron export leading to iron accumulation in macrophages [85,86]. Mutations affecting the interaction with hepcidin prevent its proper functioning and cause hereditary iron overloads [84]. Because reduction of cellular iron inhibits HIV-1 transcription, depletion of cellular iron through the expression of ferroportin is thought to inhibit HIV-1 gene expression [14]. Expression of ferroportin is negatively regulated by hepcidin, a hepatocyte peptide hormone, resulting in the hepatic iron overload condition similar to hemochromatosis [87]. Hepcidin is secreted by hepatocytes and macrophages, and interacts with ferroportin, leading to its internalization and degradation by lysosomes [84].
The ferroportin mutants Y64N, N144D, N144H, and C326Y, which have a reduced sensitivity to hepcidin, lead to a higher dietary iron uptake, more iron recycling by macrophages and higher serum transferrin saturation [85]. Hepcidin has recently been shown to induce HIV-1 transcription in 293 T cells expressing WT ferroportin, but not the C326Y mutant [14]. In addition, hepcidin treatment of HIV-1-infected primary macrophages and T cells cultured in the presence of iron significantly induced HIV-1 replication [14]. These findings suggest that the interplay between ferroportin expression and its degradation by hepcidin may play a regulatory role in HIV-1 transcription and that HIV-1 transcription can be transiently activated by hepcidin. However, the role of ferroportin or hepcidin in the progression of HIV-1 disease has remained undefined. Hepcidin expression is facilitated by IL-6 and other proinflammatory cytokines that are elevated during inflammation [88] and constitute a risk factor for the progression of HIV-1 disease and pathogenesis [89].
Effect of hypoxia on HIV-1
The role of physiological factors in the regulation of HIV-1 transcription is not well defined. Our knowledge about the activation of HIV-1 transcription is primarily based on observations from cell culture experiments, which are typically conducted at atmospheric oxygen level (21% O2). However, the physiologic level of O2 is significantly less than that present in these in vitro experimental conditions. For example, in lungs, liver, kidneys and heart, O2 varies between 4 and 14%; in the brain, from 0.5 to 7%; in the eye, from 1 to 5%; and in the bone marrow, from 0 to 4% [90]. Thus, the function of cellular and viral proteins may differ under the lower-than-atmospheric oxygen tension. This is exemplified by the iron-responsive proteins, IRP-1 and IRP-2, where IRP-1 is the major sensor of intracellular labile iron in tissue culture experiments at 21% O2 but IRP-2 is the main iron sensor in vivo and in cells cultured at 3% O2 [91].
Hypoxia can induce some viruses and inhibit the others. Hypoxia (1% O2) induces lytic replication of EBV [92] and Kaposi sarcoma-associated herpesvirus [93], but suppresses replication of oncolytic parvovirus Minute virus of mice [94], adenovirus [95] or Moloney murine leukemia virus [96]. Primary T cells cultured at 3–6% O2 maintain an intracellular redox environment similar to the in vivo situation, as opposed to T cells cultured at atmospheric 21% O2, in which the intracellular redox state is significantly altered [97]. Peripheral blood mononuclear cells (PBMCs) cultured at 5% O2 are efficiently activated by extracellular HIV-1 Tat protein, which mimicks the activation by IL-2 and PMA, and the activated cells support HIV 1 replication [98].
Iron chelators deplete iron from cellular enzymes including prolyl hydroxylase, which decreases prolyl hydroxylase activity and increases the protein level of HIF-1α, thus mimicking the effect of hypoxia [99]. Therefore, it may be argued that HIV-1 transcription and replication could also be reduced in cells cultured in lower levels of oxygen as compared with the atmospheric oxygen concentration [4]. Interestingly, HIF-1α expression is seen only at low 1% O2, but not at physiological 3% O2, and CDK2 activity remains unchanged at 3% O2 [4]. In contrast, CDK9/cyclin T1 activity is significantly reduced in T cells cultured at 3% oxygen [4]. This is due to reduced association of CDK9 with cyclin T1, and not because of the reduced expression of CDK9 or cyclin T1 [4]. Accordingly, the CDK9 inhibitor ARC, which efficiently inhibits HIV-1 transcription [100], has significantly less effect on HIV-1 transcription in cells cultured at 3% O2 [4], suggesting that CDK9 activity may be decreased and less critical for the regulation of HIV-1 transcription in the cells cultured at 3% O2.
CDK9 activity is regulated in part by PP1 [53,54,101]. Previously, PP1 has been shown to be negatively affected by hypoxia either through a decrease of mRNA expression [102] or increased association with its regulatory subunit, nuclear inhibitor of PP1 [103]. Thus, PP1 inhibition may lead to changes in CDK9 phosphorylation and modulation of its activity. Hypoxia also modulates the activity of CDK9 through the expression of p27 protein [104], which may also have a downstream effect on CDK9.
Low prevalence of HIV-1 infection in sickle cell disease
In sickle cell disease (SCD), a hereditary disorder that affects approximately 100,000 people in the USA, primarily of African descent [105], a single V6G mutation in the β-globin gene can lead to the production of hemoglobin S, development of chronic hemolytic anemia, adhesion of sickle erythrocytes to endothelium, immune activation and abnormal cytokine production [106]. Despite frequent blood transfusion, SCD patients have lower risk for HIV-1 infection in comparison with Human T-lymphotropic virus type I (HTLV-1) infection (0% HIV-1 + vs 7.9% HTLV-1+) [107] and higher frequency of HIV-1 long-term nonprogressors compared with the general HIV+ population (44% in SCD patients vs. 13.9% in controls) [108]. Out of 116 SCD patients, 88 patients received an average 18 transfusions between 1978 and 1985, and negative for the HIV antibody. Among the same patient population, nine (7.8%) were tested positive for HTLV-I antibodies. A recent study conducted among five University Centers, including Howard University, revealed a higher frequency of HIV-1 long-term nonprogressors among HIV-1-infected SCD patients (44%) as compared with HIV-1-infected controls (13.9%) [108]. Another recently published report from Republic of Congo indicated that frequency of HIV among 127 SCD patients was lower than among 3390 blood donors [109,110]. Our recent analysis of national hospital discharge surveys (1997–2009) showed that SCD is associated with lower frequency of HIV-1 diagnosis (odds ratio 0.33) as compared with other diagnosis [111].
The level of hepcidin that regulates internalization and degradation of the iron export protein ferroportin is reduced in some SCD patients [112]. Gene expression analysis showed increased expression of HO-1, biliverdin reductase and p21 in PBMCs from SCD patients in steady-state SCD [113]. Our unpublished observations show that HIV-1 replication is reduced in PBMCs obtained from SCD patients, suggesting that chronic hemolysis and local ischemia may contribute to protection against HIV-1 infection in SCD.
Conclusion
In this review, the authors have discussed cellular factors involved in the iron and oxygen homeostasis that affect HIV-1. Understanding the role of these cellular factors in HIV-1 replication may provide new targets for HIV-1 therapy. Also, information on the emerging roles of iron and oxygen-regulatory pathways in antiviral immune response may provide a supplemental strategy for controlling HIV-1 infection in addition to the existing antiretroviral therapy or vaccination trials.
Future perspective
As outlined in this review, iron- and oxygen-related metabolic pathways emerge as important regulatory players in HIV-1 infection. With the failure of anti-HIV-1 vaccines targeted to gp120 and HIV-1 Gag, Pol and Nef proteins, and the low (31%) efficacy of a combined vaccine recently tested among community-risk Thai participants [114], additional strategies are needed to curb HIV-1 epidemics. As proteins regulating cellular iron metabolism, such as ferroportin and hepcidin, emerge as major regulators of innate immune response [115] , therapeutic strategies that provide iron chelation or control iron absorption could be developed as supplemental approaches that may be combined with and augment vaccination. Iron supplementation strategy has to be taken with caution in this situation, especially because of the high rates of HIV-1 coinfection with tuberculosis [116], HBV and HCV [117], and thus the possibility of increased adverse effects dues to the augmentation of coinfections [118,119].
Executive summary.
Increased iron levels associate with severe HIV-1 disease progression
A number of studies showed association between increased iron stores and severity of HIV-1 infection.
Cellular iron affects HIV-1 replication
Cellular iron affects HIV-1 transcription through the control of CDK 2 activity; splicing through Rev and eIF5α; and viral assembly through Nef, HFE, and ABCE1.
Regulation of HIV-1 transcription
HIV-1 Tat engages CDK2/cyclin E and protein phosphatase-1 that phosphorylates and dephosphorylates CDK9 on Ser-90 and Ser-175 respectively and which are subject to a regulation by cellular iron and hypoxia.
HIV-1 infection and iron metabolism regulation in macrophages
Heme processing by macrophages leads to induction of HO-1 and ferroportin, inhibiting HIV-1. The negative regulator of ferroportin expression, hepcidin, induces HIV-1 transcription and replication.
Effect of hypoxia on HIV-1
Hypoxia inhibits HIV-1 by decreasing CDK9/cyclin T1 activity. The effect of hypoxia could be due to deregulation of protein phosphatase-1 and/or increased expression of p27.
Low prevalence of HIV-1 in sickle cell disease
Sickle cell disease associates with lower rates or slower progression of HIV-1 infection. Decreased hepcidin levels and increased levels of HO-1 and p21 may reduce HIV-1 replication.
Footnotes
Disclaimer
The work described in this article has not been formally disseminated by the US FDA and should not be construed to represent any Agency determination or policy.
Financial & competing interests disclosure
Work in S Nekhai’s laboratory is supported by extramural NIH grants 1SC1GM082325, 1P30HL107253- 01, 5UH1 HL03679, AI043894, District of Columbia Developmental Center for AIDS Research (P30AI087714), 8G12MD0 07597, GW Project No. : 31440-16-CCLS90469F and intramural funding from Howard University. Work in S Dhawan’s laboratory is supported by intramural funding from the US FDA. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Lafeuillade A, Stevenson M. The search for a cure for persistent HIV reservoirs. AIDS Rev. 2011;13(2):63–66. [PubMed] [Google Scholar]
- 2.Gordeuk VR, Delanghe JR, Langlois MR, Boelaert JR. Iron status and the outcome of HIV infection: an overview. J Clin Virol. 2001;20(3):111–115. doi: 10.1016/s1386-6532(00)00134-7. [DOI] [PubMed] [Google Scholar]
- 3▪▪.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6(7):541–552. doi: 10.1038/nrmicro1930. Excellent review describing iron-regulatory pathways in HIV-1 replication. [DOI] [PubMed] [Google Scholar]
- 4▪.Charles S, Ammosova T, Cardenas J, et al. Regulation of HIV-1 transcription at 3% versus 21% oxygen concentration. J Cell Physiol. 2009;221(2):469–479. doi: 10.1002/jcp.21882. First evidence that HIV-1 is inhibited at hypoxia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drakesmith H, Chen N, Ledermann H, Screaton G, Townsend A, Xu XN. HIV-1 Nef down-regulates the hemochromatosis protein HFE, manipulating cellular iron homeostasis. Proc Natl Acad Sci USA. 2005;102(31):11017–11022. doi: 10.1073/pnas.0504823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boelaert JR, Weinberg GA, Weinberg ED. Altered iron metabolism in HIV infection: mechanisms, possible consequences, and proposals for management. Infect Agents Dis. 1996;5(1):36–46. [PubMed] [Google Scholar]
- 7.Rawat R, Humphrey J, Ntozini R, Mutasa K, Iliff P, Stoltzfus R. Elevated iron stores are associated with HIV disease severity and mortality among postpartum women in Zimbabwe. Public Health Nutr. 2008;12(9):1321–1329. doi: 10.1017/S136898000800390X. [DOI] [PubMed] [Google Scholar]
- 8.McDermid JM, Jaye A, Schim Van Der Loeff MF, et al. Elevated iron status strongly predicts mortality in West African adults with HIV infection. J Acquir Immune Defic Syndr. 2007;46(4):498–507. doi: 10.1097/qai.0b013e31815b2d4b. [DOI] [PubMed] [Google Scholar]
- 9.McDermid JM, Van Der Loeff MF, Jaye A, et al. Mortality in HIV infection is independently predicted by host iron status and SLC11a1 and hp genotypes, with new evidence of a gene-nutrient interaction. Am J Clin Nutr. 2009;90(1):225–233. doi: 10.3945/ajcn.2009.27709. [DOI] [PubMed] [Google Scholar]
- 10.Tsimberidou AM, Alvarado Y, Giles FJ. Evolving role of ribonucleoside reductase inhibitors in hematologic malignancies. Expert Rev Anticancer Ther. 2002;2(4):437–448. doi: 10.1586/14737140.2.4.437. [DOI] [PubMed] [Google Scholar]
- 11.Nekhai S, Jeang KT. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 2006;1(4):417–426. doi: 10.2217/17460913.1.4.417. [DOI] [PubMed] [Google Scholar]
- 12▪.Debebe Z, Ammosova T, Breuer D, et al. Iron chelators of the di-2-pyridylketone thiosemicarbazone and 2-benzoylpyridine thiosemicarbazone series inhibit HIV-1 transcription: identification of novel cellular targets - iron, cyclin-dependent kinase (CDK) 2, and CDK9. Mol Pharmacol. 2011;79(1):185–196. doi: 10.1124/mol.110.069062. Research article describing inhibition of HIV-1 transcription by iron chelators and poiting to the inhibition of CDK2 and CDK9 as the cause of inhibition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debebe Z, Ammosova T, Jerebtsova M, et al. Iron chelators ICL670 and 311 inhibit HIV-1 transcription. Virology. 2007;367(2):324–333. doi: 10.1016/j.virol.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14▪.Xu M, Kashanchi F, Foster A, et al. Hepcidin induces HIV-1 transcription inhibited by ferroportin. Retrovirology. 2010;7:104. doi: 10.1186/1742-4690-7-104. First evidence that ferroportin inhibits and hepcidin activates HIV-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28(3):663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Xiong S, She H, Lin SW, Wang J, Tsukamoto H. Iron causes interactions of TAK1, p21RAS, and phosphatidylinositol 3-kinase in caveolae to activate IκB kinase in hepatic macrophages. J Biol Chem. 2007;282(8):5582–5588. doi: 10.1074/jbc.M609273200. [DOI] [PubMed] [Google Scholar]
- 17.Xiong S, She H, Takeuchi H, et al. Signaling role of intracellular iron in NF-κB activation. J Biol Chem. 2003;278(20):17646–17654. doi: 10.1074/jbc.M210905200. [DOI] [PubMed] [Google Scholar]
- 18.Xiong S, She H, Tsukamoto H. Signaling role of iron in NF-kappa B activation in hepatic macrophages. Comp Hepatol. 2004;3(Suppl 1):S36. doi: 10.1186/1476-5926-2-S1-S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YS, Kang KR, Wolff EC, Bell JK, Mcphie P, Park MH. Deoxyhypusine hydroxylase is a Fe(II)-dependent, heat-repeat enzyme Identification of amino acid residues critical for Fe(II) binding and catalysis. J Biol Chem. 2006;281(19):13217–13225. doi: 10.1074/jbc.M601081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoque M, Hanauske-Abel HM, Palumbo P, et al. Inhibition of HIV -1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology. 2009;6:90. doi: 10.1186/1742-4690-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barthelme D, Scheele U, Dinkelaker S, et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J Biol Chem. 2007;282(19):14598–14607. doi: 10.1074/jbc.M700825200. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman C, Klein KC, Kiser PK, et al. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415(6867):88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 23.Berkhout B, Silverman RH, Jeang KT. Tat transactivates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59(2):273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 24.Wilhelm E, Doyle MC, Nzaramba I, Magdzinski A, Dumais N, Bell B. CTGC motifs within the HIV core promoter specify Tat-responsive pre-initiation complexes. Retrovirology. 2012;9:62. doi: 10.1186/1742-4690-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura M, Basavarajaiah P, Rousset E, et al. Spt6 levels are modulated by PAAF1 and proteasome to regulate the HIV-1 LTR. Retrovirology. 2012;9:13. doi: 10.1186/1742-4690-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 28.He N, Jahchan NS, Hong E, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29(5):588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krueger BJ, Jeronimo C, Roy BB, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HAXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36(7):2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markert A, Grimm M, Martinez J, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9(6):569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci USA. 2009;106(19):7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeronimo C, Forget D, Bouchard A, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27(2):262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michels AA, Fraldi A, Li Q, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (Cdk9/cyclin T) inhibitor. EMBO J. 2004;23(13):2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HAXIM1 to TAR. Nucleic Acids Res. 2007;35(13):4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’orso I, Frankel AD. HIV-1 Tat: its dependence on host factors is crystal clear. Viruses. 2010;2(10):2226–2234. doi: 10.3390/v2102226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobhian B, Laguette N, Yatim A, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38(3):439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He N, Liu M, Hsu J, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38(3):428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 40.Kashanchi F, Agbottah ET, Pise-Masison CA, et al. Cell cycle-regulated transcription by the human immunodeficiency virus type 1 Tat transactivator. J Virol. 2000;74(2):652–660. doi: 10.1128/jvi.74.2.652-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nekhai S, Shukla RR, Fernandez A, Kumar A, Lamb NJ. Cell cycle-dependent stimulation of the HIV -1 promoter by Tat-associated CAK activator. Virology. 2000;266(2):246–256. doi: 10.1006/viro.1999.0035. [DOI] [PubMed] [Google Scholar]
- 42.Deng L, Ammosova T, Pumfery A, Kashanchi F, Nekhai S. HIV-1 Tat interaction with RNA polymerase II C-terminal domain (CTD) and a dynamic association with CDK2 induce CTD phosphorylation and transcription from HIV-1 promoter. J Biol Chem. 2002;277(37):33922–33929. doi: 10.1074/jbc.M111349200. [DOI] [PubMed] [Google Scholar]
- 43.Nekhai S, Zhou M, Fernandez A, et al. HIV-1 Tat-associated RNA polymerase C-terminal domain kinase, CDK2, phosphorylates CDK7 and stimulates Tat-mediated transcription. Biochem J. 2002;364(Pt 3):649–657. doi: 10.1042/BJ20011191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agbottah E, De La Fuente C, Nekhai S, et al. Antiviral activity of cyc202 in HIV -1-infected cells. J Biol Chem. 2005;280(4):3029–3042. doi: 10.1074/jbc.M406435200. [DOI] [PubMed] [Google Scholar]
- 45.Pumfery A, De La Fuente C, Berro R, Nekhai S, Kashanchi F, Chao SH. Potential use of pharmacological cyclin-dependent kinase inhibitors as anti- HIV therapeutics. Curr Pharm Des. 2006;12(16):1949–1961. doi: 10.2174/138161206777442083. [DOI] [PubMed] [Google Scholar]
- 46.Biglione S, Byers SA, Price JP, et al. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, Seliciclib and Flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galons H, Oumata N, Meijer L. Cyclin-dependent kinase inhibitors: a survey of recent patent literature. Expert Opin Ther Pat. 2010;20(3):377–404. doi: 10.1517/13543770903524284. [DOI] [PubMed] [Google Scholar]
- 48.Carpio L, Klase Z, Coley W, et al. MicroRNA machinery is an integral component of drug-induced transcription inhibition in HIV-1 infection. J RNAi Gene Silencing. 2010;6(1):386–400. [PMC free article] [PubMed] [Google Scholar]
- 49.Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279(6):4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing CDK9 phosphorylated at threonine 186. J Biol Chem. 2005;280(31):28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 51.Chen R, Liu M, Li H, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22(10):1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou M, Lu H, Park H, Wilson-Chiru J, Linton R, Brady JN. Tax interacts with P-TEFb in a novel manner to stimulate human t-lymphotropic virus type 1 transcription. J Virol. 2006;80(10):4781–4791. doi: 10.1128/JVI.80.10.4781-4791.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ammosova T, Obukhov Y, Kotelkin A, et al. Protein phosphatase-1 activates CDK9 by dephosphorylating ser175. PLOS One. 2011;6(4):e18985. doi: 10.1371/journal.pone.0018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ammosova T, Yedavalli VR, Niu X, et al. Expression of a protein phosphatase 1 inhibitor, cdNIPP1, increases CDK9 threonine 186 phosphorylation and inhibits HIV-1 transcription. J Biol Chem. 2011;286(5):3798–3804. doi: 10.1074/jbc.M110.196493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ammosova T, Platonov M, Yedavalli VR, et al. Small molecules targeted to a non-catalytic ‘RVxF’ binding site of protein phosphatase-1 inhibit HIV-1. PLoS One. 2012;7(6):e39481. doi: 10.1371/journal.pone.0039481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breuer D, Kotelkin A, Ammosova T, et al. CDK2 regulates HIV-1 transcription by phosphorylation of CDK9 on serine 90. Retrovirology. 2012;9(1):94. doi: 10.1186/1742-4690-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traore HN, Meyer D. The effect of iron overload on in vitro HIV-1 infection. J Clin Virol. 2004;31(Suppl 1):S92–S98. doi: 10.1016/j.jcv.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 58.Georgiou NA, Van Der Bruggen T, Oudshoorn M, Nottet HS, Marx JJ, Van Asbeck BS. Inhibition of human immunodeficiency virus type 1 replication in human mononuclear blood cells by the iron chelators deferoxamine, deferiprone, and bleomycin. J Infect Dis. 2000;181(2):484–490. doi: 10.1086/315223. [DOI] [PubMed] [Google Scholar]
- 59.Pahl PM, Reese SM, Horwitz LD. A lipid-soluble iron chelator alters cell cycle regulatory protein binding in breast cancer cells compared to normal breast cells. J Exp Ther Oncol. 2007;6(3):193–200. [PubMed] [Google Scholar]
- 60.Gao J, Richardson DR. The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents, IV: the mechanisms involved in inhibiting cell-cycle progression. Blood. 2001;98(3):842–850. doi: 10.1182/blood.v98.3.842. [DOI] [PubMed] [Google Scholar]
- 61.Lucas JJ, Szepesi A, Domenico J, et al. Effects of iron-depletion on cell cycle progression in normal human t lymphocytes: selective inhibition of the appearance of the cyclin A-associated component of the p33CDK2 kinase. Blood. 1995;86(6):2268–2280. [PubMed] [Google Scholar]
- 62.Pahl PM, Yan XD, Hodges YK, Rosenthal EA, Horwitz MA, Horwitz LD. An exochelin of mycobacterium tuberculosis reversibly arrests growth of human vascular smooth muscle cells in vitro. J Biol Chem. 2000;275(23):17821–17826. doi: 10.1074/jbc.M909918199. [DOI] [PubMed] [Google Scholar]
- 63.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 64.Coulonval K, Bockstaele L, Paternot S, Roger PP. Phosphorylations of cyclin-dependent kinase 2 revisited using two-dimensional gel electrophoresis. J Biol Chem. 2003;278(52):52052–52060. doi: 10.1074/jbc.M307012200. [DOI] [PubMed] [Google Scholar]
- 65.Richardson DR. Molecular mechanisms of iron uptake by cells and the use of iron chelators for the treatment of cancer. Curr Med Chem. 2005;12(23):2711–2729. doi: 10.2174/092986705774462996. [DOI] [PubMed] [Google Scholar]
- 66.Wang G, Miskimins R, Miskimins WK. Regulation of p27(kip1) by intracellular iron levels. Biometals. 2004;17(1):15–24. doi: 10.1023/a:1024417309370. [DOI] [PubMed] [Google Scholar]
- 67.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;(407):cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 68.Guendel I, Carpio L, Easley R, et al. 9-aminoacridine inhibition of HIV-1 Tat dependent transcription. Virol J. 2009;6:114. doi: 10.1186/1743-422X-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu W, Kehn-Hall K, Pedati C, et al. Drug 9aa reactivates p21/WAF1 and inhibits HIV -1 progeny formation. Virol J. 2008;5:41. doi: 10.1186/1743-422X-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferreira A, Marguti I, Bechmann I, et al. Sickle hemoglobin confers tolerance to plasmodium infection. Cell. 2011;145(3):398–409. doi: 10.1016/j.cell.2011.03.049. [DOI] [PubMed] [Google Scholar]
- 71.Ammosova T, Berro R, Jerebtsova M, et al. Phosphorylation of HIV-1 Tat by CDK2 in HIV -1 transcription. Retrovirology. 2006;3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol. 2008;62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 73.Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci USA. 2005;102(5):1324–1328. doi: 10.1073/pnas.0409409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74▪▪.Beaumont C. Multiple regulatory mechanisms act in concert to control ferroportin expression and heme iron recycling by macrophages. Haematologica. 2010;95(8):1233–1236. doi: 10.3324/haematol.2010.025585. Excellent review discussing new findings in ferroportin transcription regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams TN, Mwangi TW, Wambua S, et al. Sickle cell trait and the risk of plasmodium falciparum malaria and other childhood diseases. J Infect Dis. 2005;192(1):178–186. doi: 10.1086/430744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aidoo M, Terlouw DJ, Kolczak MS, et al. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359(9314):1311–1312. doi: 10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- 77▪.Devadas K, Dhawan S. Hemin activation ameliorates HIV-1 infection via heme oxygenase-1 induction. J Immunol. 2006;176(7):4252–4257. doi: 10.4049/jimmunol.176.7.4252. First evidence that HO-1 expression inhibits HIV-1. [DOI] [PubMed] [Google Scholar]
- 78.Devadas K, Hewlett IK, Dhawan S. Lipopolysaccharide suppresses HIV-1 replication in human monocytes by protein kinase c-dependent heme oxygenase-1 induction. J Leukoc Biol. 2010;87(5):915–924. doi: 10.1189/jlb.0307172. [DOI] [PubMed] [Google Scholar]
- 79.Finn AV, Nakano M, Polavarapu R, et al. Hemoglobin directs macrophage differentiation and prevents foam cell formation in human atherosclerotic plaques. J Am Coll Cardiol. 2012;59(2):166–177. doi: 10.1016/j.jacc.2011.10.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donovan A, Lima CA, Pinkus JL, et al. The iron exporter ferroportin/slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1(3):191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 81.Donovan A, Brownlie A, Zhou Y, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403(6771):776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 82.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275(26):19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- 83.Mckie AT, Marciani P, Rolfs A, et al. A novel duodenal iron-regulated transporter, ireg1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5(2):299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- 84.Ganz T. Molecular control of iron transport. J Am Soc Nephrol. 2007;18(2):394–400. doi: 10.1681/ASN.2006070802. [DOI] [PubMed] [Google Scholar]
- 85.Drakesmith H, Schimanski LM, Ormerod E, et al. Resistance to hepcidin is conferred by hemochromatosis-associated mutations of ferroportin. Blood. 2005;106(3):1092–1097. doi: 10.1182/blood-2005-02-0561. [DOI] [PubMed] [Google Scholar]
- 86.Schimanski LM, Drakesmith H, Merryweather-Clarke AT, et al. In vitro functional analysis of human ferroportin (Fpn) and hemochromatosis-associated Fpn mutations. Blood. 2005;105(10):4096–4102. doi: 10.1182/blood-2004-11-4502. [DOI] [PubMed] [Google Scholar]
- 87.Nicolas G, Bennoun M, Devaux I, et al. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci USA. 2001;98(15):8780–8785. doi: 10.1073/pnas.151179498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ganz T, Nemeth E. Iron sequestration and anemia of inflammation. Semin Hematol. 2009;46(4):387–393. doi: 10.1053/j.seminhematol.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Appay V, Sauce D. Immune activation and inflammation in HIV-1 infection: causes and consequences. J Pathol. 2008;214(2):231–241. doi: 10.1002/path.2276. [DOI] [PubMed] [Google Scholar]
- 90.Ivanovic Z. Hypoxia or in situ normoxia: the stem cell paradigm. J Cell Physiol. 2009;219(2):271–275. doi: 10.1002/jcp.21690. [DOI] [PubMed] [Google Scholar]
- 91.Meyron-Holtz EG, Ghosh MC, Rouault TA. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306(5704):2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 92.Jiang JH, Wang N, Li A, et al. Hypoxia can contribute to the induction of the Epstein-Barr virus (EBV) lytic cycle. J Clin Virol. 2006;37(2):98–103. doi: 10.1016/j.jcv.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 93.Davis DA, Rinderknecht AS, Zoeteweij JP, et al. Hypoxia induces lytic replication of kaposi sarcoma-associated herpesvirus. Blood. 2001;97(10):3244–3250. doi: 10.1182/blood.v97.10.3244. [DOI] [PubMed] [Google Scholar]
- 94.Servais C, Caillet-Fauquet P, Draps ML, Velu T, De Launoit Y, Brandenburger A. Hypoxic-response elements in the oncolytic parvovirus minute virus of mice do not allow for increased vector production at low oxygen concentration. J Gen Virol. 2006;87(Pt 5):1197–1201. doi: 10.1099/vir.0.81754-0. [DOI] [PubMed] [Google Scholar]
- 95.Shen BH, Bauzon M, Hermiston TW. The effect of hypoxia on the uptake, replication and lytic potential of group b adenovirus type 3 (Ad3) and type 11p (Ad11p) Gene Ther. 2006;13(12):986–990. doi: 10.1038/sj.gt.3302736. [DOI] [PubMed] [Google Scholar]
- 96.Puppo M, Bosco MC, Federico M, Pastorino S, Varesio L. Hypoxia inhibits moloney murine leukemia virus expression in activated macrophages. J Leukoc Biol. 2007;81(2):528–538. doi: 10.1189/jlb.0506361. [DOI] [PubMed] [Google Scholar]
- 97.Atkuri KR, Herzenberg LA, Niemi AK, Cowan T. Importance of culturing primary lymphocytes at physiological oxygen levels. Proc Natl Acad Sci USA. 2007;104(11):4547–4552. doi: 10.1073/pnas.0611732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sahaf B, Atkuri K, Heydari K, et al. Culturing of human peripheral blood cells reveals unsuspected lymphocyte responses relevant to HIV disease. Proc Natl Acad Sci USA. 2008;105(13):5111–5116. doi: 10.1073/pnas.0712363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sargent PJ, Farnaud S, Evans RW. Structure/function overview of proteins involved in iron storage and transport. Curr Med Chem. 2005;12(23):2683–2693. doi: 10.2174/092986705774462969. [DOI] [PubMed] [Google Scholar]
- 100.Nekhai S, Bhat UG, Ammosova T, et al. A novel anticancer agent ARCc antagonizes HIV-1 and HCV. Oncogene. 2007;26(26):3899–3903. doi: 10.1038/sj.onc.1210158. [DOI] [PubMed] [Google Scholar]
- 101.Nekhai S, Jerebtsova M, Jackson A, Southerland W. Regulation of HIV-1 transcription by protein phosphatase 1. Curr HIV Res. 2007;5(1):3–9. doi: 10.2174/157016207779316279. [DOI] [PubMed] [Google Scholar]
- 102.Taylor CT, Furuta GT, Synnestvedt K, Colgan SP. Phosphorylation-dependent targeting of camp response element binding protein to the ubiquitin/proteasome pathway in hypoxia. Proc Natl Acad Sci USA. 2000;97(22):12091–12096. doi: 10.1073/pnas.220211797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Comerford KM, Leonard MO, Cummins EP, et al. Regulation of protein phosphatase 1gamma activity in hypoxia through increased interaction with NIPP1: implications for cellular metabolism. J Cell Physiol. 2006;209(1):211–218. doi: 10.1002/jcp.20726. [DOI] [PubMed] [Google Scholar]
- 104.Horree N, Gort EH, Van Der Groep P, Heintz AP, Vooijs M, Van Diest PJ. Hypoxia-inducible factor 1 alpha is essential for hypoxic p27 induction in endometrioid endometrial carcinoma. J Pathol. 2008;214(1):38–45. doi: 10.1002/path.2244. [DOI] [PubMed] [Google Scholar]
- 105.Brousseau DC, Panepinto JA, Nimmer M, Hoffmann RG. The number of people with sickle-cell disease in the United States: national and state estimates. Am J Hematol. 85(1):77–78. doi: 10.1002/ajh.21570. [DOI] [PubMed] [Google Scholar]
- 106.Niu X, Nouraie M, Campbell A, et al. Angiogenic and inflammatory markers of cardiopulmonary changes in children and adolescents with sickle cell disease. PLoS One. 2009;4(11):e7956. doi: 10.1371/journal.pone.0007956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Castro O, Saxinger C, Barnes S, Alexander S, Flagg R, Frederick W. Prevalence of antibodies to human immunodeficiency virus and to human T cell leukemia virus type I in transfused sickle cell disease patients. J Infect Dis. 1990;162(3):743–745. doi: 10.1093/infdis/162.3.743. [DOI] [PubMed] [Google Scholar]
- 108.Bagasra O, Steiner RM, Ballas SK, et al. Viral burden and disease progression in HIV-1-infected patients with sickle cell anemia. Am J Hematol. 1998;59(3):199–207. doi: 10.1002/(sici)1096-8652(199811)59:3<199::aid-ajh4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 109.Batina Agasa S, Dupont E, Kayembe T, et al. Multiple transfusions for sickle cell disease in the democratic republic of congo: the importance of the hepatitis C virus. Transfus Clin Biol. 17(4):254–259. doi: 10.1016/j.tracli.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 110.Batina A, Kabemba S, Malengela R. Iinfectious markers among blood donors in Democratic Republic of Congo (DRC) Rev Med Brux. 2007;28(3):145–149. [PubMed] [Google Scholar]
- 111▪.Nouraie M, Nekhai S, Gordeuk VR. Sickle cell disease is associated with decreased HIV but higher HBV and HCVv comorbidities in US hospital discharge records: a cross-sectional study. Sex Transm Infect. 2012;88(7):528–533. doi: 10.1136/sextrans-2011-050459. Comprehensive analysis of discharge records showing lower odds for association of HIV-1 and sickle cell disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kroot JJ, Laarakkers CM, Kemna EH, Biemond BJ, Swinkels DW. Regulation of serum hepcidin levels in sickle cell disease. Haematologica. 2009;94(6):885–887. doi: 10.3324/haematol.2008.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jison ML, Munson PJ, Barb JJ, et al. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood. 2004;104(1):270–280. doi: 10.1182/blood-2003-08-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O’connell RJ, Kim JH, Corey L, Michael NL. Human immunodeficiency virus vaccine trials. Cold Spring Harb Perspect Med. 2012;2(12):a007351. doi: 10.1101/cshperspect.a007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115▪▪.Drakesmith H, Prentice AM. Hepcidin and the iron-infection axis. Science. 2012;338(6108):768–772. doi: 10.1126/science.1224577. Excellent review discussing recent findings in iron regulatory proteins and their effect on parasitic infection. [DOI] [PubMed] [Google Scholar]
- 116.Harries AD, Hargreaves NJ, Kemp J, et al. Deaths from tuberculosis in sub-Saharan African countries with a high prevalence of HIV-1. Lancet. 2001;357(9267):1519–1523. doi: 10.1016/S0140-6736(00)04639-0. [DOI] [PubMed] [Google Scholar]
- 117.Parikh N, Nonnemacher MR, Pirrone V, Block T, Mehta A, Wigdahl B. Substance abuse, HIV-1 and hepatitis. Curr HIV Res. 2012;10(7):557–571. doi: 10.2174/157016212803306023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murray MJ, Murray AB, Murray MB, Murray CJ. The adverse effect of iron repletion on the course of certain infections. BMJ. 1978;2(6145):1113–1115. doi: 10.1136/bmj.2.6145.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McDermid JM, Prentice AM. Iron and infection: effects of host iron status and the iron-regulatory genes Haptoglobin and NR AMP1 (SLC11a1) on host-pathogen interactions in tuberculosis and HIV. Clin Sci (Lond) 2006;110(5):503–524. doi: 10.1042/CS20050273. [DOI] [PubMed] [Google Scholar]