Abstract
By constantly selecting for novel genotypes, coevolution between hosts and parasites can favour elevated mutation rates. Models of this process typically assume random encounters. However, offspring are often more likely to encounter their mother's parasites. Because parents and offspring are genetically similar, they may be susceptible to the same parasite strains and thus, in hosts, maternal transmission should select for mechanisms that decrease intergenerational genetic similarity. In parasites, however, maternal transmission should select for genetic similarity. We develop and analyse a model of host and parasite mutation rate evolution when parasites are maternally inherited. In hosts, we find that maternal transmission has two opposing effects. First, it eliminates coevolutionary cycles that previous work shows select for higher mutation. Second, it independently selects for higher mutation rates, because offspring that differ from their mothers are more likely to avoid infection. In parasites, however, the two effects of maternal transmission act in the same direction. As for hosts, maternal transmission eliminates coevolutionary cycles, thereby reducing selection for increased mutation. Unlike for hosts, however, maternal transmission additionally selects against higher mutation by favouring parasite offspring that are the same as their mothers.
Keywords: mutation rate, parasites, coevolution, modifier model, maternal effects
1. Introduction
Antagonistic coevolution between hosts and parasites has been shown to select for higher mutation rates in either or both species [1–3]. By constantly evolving new mechanisms to invade or resist one another, hosts and parasites impose fluctuating selection pressures on one another. A host individual with a novel immunity allele will evade the most common infective parasites in the population and, consequently, experience higher fitness. Similarly, a parasite with a novel allele that facilitates infection will gain an advantage. In a microcosm experiment, Pal et al. [4] showed that populations of the bacterium Pseudomonas fluorescens, evolving in the presence of a phage, evolved elevated mutation rates compared with control populations. Furthermore, this elevated mutation rate was shown to be genetically based.
The genetic basis of mutation rate [4–9] is the foundation for much theory [10–13]. These models typically consider a modifier allele that changes the mutation rate at other fitness-affecting loci (e.g. immunity loci) and then identify the conditions under which such a modifier spreads. In this way, theoretical models may help explain the variation in mutation rates across taxa [14].
In many species, offspring are more likely to encounter their mothers' parasites. For example, many plants transmit diseases via their seeds [15,16]; several human pathogens, such as HIV [17] and the malaria-causing Plasmodium species [18], are transmitted from mother to foetus; in insects, many sexually transmitted diseases can be vertically transmitted [19]. Moreover, in species with maternal care, offspring are more exposed to their mothers' parasites than parasites from other individuals. Because parents and offspring are genetically similar, they may be susceptible to the same parasite strains. Consequently, maternal transmission of parasites should select for mechanisms that enable host offspring to differ from their parents. Indeed, theoretical work has shown that increased allocation to sexual reproduction is advantageous in hosts when parasites are maternally transmitted [20]. Here, we ask whether maternal transmission also selects for higher mutation rates at host immunity loci. Conversely, we ask whether maternal transmission selects for lower mutation rates in parasite infectivity loci, which we expect because maternally transmitted parasites that mutate would less effectively target host offspring.
We find that higher maternal transmission of parasites can select for higher mutation rates in hosts, as expected. However, we also find that maternal transmission can eliminate coevolutionary fluctuations in allele frequency that previous work has found are essential for the evolution of elevated mutation rates [3]. Counterintuitively, maternal infection can thus also hamper the evolution of increased host mutation rates. Which of these two conflicting effects predominates depends on the strength of maternal transmission and the rate of recombination between the selected locus and the modifier. In parasites, we find that maternal transmission weakens selection for higher mutation rates by diminishing coevolutionary cycling, and selects against higher mutation rate because this favours parasites that produce non-mutated offspring.
2. Simulation model
To study mutation rate evolution with maternal transmission of parasites, we construct and analyse a stochastic simulation model. In the electronic supplementary material, table S1, we provide a list of model parameters. We consider a population of N sexually reproducing haploid hosts, some of whom are infected by parasites (also haploid). Hosts and parasites are each characterized by an antigen A-locus, with alleles a and A, and a ‘modifier’ M-locus (described below). The antigen locus affects fitness via parasite infectivity. As there are no free-living parasites in our model, parasite population size is determined by the number of infected hosts, with each infected host carrying only one parasite type. Because parasite reproduction takes place within the host and because hosts carry parasites with only one genotype at the M-locus, sexual and asexual parasite reproduction are equivalent. Although parasite extinction is possible in our model, it was not observed in any runs reported here, except occasionally when examining biologically implausible scenarios with mutation rates equal to or very near 1. We assume a matching-alleles model of host immunity; parasites with an A-allele (a-allele) can only infect hosts with an A-allele (a-allele). Because our model includes only one antigen locus with two alleles, the matching-alleles model is equivalent to the inverse-matching-alleles model, in which parasites with an A-allele (a-allele) can only infect hosts with an a-allele (A-allele). Matching-alleles models, which are thought to characterize components of the vertebrate major histocompatibility complex (MHC) system [21], require that hosts have a self versus non-self recognition system and only parasites bearing an antigen that mimics the host cells can evade the immune system.
Each of the discrete, non-overlapping host generations comprises the following steps: reproduction, infection and selection. Each of these steps is deterministic, and collectively, they determine the probabilities with which N host offspring are randomly drawn to found the next generation. We next describe each step in more detail.
First, host reproduction occurs. Mating is random and inheritance is Mendelian. Within a transient diploid phase, recombination occurs between the antigen and modifier loci at rate r, producing haploid offspring. In hosts, mutation occurs at the antigen locus during gamete production with mutation probability depending on an individual's genotype at the modifier locus (hosts with genotype M (m) mutate at rate 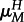 (
(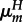 )). Mutation in parasites occurs in infected hosts at a rate governed by the parasite's genotype at the modifier locus (parasites with genotype M (m) mutate at rate
)). Mutation in parasites occurs in infected hosts at a rate governed by the parasite's genotype at the modifier locus (parasites with genotype M (m) mutate at rate 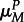 (
(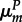 )) so that each host can transmit both mutant and non-mutant parasites. For simplicity, we investigate evolution at the M-locus in hosts and parasites separately.
)) so that each host can transmit both mutant and non-mutant parasites. For simplicity, we investigate evolution at the M-locus in hosts and parasites separately.
Newborn hosts are uninfected, but can become infected in one of two rounds of parasite exposure. First, offspring with infected mothers are exposed to their mothers' parasite pools, which comprise mutant and non-mutant parasites in a ratio of 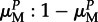 (respectively,
(respectively, 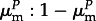 ) for parasites of genotype M (respectively, m). Second, all remaining uninfected offspring are exposed to parasites from the total parent population (global infection). During both infection stages, hosts can only be infected by compatible parasites (i.e. ones with genotypes that match). This occurs with probability ϕf in the maternal infection phase and θf in the global infection phase (0 < ϕ, θ < 1), where f denotes the frequency of that parasite type in the relevant pool (i.e. in the mother during the maternal phase, and in the whole population during the global phase), and ϕ (respectively, θ) denotes the probability of successful infection by a compatible parasite in the maternal (respectively, global) infection stage. The relative values of ϕ and θ thus reflect the relative strengths of maternal and global infection.
) for parasites of genotype M (respectively, m). Second, all remaining uninfected offspring are exposed to parasites from the total parent population (global infection). During both infection stages, hosts can only be infected by compatible parasites (i.e. ones with genotypes that match). This occurs with probability ϕf in the maternal infection phase and θf in the global infection phase (0 < ϕ, θ < 1), where f denotes the frequency of that parasite type in the relevant pool (i.e. in the mother during the maternal phase, and in the whole population during the global phase), and ϕ (respectively, θ) denotes the probability of successful infection by a compatible parasite in the maternal (respectively, global) infection stage. The relative values of ϕ and θ thus reflect the relative strengths of maternal and global infection.
Finally, hosts survive with probabilities that depend on infection status. Uninfected and infected hosts survive with probability 1 and 1 – v, respectively (note that we call v the ‘virulence’ of the parasite). Parasite population dynamics are thus affected by the deaths of their hosts.
3. Simulation results
(a). Coevolutionary dynamics
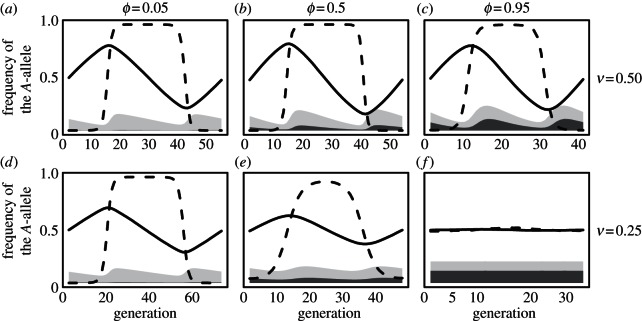
Over a range of parameters, allele frequencies cycle at the antigen locus (figure 1). Such dynamics are typical of host–parasite models when selection is frequency-dependent and there is no maternal transmission [1,3,22,23]. We find that the amplitude and period of these cycles decrease as more parasites are transmitted maternally (figure 2). By enabling parasites with rare but beneficial alleles to gain a foothold, maternal infection allows the parasite population to respond more quickly to allele frequency changes in the hosts and thus to outpace host evolution. When parasite evolution significantly outpaces host evolution, hosts essentially ‘give up’ the arms race and converge to an intermediate allele frequency [3]. Interestingly, maternal transmission's effect on cycle dynamics is most dramatic when parasites are weakly virulent (compare figure 1(a–c) to (d–f)). Because more infected mothers survive to produce and infect offspring when parasites are weakly virulent, maternal infection plays a comparatively larger role than when parasites are strongly virulent and most infected mothers die before reproducing (compare the sizes of the dark grey shaded regions between figure 1(a–c) and (d–f)).
Figure 1.
(a-f) Dependence of host–parasite coevolutionary dynamics on rates of maternal infection, ϕ, and virulence, v. Curves indicate the frequency of the A-allele in hosts (solid lines) and parasites (dashed lines). The shaded regions indicate the fraction of hosts that become infected during each generation in the maternal stage (dark grey) and the global stage (light grey). The sum of these two regions indicates the total fraction of hosts that are infected each generation. The population size of hosts is 106, mutation rate is fixed at 10–5 in both species and θ = 0.2.
Figure 2.
(a) Amplitude and (b) period of coevolutionary cycles at the A-locus in hosts when mutation rate is fixed at 10–5 in both species. Amplitude is calculated by measuring the difference between the maximum and minimum allele frequencies over 1000 generations of a model run, after an initial burn-in period. Cells are shaded white in regions where amplitude is less than 0.1 and cycles are largely absent. Letters indicate parameter combinations used to generate the six panels in figure 1. All other parameters are as in figure 1.
(b). Mutation rate evolution in hosts
Here, we investigate evolution at the modifier locus (M-locus) in hosts. We take two approaches. First, we let evolution proceed for a fixed amount of time, starting from mutation rates of zero, so that we may investigate relative rates of evolution across varying conditions and, second, we find evolutionarily stable (ESS) mutations rates by evaluating relative fitnesses of successive modifier alleles in order to make inferences about longer-term outcomes. Interestingly, we find that evolution becomes extremely slow, even when mutation rates are still far from the ESS. We therefore focus on understanding rates of evolution first and then provide a short investigation of ESS mutation rates.
By letting host mutation rates evolve (while holding parasite mutation rates fixed), we investigate how the rate of modifier evolution varies with strength of maternal transmission. In accordance with previous results [3], we find that when maternal transmission is weak, appreciable increases in mutation rate only occur when coevolutionary cycles characterize dynamics at the A-locus (compare left halves of figures 2a and 3a). By contrast, with high maternal transmission, large mutation rates evolve more readily (compare right halves of figures 2a and 3a). Here, the advantage to differing from one's mother creates a direct benefit to higher mutation. Thus mutation rates evolve to similar levels, regardless of cycle dynamics. In addition, high virulence favours high mutation rates, regardless of maternal infection (figure 3).
Figure 3.
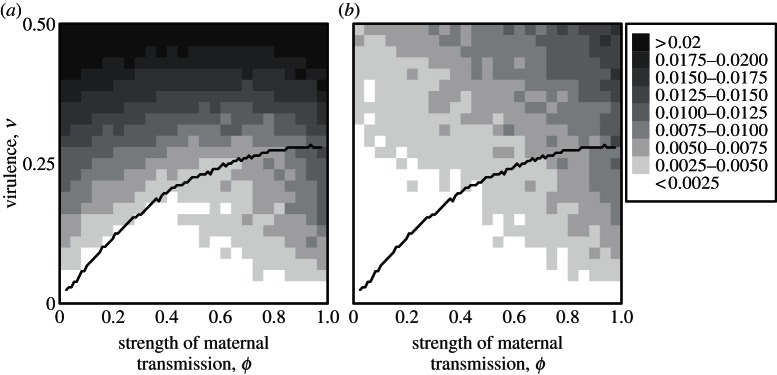
Evolved mutation rate in hosts after 107 generations with (a) complete linkage (r = 0) and (b) free recombination (r = 0.5) between the modifier and antigen loci. The value in each cell is calculated as the mean mutation rate that evolves over 10 replicate runs, with darker shading indicating higher mutation rates. Mutation rates in hosts were initialized to zero and a novel modifier allele was introduced into the population whenever fixation occurred at this locus. The mutation rate associated with this novel allele was drawn from a normal distribution with mean equal to the current population-wide mutation rate and standard deviation 0.001. The solid curve indicates the threshold, below which, cycle amplitude in hosts was negligible for the duration of the evolution runs (this corresponds to the white region in figure 2). All other parameters are as in figure 1.
Also in accordance with previous work, we find that mutation rate evolution is much slower when there is recombination between the mutation modifier and the antigen locus, as long as maternal transmission is weak (see left side of figure 3b). This effect of recombination on mutation rate modifiers has been discussed in detail by others [3,24]. Briefly, with high recombination, high-mutator lineages ‘share’ the benefits of advantageous mutations they create with low-mutator lineages, while bearing most of the costs of the deleterious mutations they produce.
When maternal transmission is high, however, rates of modifier evolution depend much less sensitively on recombination (compare right sides of panels in figure 3 and see the electronic supplementary material, figure S1). When host–parasite cycles are absent, mutation rates evolve to only a slightly higher level with complete linkage. By contrast, when host–parasite cycles are present, the evolved mutation rate is dramatically higher with complete linkage (see the electronic supplementary material, figures S1 and S2). Intuitively, a new mutation can be beneficial for two non-exclusive reasons: (i) because it can render an offspring genotypically different from its mother and, thus, immune to her parasites (the maternal infection effect), and (ii) because it can confer resistance to the majority of the current parasites (the coevolutionary dynamics effect). A mutation rate-increasing modifier allele only experiences indirect positive selection if it remains linked to the beneficial mutant it produces. In both mechanisms described above, recombination hampers indirect positive selection on the modifier. However, in mechanism (i), the benefit of the mutation only exists for one generation and thus recombination has only one chance to disassociate the modifier from the beneficial mutant it creates. This contrasts with mechanism (ii), where the benefits of novel mutants persist for many generations. Consequently, the negative effect of recombination is much more pronounced when maternal transmission is weak, and coevolutionary cycles are the predominant process creating selection for higher mutation rates.
As higher mutation rates evolve, cycles decrease in size until they eventually disappear altogether (see the electronic supplementary material, figure S3a). This finding is consistent with previous work [1,3]. M'Gonigle et al. [3] showed that, once mutation rates are high enough that cycles disappear, modifier evolution ceases, as new mutants at the antigen locus no longer confer any benefit. With maternal transmission, however, we find that host mutation rates can reach even higher levels than those where cycles disappear (see the electronic supplementary material, figure S4).
We next turn our attention to evolutionarily stable mutation rates (i.e. those that would be expected to evolve, given enough time). Not surprisingly, we find that, for most strengths of maternal transmission, ϕ, and virulence, v, the ESS mutation rate in hosts is 1 (figure 4). This is because maternal transmission of parasites always selects for higher mutation rates in hosts. While evolution should eventually lead populations to these ESS values, the rate at which evolution proceeds declines dramatically as higher mutation rates evolve, suggesting that these ESS values might not be attainable over biologically realistic time-scales (compare figure 3 with figure 4). The time-course of evolution shows that, except when maternal transmission is very strong, little evolutionary change is happening by generation 107, even though the populations remain relatively far from their ESS values (see the electronic supplementary material, figure S5). This is because there is negative feedback when coevolutionary cycles are the main force selecting for higher mutation rates; higher mutation rates reduce cycle size, thereby reducing further selection for higher mutation rates (see the electronic supplementary material, figure S4). The effect of maternal transmission, however, does not attenuate as mutation rates increase and, hence, mutation rates keep evolving (see the electronic supplementary material, figure S4).
Figure 4.
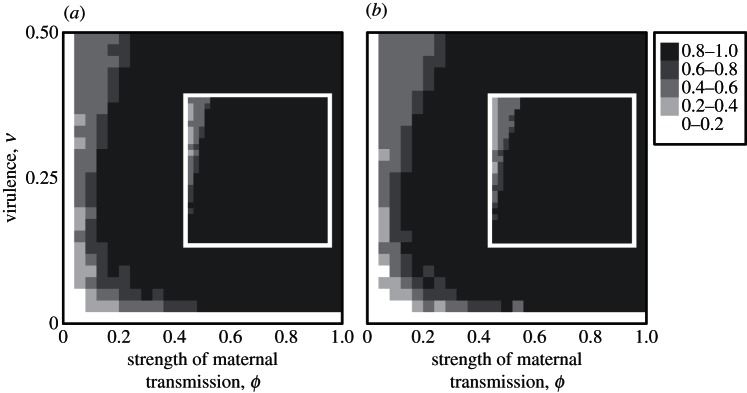
The evolutionarily stable mutation rate in hosts with (a) complete linkage (r = 0) and (b) free recombination (r = 0.5) between the modifier and antigen loci. Main panels show final values obtained when started from initial mutation rates of 0, and inset panels show final values obtained when started from initial mutation rates near 1. To find the ESS, we used an ‘accelerated’ version of the procedure described in the figure 3 caption. This method simply evaluates the sign of the fitness differences between resident and mutant alleles before substituting an allele. In addition, novel mutation rates were drawn from a normal distribution with much larger standard deviation than was used in our standard evolution runs (we used σM = 0.1 for the first half of each run and σM = 0.01 for the second half, compared with σM = 0.001 in figure 3; reducing σM in the second half of each run helps reduce variation about the ESS across replicates). With these modifications, convergence to the ESS required many fewer generations than it would in our standard evolution runs (results presented above are for 105 generations). All other parameters are as in figure 3.
In conclusion, we summarize two factors that select for higher mutation rates in hosts. First, frequency-dependent interactions at the A-locus select for higher mutation rates whenever there are host–parasite cycles, but this effect becomes weak when there is recombination (see the electronic supplementary material, figure S1). Second, maternal transmission of parasites selects for a higher mutation rate by creating an advantage to differing from one's mother. Because this benefit only lasts for a single generation, it is much less sensitive to recombination. In the absence of host–parasite coevolutionary cycles, only the latter process acts and we see that evolved mutation rates are largely the same, with or without recombination. When there are host–parasite cycles, however, mutation rates evolve to much higher levels with complete linkage.
(c). Mutation rate evolution in parasites
Here, we investigate evolution at the modifier locus (M-locus) in parasites while holding host mutation rates fixed. For parasites, ESS values were obtained during evolution runs and we do not, therefore, present or discuss these results separately (figure 5). As occurred in hosts, coevolutionary cycles select for higher mutation rates in parasites. In the absence of maternal transmission, mutation rates in parasites increase until they reach the point at which cycles disappear [3]. By reducing the size of coevolutionary cycles, maternal transmission, therefore, weakens selection for increased mutation rates in parasites, as it did for hosts (figure 5). In contrast to hosts, however, maternal transmission selects against higher mutation rates by favouring those offspring that express the same antigen allele as their mother. Therefore, with moderately strong maternal transmission, evolved mutation rates are lower than they would be if frequency-dependent interactions were the only selection pressure (see the electronic supplementary material, figure S6). This differs from hosts, where maternal transmission causes evolved mutation rates to exceed this value (see the electronic supplementary material, figure S4). When maternal transmission is very weak, however, we find mutation rates evolve to higher levels than expected (left side of the electronic supplementary material, figure S6). As shown by M'Gonigle et al. [3], this is expected in finite populations where stochastic fluctuations in host allele frequencies continue to create periodic selection for increased mutation rates in parasites, even when coevolutionary cycling no longer occurs [3]. When rates of maternal transmission are higher, however, this effect becomes marginal.
Figure 5.

(a) Evolved mutation rate in parasites after 107 generations and (b) the evolutionarily stable mutation rate in parasites. The main panel of (b) shows final values obtained when started from initial mutation rates of 0, and the inset panel shows final values obtained when started from initial mutation rates near 1. The method for finding the ESS is the same as was used for figure 4. All else, including the definition of the solid curve, is as in figure 3.
4. Analytical model
Here, we make connections between our model and that of Agrawal [20], the only previous model to investigate the impact of maternal transmission of parasites on the evolution of a genetic modifier (in his case, a modifier of the rate of sexual reproduction; [20]). Because an individual's fitness depends not only on its own genotype, but also on the infection status of its mother, an analytical treatment of our above simulation model would be challenging. Therefore, following Agrawal [20], we construct a simplified model of host dynamics that approximates the fitness effects of maternal transmission without explicitly modelling parasite dynamics by computing selection coefficients based on the existence of a hypothetical parasite population. In particular, we assume that an individual suffers a fitness reduction, γ, if it shares the same genotype as its mother. This selection is referred to as ‘similarity selection’, because each offspring experiences a parasite community that is ‘similar’ to the parasite community infecting its mother, and would be well targeted for infection if it were genetically similar as well. Agrawal [20] justified the use of this method by showing that maternal transmission of parasites indeed generates similarity selection. We also consider the effect of selection imposed by the global parasite pool by assuming that one of the two alleles at the antigen locus is deleterious, with selection coefficient α. While such an assumption does not enable us to consider fluctuations in the direction of selection, as would occur when there are coevolutionary cycles, by varying γ relative to α, we are able to assess the importance of similarity selection compared with directional selection, as would be imposed by the global parasite pool during a small interval of time. In the electronic supplementary material, we provide a fuller description of our analytical model.
5. Analytical results
As in Agrawal [20], we performed a quasi-linkage equilibrium (QLE) analysis of our model. The QLE analysis assumes that selection and mutation are weak relative to recombination and segregation and, therefore, that association measures (e.g. linkage disequilibrium) change faster than allele frequencies [25]. Because of this, the association measures can be assumed to be at their steady-state values, whereas allele frequencies continue to change (see the electronic supplementary information). We assume that the strength of directional selection, α, and the strength of similarity selection, γ, are both of order ζ, where  We additionally assume that mutation rate, μ, and the effect of the modifier, Δμ, are both of order ζ2. Assuming such weak selection and mutation is necessary for carrying out the QLE. Below, we assume that the A-allele is favoured relative to the a-allele by genotypic selection, and that the M-allele encodes a higher mutation rate than the m-allele. We find that the rate of change in frequency of the A-allele to leading order is
We additionally assume that mutation rate, μ, and the effect of the modifier, Δμ, are both of order ζ2. Assuming such weak selection and mutation is necessary for carrying out the QLE. Below, we assume that the A-allele is favoured relative to the a-allele by genotypic selection, and that the M-allele encodes a higher mutation rate than the m-allele. We find that the rate of change in frequency of the A-allele to leading order is
| 5.1 |
where VA = PA(1 − PA) is the variance at the A-locus, and the change in frequency of the M-allele is
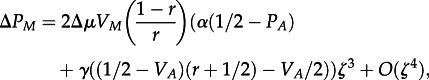 |
5.2 |
where VM = PM(1−PM) is the variance at the M-locus.
From equation (5.2), we see that α and γ affect dynamics at the M-locus to the same order. Thus, genotypic and similarity selection are similarly potent forces affecting the evolution of mutation rate. This contrasts with what has been found for the evolution of sex, where similarity selection is more potent than genotypic selection (see the electronic supplementary information; [20]). From equation (5.2), we can also see that the rate of change in frequency at the M-locus is lower with higher recombination. This is expected, as there is no direct selection at the M-locus and dynamics can only occur as a correlated response to dynamics at the A-locus. Although recombination's overall effect is to slow the evolution of higher mutation rate, equation (5.2) shows that this effect is weaker when similarity selection is stronger (higher γ). We can see that, if the favoured A-allele is at frequency greater than 0.5, then similarity selection must be sufficiently strong compared with genotypic selection for increased mutation rates to evolve. Furthermore, when increased mutation rates do evolve, they do so more quickly when similarity selection is strong and/or genetic variance at the A-locus is low. When the latter condition is met, most individuals have the same antigen allele as their mothers and thus a mutant that differs from its mother will have a higher relative fitness advantage. Not surprisingly, the rate of change at the M-locus is also higher when the effect size of the modifier is large and when there is greater genetic variance at the M-locus.
Importantly, from equation (5.2), we can see that stronger similarity selection (higher γ, our proxy for strength of maternal transmission) promotes the evolution of mutation rate. While maternal transmission in our simulation model could either strengthen or weaken selection for higher mutation rates, the weakening is always an indirect consequence of its tendency to reduce cycles which, for tractability, are absent from the analytical model. When cycles are absent or not important for modifier evolution in our simulation model, the effect of maternal transmission is always to select for higher mutation (figure 3). Consequently, we found the ESS mutation rate in hosts to be 1 for all but the weakest strengths of maternal transmission, a result consistent with equation (5.2), which predicts that higher mutation rates should evolve whenever similarity selection is sufficiently strong. Hence, our simplified analytical model captures the strengthening effect of maternal transmission that we observed in our simulation. It is also apparent from equation (5.2) that recombination has less of an effect in slowing the evolution of higher mutation when similarity selection is strong. This agrees with our simulation model, where we also see a reduced effect of recombination on mutation rate evolution when maternal transmission is strong (see the electronic supplementary material, figure S1).
Extending to parasites requires modifying the fitness functions so that it is advantageous for a parasite to be the same as its mother (see the electronic supplementary material, equation S6). This follows from the assumption that, with more maternal transmission, parasites are more likely to infect hosts with the same antigen genotype as the host that their mother infected. With this change, equation (5.2) becomes
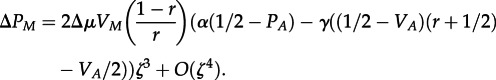 |
5.3 |
For parasites, maternal transmission tends to select against higher mutation rates. This is also what we found in the simulation model. The ESS values obtained in our simulation (figure 5) show a balance between this effect and the effect of coevolutionary cycles, which select for higher mutation rate in parasites [3]. Coevolutionary cycles, however, are absent from our simple analytical model.
6. Discussion
By constantly seeking novel ways to invade or defend against each other, hosts and their parasites create an ever-changing environment for one another [1,26]. Previous models have shown that these antagonistic interactions can select for high mutation rates in both species [1–3]. This idea has also been confirmed in experiments with P. fluorescens, which evolved higher mutation rates when coevolving with viral parasites [4].
Theoretical work on mutation rate evolution in host–parasite systems has assumed that host and parasite encounters occur at random. However, there are numerous examples of parasites that are transmitted non-randomly (e.g. from parents to their offspring [15–19]). Here, we investigated the effect of maternal transmission of parasites on mutation rate evolution in both hosts and parasites. In hosts with no recombination and little to no maternal infection, we found that higher mutation rates evolved whenever coevolutionary cycles occurred and when parasites were more virulent (figure 3). Both these findings are consistent with previous theoretical work [3]. When maternal transmission was common, however, high mutation rates evolved in hosts, even when parasites were weakly virulent and cycles were absent. In parasites, coevolutionary cycles and high virulence also led to the evolution of higher mutation rates. In contrast to hosts, however, maternal transmission always favours parasites that are the same as their mothers and, consequently, selects against higher mutation rates. Hence, we predict that, in systems with substantial maternal transmission (i.e. a high vertical : horizontal transmission ratio), there should be a greater difference between the mutation rates of hosts and parasites compared with systems with little or no maternal transmission (i.e. low vertical : horizontal transmission ratio). Moreover, we expect that loci in hosts involved in immunity against vertically transmitted parasites should exhibit higher mutation rates than loci involved in immunity against horizontally transmitted parasites.
Provided that virulence was not too high, even a small amount of maternal transmission caused cycles to disappear (figure 2). Although much theory predicts cyclical fluctuations in host and parasite gene frequencies [1,3], few studies have provided corroborating empirical evidence [27–29]. Our findings suggest that cyclical dynamics might not occur in coevolving host–parasite systems, if even a small fraction of parasites are transmitted maternally (figure 2). It follows that predictions based on previous theoretical models, which implicitly assume the existence of such cycles, might not apply to cases when parasites can be transmitted maternally. In parasites, this is a crucial point, because we have shown that in the absence of cycles, parasite mutation rate should evolve downwards. In hosts, however, maternal transmission itself might provide an alternative explanation for how host–parasite interactions can select for increased mutation rates; the advantage to differing genetically from one's mother can select for modifier alleles that increase mutation rate. Furthermore, in our model, the effect of maternal transmission on cycle dynamics and mutation rate evolution is most pronounced when parasites are weakly virulent—a phenotype that is expected to be common in maternally transmitted (relative to horizontally transmitted) parasites [30].
With high rates of recombination, evolved host mutation rates were not as high as with complete linkage (figure 3), a result that is consistent with earlier findings on the effect of recombination [3]. Recombination slows mutation rate evolution because it disassociates dynamics at the antigen locus from dynamics at the modifier locus. With high recombination, effects of maternal transmission were simple; higher rates of maternal transmission led to the evolution of higher mutation rates. Because sexual and asexual reproduction are equivalent in parasites in our model, recombination did not affect mutation rate evolution in parasites.
Recombination is expected to slow the evolution of higher mutation rates when the benefit of new mutations persists for many generations [3]. During this time, recombination dissociates the benefit of the high mutation modifier from the modifier itself. While the benefit associated with producing a globally advantageous allele can last for many generations, the benefit associated with producing an allele that causes a host to differ from its mother only lasts for one generation. Hence, recombination has only one opportunity to separate the modifier and the beneficial allele and it thus has a much reduced effect. This highlights an important aspect of maternal transmission: although the effect of parasite-induced frequency-dependent selection on mutation rate evolution diminishes with increasing recombination, the effect of maternal transmission remains largely constant. Previous theory has shown that while adaptive mutations are an important factor promoting mutation rate evolution in asexuals, they are much less relevant in sexual species that undergo recombination [3,24,31]. Here, we have identified a mechanism that is apparently much less affected by recombination.
In accordance with earlier models [3,32], we predict evolution of mutation rates well above current empirical estimates [14,33]. However, mutation rate has been found to vary across the genome [34] and, in particular, loci involved in immunity are known to have higher than average mutation rates [21,35]. We have neglected to include any costs associated with higher mutation rate (e.g. in the form of unconditionally deleterious alleles), the inclusion of which would probably lower evolved mutation rates dramatically. However, we would not expect the qualitative effects of maternal transmission to change with such costs.
We have shown that maternal transmission of parasites can select for increased mutation rates in hosts. For infections with a genetic basis, mutation represents only one means to evade infection. Previous models have investigated the evolution of other infection-resistance strategies, including parasite avoidance, immunity and transgenerational transfer of immunity ([36] and references therein). Including maternal transmission in such models would probably have important consequences for their dynamics. For example, a recent study of the evolution of maternal transmission of immunity, which did not include maternal transmission of parasites [36], found that this trait would be more likely to evolve if it protected against not just the parasite strain of the mother, but also the alternate parasite strain in the population (i.e. if there was high cross-immunity). This relationship would probably become weaker if offspring are preferentially infected by the parasite strain of their mother, because cross infection would occur less frequently.
There is evidence that higher mutation rates can be induced by physiological stress [37,38]. Although parasite infection has not been studied as a mutation-inducing stressor, it is plausible that infected hosts would have higher mutation rates owing to the stress of being infected. If our model included costly mutation, condition-dependent mutation would be expected to replace facultatively high mutation, because mutations would occur more often when they are advantageous, and less often when they are merely deleterious [39]. Further theoretical studies investigating such extensions are needed to help us fully appreciate the role played by maternal transmission in host–parasite coevolutionary dynamics, and further empirical studies are needed to quantify the pervasiveness of maternal transmission of parasites.
Acknowledgements
We would like to thank Marcus Feldman, Sally Otto, Florence Débarre, the Feldman and Otto laboratories and Robert Furrow for helpful discussions and comments on the manuscript. We are grateful to two anonymous reviewers who greatly improved this manuscript. This work was financially supported by the Natural Sciences and Engineering Research Council of Canada (PGS-D to P.B.G. and PDF to L.K.M.) and the National Institute of Health (GM 28016 to Marcus Feldman). Mathematica code available upon request to the corresponding author.
References
- 1.Nee S. 1989. Antagonistic co-evolution and the evolution of genotypic randomization. J. Theor. Biol. 140, 499–518 10.1016/S0022-5193(89)80111-0 (doi:10.1016/S0022-5193(89)80111-0) [DOI] [PubMed] [Google Scholar]
- 2.Haraguchi Y, Sasaki A. 1996. Host–parasite arms race in mutation modifications: indefinite escalation despite a heavy load? J. Theor. Biol. 183, 121–137 10.1006/jtbi.1996.9999 (doi:10.1006/jtbi.1996.9999) [DOI] [PubMed] [Google Scholar]
- 3.M'Gonigle LK, Shen JJ, Otto SP. 2009. Mutating away from your enemies: the evolution of mutation rate in a host–parasite system. Theor. Popul. Biol. 75, 301–311 10.1016/j.tpb.2009.03.003 (doi:10.1016/j.tpb.2009.03.003) [DOI] [PubMed] [Google Scholar]
- 4.Pal C, Maciá MD, Oliver A, Schachar I, Buckling A. 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450, 1079–1081 10.1038/nature06350 (doi:10.1038/nature06350) [DOI] [PubMed] [Google Scholar]
- 5.Sniegowski PD, Gerrish PJ, Lenski RE. 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387, 703–705 10.1038/42701 (doi:10.1038/42701) [DOI] [PubMed] [Google Scholar]
- 6.Baer CF, et al. 2005. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc. Natl Acad. Sci. USA 102, 5785–5790 10.1073/pnas.0406056102 (doi:10.1073/pnas.0406056102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ávila V, Chavarrías D, Sánchez E, Manrique A, López-Fanjul C, García-Dorado A. 2006. Increase of the spontaneous mutation rate in a long-term experiment with Drosophila melanogaster. Genetics 173, 267–277 10.1534/genetics.106.056200 (doi:10.1534/genetics.106.056200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haag-Liautard C, Dorris M, Maside X, Macaskill S, Halligan DL, Houle D, Charlesworth B, Keightley PD. 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445, 82–85 10.1038/nature05388 (doi:10.1038/nature05388) [DOI] [PubMed] [Google Scholar]
- 9.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 10.1038/nature08480 (doi:10.1038/nature08480) [DOI] [PubMed] [Google Scholar]
- 10.Leigh EG. 1970. Natural selection and mutability. Am. Nat. 104, 301–305 10.1086/282663 (doi:10.1086/282663) [DOI] [Google Scholar]
- 11.Leigh EG. 1973. The evolution of mutation rates. Genetics 73, 1–18 [PubMed] [Google Scholar]
- 12.Liberman U, Feldman MW. 1986. Modifiers of mutation rate: a general reduction principle. Theor. Popul. Biol. 30, 125–142 10.1016/0040-5809(86)90028-6 (doi:10.1016/0040-5809(86)90028-6) [DOI] [PubMed] [Google Scholar]
- 13.Ishii K, Matsuda H, Iwasa Y, Sasaki A. 1989. Evolutionarily stable mutation rate in a periodically changing environment. Genetics 121, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baer CF, Miyamoto MM, Denver DR. 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8, 619–631 10.1038/nrg2158 (doi:10.1038/nrg2158) [DOI] [PubMed] [Google Scholar]
- 15.Pearce DA. 1998. PCR as a tool for the investigation of seed-borne disease. In Applications of PCR in mycology (eds Bridge PD, Arora DK, Reddy CA, Elander RP.), pp. 309–324 Wallingford, UK: CAB International [Google Scholar]
- 16.Agarwal VK, Sinclair JB. 1997. Principles of seed pathology, 2nd edn Boca Raton, FL: CRC Press [Google Scholar]
- 17.Fowler MG, Simonds RJ, Roongpisuthipong A. 2000. Update on perinatal HIV transmission. Pediatr. Clin. North Am. 47, 21–38 10.1016/S0031-3955(05)70193-0 (doi:10.1016/S0031-3955(05)70193-0) [DOI] [PubMed] [Google Scholar]
- 18.Carlier Y, Truyens C, Deloron P, Peyron F. 2012. Congenital parasitic infections: a review. Acta Trop. 121, 55–70 10.1016/j.actatropica.2011.10.018 (doi:10.1016/j.actatropica.2011.10.018) [DOI] [PubMed] [Google Scholar]
- 19.Knell RJ, Webberley KM. 2004. Sexually transmitted diseases of insects: distribution, evolution, ecology and host behaviour. Biol. Rev. Camb. Philos. Soc. 79, 557–581 10.1017/S1464793103006365 (doi:10.1017/S1464793103006365) [DOI] [PubMed] [Google Scholar]
- 20.Agrawal AF. 2006. Similarity selection and the evolution of sex: revisiting the Red Queen. PLoS Biol. 4, e265. 10.1371/journal.pbio.0040265 (doi:10.1371/journal.pbio.0040265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank SA. 2002. Immunology and the evolution of infectious diseases. Princeton, NJ: Princeton University Press; [PubMed] [Google Scholar]
- 22.Gandon S, Otto SP. 2007. The evolution of sex and recombination in response to abiotic or coevolutionary fluctuations in epistasis. Genetics 175, 1835–1853 10.1534/genetics.106.066399 (doi:10.1534/genetics.106.066399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M'Gonigle LK, Otto SP. 2011. Ploidy and the evolution of parasitism. Proc. R. Soc. B 278, 2814–2822 10.1098/rspb.2010.2146 (doi:10.1098/rspb.2010.2146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson T. 1999. Beneficial mutations, hitchhiking and the evolution of mutation rates in sexual populations. Genetics 151, 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton NH, Turelli M. 1991. Natural and sexual selection on many loci. Genetics 127, 229–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaenike J. 1978. An hypothesis to account for the maintenance of sex within populations. Evol. Theory 3, 191–194 [Google Scholar]
- 27.Stahl EA, Dwyerm G, Mauricio R, Kreitman M, Bergelson J. 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400, 667–671 10.1038/23260 (doi:10.1038/23260) [DOI] [PubMed] [Google Scholar]
- 28.Decaestecker E, Gaba S, Raeymaekers JA, Stoks R, Van Kerckhoven L, Ebert D, De Meester L. 2007. Host–parasite ‘Red Queen’ dynamics archived in pond sediment. Nature 450, 870–873 10.1038/nature06291 (doi:10.1038/nature06291) [DOI] [PubMed] [Google Scholar]
- 29.Jokela J, Dybdahl MF, Lively CM. 2009. The maintenance of sex, clonal dynamics, and host–parasite coevolution in a mixed population of sexual and asexual snails. Am. Nat. 174, S43–S53 10.1086/599080 (doi:10.1086/599080) [DOI] [PubMed] [Google Scholar]
- 30.Ebert D, Herre EA. 1996. The evolution of parasitic diseases. Parasitol. Today 12, 96–101 10.1016/0169-4758(96)80668-5 (doi:10.1016/0169-4758(96)80668-5) [DOI] [PubMed] [Google Scholar]
- 31.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. 2000. The evolution of mutation rates: separating causes from consequences. BioEssays 22, 1057–1066 (doi:10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W) [DOI] [PubMed] [Google Scholar]
- 32.Sasaki A. 1994. Evolution of antigen drift/switching: continuously evading pathogens. J. Theor. Biol. 168, 291–308 10.1006/jtbi.1994.1110 (doi:10.1006/jtbi.1994.1110) [DOI] [PubMed] [Google Scholar]
- 33.Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148, 1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martincorena I, Seshasayee ASN, Luscombe NM. 2012. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature 485, 95–98 10.1038/nature10995 (doi:10.1038/nature10995) [DOI] [PubMed] [Google Scholar]
- 35.Chuang JH, Li H. 2004. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2, 0253–0263 10.1371/journal.pbio.0020253 (doi:10.1371/journal.pbio.0020253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garnier R, Boulinier T, Gandon S. 2012. Coevolution between maternal transfer of immunity and other resistance strategies against pathogens. Evolution 66, 3067–3078 10.1111/j.1558-5646.2012.01665.x (doi:10.1111/j.1558-5646.2012.01665.x) [DOI] [PubMed] [Google Scholar]
- 37.Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. 2011. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl Acad. Sci. USA 108, 13 659–13 664 10.1073/pnas.1104681108 (doi:10.1073/pnas.1104681108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp NP, Agrawal AF. 2012. Evidence for elevated mutation rates in low-quality genotypes. Proc. Natl Acad. Sci. USA 109, 6142–6146 10.1073/pnas.1118918109 (doi:10.1073/pnas.1118918109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ram Y, Hadany L. 2012. The evolution of stress-induced hypermutation in asexual populations. Evolution 66, 2315–2328 10.1111/j.1558-5646.2012.01576.x (doi:10.1111/j.1558-5646.2012.01576.x) [DOI] [PubMed] [Google Scholar]