Abstract
We present a new approach to combine λ dynamics with meta-dynamics (named λ-meta dynamics) to compute free energy surface with respect to λ. Particularly, the λ-meta dynamics method extends meta-dynamics to a single virtual variable λ, i.e., the coupling parameter between solute and solvent, to compute absolute solvation free energy as an exemplary application. We demonstrate that λ-meta dynamics simulations can recover the accurate potential of mean force surface with respect to λ compared to the benchmark results from traditional λ-dynamics with umbrella sampling. The solvation free energy results for five small organic molecules from λ-meta dynamics simulations using the same filling scheme show that the statistical errors are within ±0.5 kcal/mol. The new λ-meta dynamics method is general and other variables such as order parameters to describe conformational changes can be easily combined with λ-meta dynamics. This should allow for efficient samplings on high-dimension free energy landscapes.
Keywords: Meta-dynamics, λ dynamics, QM/MM, solvation free energy, λ-meta dynamics
Atomistic simulations to compute free energy difference between two states are pivotal to predict and explain molecular properties. Many simulation techniques have been developed, such as umbrella sampling1-3, thermodynamics integration4, and free energy perturbation5. All these methods use a virtual coupling parameter (i.e., λ) to drive the molecular system from one state to another one. To enhance the sampling efficiency on λ, λ dynamics6, 7 was developed to treat the coupling parameter as a virtual dynamics variable in conjunction with molecular dynamics of real system. λ dynamics has been applied to study solvation free energy and ligands binding affinity in association with umbrella sampling8, 2 and adiabatic molecular dynamics9. On the other hand, another efficient sampling method, meta-dynamics, was developed recently to escape the energy minimum10 and has been further extended to recover the reaction profile11-14. In meta-dynamics, the free energy surface is obtained efficiently by accumulating and filling history-dependent Gaussian potentials.
In this work, we developed a λ-meta dynamics by combining λ-dynamics with the meta-dynamics method12, 15, 16, to help exploring the potential mean force (PMF) surface with respect to the virtual variable λ. To demonstrate the λ-meta dynamics approach, absolute solvation free energy is taken as one exemplary property. Solvation free energy is an essential property of molecules and is indispensible in drug discovery process17. Various theoretical approaches have been proposed, ranging from the simple quantitative structure-activity relationship (QSAR) model18-20 to continuum solvent model21-27, to molecular mechanics model based on force fields28-31, and to quantum mechanics/molecular mechanics (QM/MM) model32-34. We focus on using QM/MM atomistic simulations35 to calculate absolute solvation free energy in aqueous solution. The solute molecule was described with ab initio QM while the solvent with explicit TIP3P water model.
Here, two virtual variables λele and λvdw are used to drive the solute from aqueous solution state to vacuum state. λele controls the electrostatic interactions between solute in QM and solution in MM while λvdw scales the van der Waals (VDW) between QM and MM. Using the new λ-meta dynamics approach, not only the PMFs were recovered compared to λ dynamics with umbrella sampling, but also absolute solvation free energies for five small organic molecules were computed accurately. A general filling scheme of Gaussian functions required in meta-dynamics was also developed to control the statistical errors in λ-meta dynamics. More importantly, λ-meta dynamics is a general approach for free energy simulations, which can be readily extended to multiple variables and can efficiently explore such high-dimension surfaces.
To elaborate the details of λ-meta dynamics method, we chose absolute free energy computations using the QM/MM Hamiltonian defined by
| (1) |
where HQM, HQm/MM,vdw, and HMM are the Hamiltonian of QM subsystem, empirical VDW interactions between QM and MM subsystems, and MM internal energy. HESP captures the MM point charge electrostatic potential influence on the QM subsystem. The coupling parameters λele and λvdw are used to scale the electrostatic and VDW interactions between QM and MM and drive the solute from solution to vacuum environment. To scale the VDW interactions, the soft-core potential is applied to avoid the simulation singularity and instability problems. For λ-dynamics6, eq. 1 is extended to the following equation that contains a virtual variable λ,
| (2) |
where λ̇ is velocity of λ and mλ is the virtual mass of λ. This method has been applied to compute the relative solvation free energies6, 37, 9 and binding free energies38, 39 between similar molecules.
When meta-dynamics is incorporated into eq. 2, a history-dependent term U*(λ,t) is added to fill the PMF well. Typically, U*(λ,t) can be expanded by a set of Gaussian functions as:
| (3) |
In this equation, w is the Gaussian height, δs is the Gaussian width, and tG is the filling frequency. So this external potential U* (λ, t) depends on the trajectory and filling scheme. When the width of the Gaussians is small and t is sufficiently large, the PMF surface can be recovered by U*(λ,t), i.e., ΔA(λ) = −U*(λ). Here, U* (λ,t) is simplified as U* (λ) for brevity.
To benchmark our results of λ-meta dynamics simulations, λ-dynamics combined with the classical umbrella sampling was performed. In umbrella sampling, 21 uniformly distributed windows from λ=1 to λ=0 were sampled with 5 ps relaxations and 95 ps MD simulations at T=300 K and 500 kcal/mol force constant on λ for λele and λvdw, respectively. (Note that umbrella sampling needs more computational cost to tune up the sampling efficiency such as the number of sampling windows and optimal force constants for each window. In this work, extensive umbrella samplings were applied to obtain the benchmark data.) The Weighted Histogram Analysis (WHAM)1, 2 with bin size 0.001 was applied on these trajectories to recover free energy surfaces.
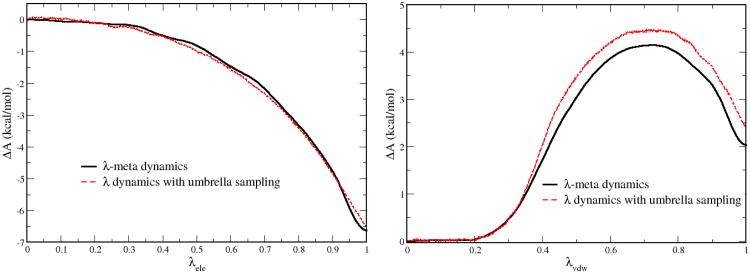
Using methanol as a model system, the PMF surfaces for λele and λvdw computed by our λ-meta dynamics approach were plotted in Figure 1 along with the umbrella sampling curves. Both curves of λele and λvdw from λ-meta dynamics agree well with umbrella sampling curves. The free energy differences between λ-meta dynamics and umbrella sampling are only 0.3 and 0.5 kcal/mol for ΔA along λele and λvdw, respectively. Therefore, the new λ-meta dynamics method can indeed recover the PMF with respect to λ and obtain accurate absolute solvation free energy of methanol compared to umbrella sampling. Note that since the mirror periodic condition was applied for λ-meta dynamics, a cusp appears when λ is close to 1. This is why a relatively small Gaussian width (0.06) should be used in the smoothing period to capture the cusp height. Although the exact cusp shape cannot be reproduced by finite Gaussian functions, these derivations from the cusp on PMF are negligible compared to the umbrella sampling results.
Figure 1.
PMF surfaces computed by λ-meta dynamics (solid black lines) and umbrella sampling (dashed red lines) for methanol in aqueous solution for λele and λvdw, respectively.
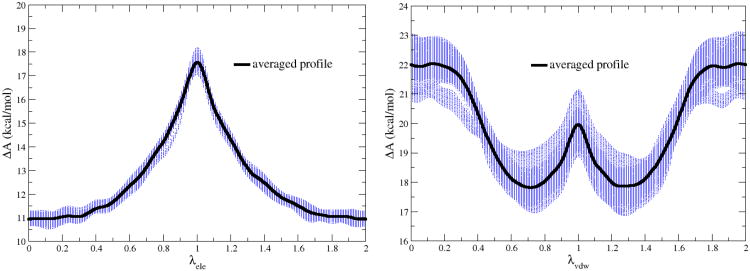
To understand the fluctuations of filling procedure in λ-meta dynamics, Figure 2 shows the PMF profiles (150 curves for λele and 390 curves for λvdw in dashed blue lines) from the smoothing period along with their averaged profiles in solid black lines. For λele, the ΔAele fluctuations are within 0.5 kcal/mol, while relatively large ΔAvdw fluctuations of ±1.2 kcal/mol for λvdw were observed. Note that these fluctuations during filling procedure are not the statistical errors of λ-meta dynamics. To estimate the statistical errors of λ-meta dynamics, we further carried out eight independent simulations for λele and λvdw with different initial velocities in λ-meta dynamics. As list in Table 1, for methanol, the standard deviation for ΔAele is 0.17 kcal/mol and is 0.36 kcal/mol for ΔAvdw Therefore, the absolute solvation free energies can be computed by the new λ-meta dynamics method with small statistical errors (<0.5 kcal/mol for methanol compared to the benchmark result from umbrella sampling). Compared to experimental data in Table 1, theoretical computations (both λ-meta dynamics and umbrella sampling) show the errors around 2 kcal/mol for acetamide and acetic acid. These errors are derived from the interactions between QM and MM subsystems such as vdW, which are beyond the scope of this letter.
Figure 2.
PMF profiles (dotted blue lines) and the averaged PMFs (solid black lines) of λele and λvdw for methanol.
Table 1.
The computed solvation free energies from λ-meta dynamics and λ dynamics with umbrella sampling (i.e., λ-US) for five compounds.
| Molecule | ΔAele | ΔAvdw | ΔAsolv | ΔAexpa | |
|---|---|---|---|---|---|
|
| |||||
| Methanol | λ-meta | −6.79 ±0.17 | 1.87 ±0.36 | −4.92 ±0.46 | |
|
|
λ-US | −6.50±0.06 | 2.46±0.10 | −4.04±0.08 | −5.11 |
|
| |||||
| Methane | λ-meta | −0.23 ±0.08 | 1.77 ±0.30 | 1.53±0.32 | |

|
λ -US | −0.28±0.06 | 2.21±0.10 | 1.93±0.06 | 2.0 |
|
| |||||
| Methylamine | λ-meta | −6.58 ±0.22 | 1.76 ±0.19 | −4.82 ±0.28 | |
|
|
λ -US | −6.17±0.10 | 2.05±0.16 | −4.12±0.19 | −4.56 |
|
| |||||
| Acetamide | λ-meta | −10.14 ±0.30 | 2.10 ±0.49 | −8.04 ±0.70 | |
|
λ -US | −9.90±0.05 | 2.34±0.10 | −7.56±0.14 | −9.71 |
|
| |||||
| Acetic acid | λ-meta | −6.72 ±0.23 | 2.11 ±0.31 | −4.76 ±0.33 | |
|
λ -US | −6.46±0.06 | 2.56±0.17 | −3.90±0.14 | −6.7 |
Experimental values from Ref. 31
It is worth discussing why ΔAvdw has the relatively larger error compared to ΔAele, which is shown in Figure 1 when comparing Δ-meta dynamics with umbrella sampling and is also observed in Figure 2. Such large errors of ΔAvdw are caused by the broad VDW force distributions of dHQM/MM, vdw(λvdw)/d λvdw, especially when the solute VDW sphere is in contact with the solvent molecules for λvdw between 0.4 and 1.0. Even though the soft-core potential approach for the VDW part is applied here, as shown in Figure S1 of the supporting information (SI), the inertial movements in VDW make λ dynamics less diffusive for λvdw than for λele when the PMF wells are filled. We also plot the curves from eight independent λ-meta dynamics simulations along with the umbrella sampling result in Figure S2. The relatively large derivations (within 1 kcal/mol) between λ-meta dynamics and umbrella sampling appear when λvdw is larger than 0.4, and this problem is caused by the empirical VDW energy expression. Nevertheless, the computed solvation free energies still agree well with the umbrella sampling results as shown in Table 1.
The computed solvation free energies for all five small organic molecules are listed in Table 1 with the estimated statistical errors from eight independent simulations for both λele and λvdw. For methane, methylamine, acetamide, and acetic acid, the PMF surfaces were plotted in Figures S3-S6 from one of eight independent λ-meta dynamics simulations along with umbrella sampling results. The PMF comparisons between umbrella sampling and λ-meta dynamics demonstrate that λ-meta dynamics with a general filling scheme is robust to obtain the absolute solvation free energies for most five diverse molecules (from large to small solvation free energies) with the errors within 1.0 kcal/mol compared to umbrella sampling results. These deviations of absolute solvation free energies computed by λ-meta dynamics are mainly caused by the cusp when λ=1.
In summary, we developed a new λ-meta dynamics method, which combines λ dynamics with meta-dynamics to enhance the samplings of virtual variables. To our best knowledge, λ-meta dynamics is the first method to demonstrate meta-dynamics can be used to recover PMF surfaces composed of virtual variables. The general filling scheme to accumulate and add Gaussian functions to fill the PMF wells can be used for all five diverse molecules. Our results for solvation free energy calculations show that λ-meta dynamics can reproduce the accurate PMF surface with respect to λ efficiently (120 ps for λele and 240 ps for and λvdw). The corresponding solvation free energies are all within 1.0 kcal/mol errors compared to umbrella sampling results. This suggests that the computational cost of solvation free energy calculations can be reduced significantly since one single simulation for λele and λvdw is sufficient, respectively. Finally, λ-meta dynamics is a general approach to enhance the samplings on virtual variables, which can be also further applied in other free energy computations such as protein ligand binding free energy. λ-meta dynamics can readily incorporate other order parameters and explore the corresponding high-dimension free energy landscapes efficiently.
Computational Details
The new λ-meta dynamics approach was implemented in our in-house program40. In QM/MM simulations, only the solute is included in the QM subsystem at the B3LYP/3-21G level. Since this work focuses on methodology demonstrations, a small Gaussian basis set of QM is applied to save the computational cost in QM/MM simulations. The VDW parameters were chosen for each atom element: C, H, O, and N from CHARMM 22 force field41 (i.e., atom type C, HA, O, and N). The MD integration step is 1 fs for all forces. The pairlist was updated every 10 fs with electrostatic cutoff as 10 Å. The solute is solvated and centered in a 40×40×40Å3 water box with more than 2000 TIP3P water molecules depending on the specific solute molecules. NVT ensemble at 300 K was applied in simulations with the Berendsen stat42 . For the virtual coordinate, mλ is chosen as 12 a.m.u., which is same as a carbon atom.
The thermo ensemble on λ is generated using Nose-Hoover Chain technique 43 with 4 chains and the temperature is maintained at 60 K for meta-dynamics.
In λ-meta dynamics, the filling scheme for Gaussian functions in eq. 3 is crucial to reproduce accurate solvation free energy. To accomplish the balance between accuracy and efficiency, a special boundary condition is applied: λ is confined in a [0, 2] region. Particularly, when λ is between 1 and 2, the value of λ is replaced by 2 − λ, which is then used to compute QM/MM energies and the corresponding forces with reverse sign. When λ is larger than 2 or smaller than 0, it is immediately mirrored by λ−2 or λ+2, respectively, to guarantee λ is within [0, 2] region. This mirror periodic condition can help Gaussian functions fill the wells of two end points at 0 and 1, which are essential to compute the accurate solvation free energy.
The entire Gaussian filling procedure is split into two parts: the coarse-filling period and the smoothing period. In the coarse-filling period, Gaussian functions with large Gaussian height from 0.16 to 0.08 in U*(λ) are applied to construct the rough PMF surface. In the smoothing period, Gaussian functions with height 0.04 are used to refine U*(λ). All Gaussian functions have the fixed width of 0.06. At every 50 MD steps, a new Gaussian function is added to U*(λ) and its Gaussian height is gradually changed according to the filling procedure. The detailed scheme to fill the PMF wells and calculate solvation free energies for all five organic molecules is the following:
For first 300 Gaussian functions, the Gaussian height is 0.16, then it is decreased to 0.12 for next 300 peaks and to 0.08 for last 300 peaks to accomplish the coarse-filling period with the initial 45 ps MD simulations;
The Gaussian height with 0.04 is added to refine U*(λ) obtained in step a). For λele-meta dynamics, 1,500 Gaussian functions are added in 75 ps simulation. For λvdw-meta dynamics, 3,900 functions are added in 195 ps simulations.
In meta dynamics, the free energy profile should be computed by using arithmetic average of profiles after collective variable starts diffusing [Ref 11]. The final solvation free energy surface is constructed by only averaging the curves in the smoothing period after every 10 Gaussian functions are added. Totally, for the λele part, 150 curves, and for λvdw 390 curves are generated to reproduce the PMF surface from λ=0 to λ=1. Note that the average value of [0, 1] and [1, 2] regions is also applied here. The solvation free energy (ΔAsolv= ΔAele + ΔAvdw) can be readily computed by two independent terms using the values at λ=0 and λ=1, i.e., ΔAele = −U*(λele=1) +U*(λele=0) and ΔAvdw = −U*(λvdw=1) +U*(λvdw=0), respectively.
Supplementary Material
Acknowledgments
Support from the National Institute of Health (NIH R01-GM061870) is greatly appreciated..
Footnotes
Supporting Information Available. The Gaussian filling trajectories for λele and for λvdw are shown in Figure S1. The PMF comparison between eight curves from λ-meta dynamics and umbrella sampling of methanol is shown in Figure S2. λ-meta dynamics curves for methane, methylamine, acetamide, and acetic acid along with the corresponding umbrella sampling curves are shown in Figures S3-S6. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Xiangqian Hu, Email: xqhu@duke.edu.
Weitao Yang, Email: weitao.yang@duke.edu.
References
- 1.Roux B. The Calculation of the Potential of Mean Force Using Computer Simulations. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 2.Souaille M, Roux B. Extension to the Weighted Histogram Analysis Method: Combining Umbrella Sampling With Free Energy Calculations. Comput Phys Commun. 2001;135:40–57. [Google Scholar]
- 3.Hub JS, de Groot BL, van der Spoel D. g_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J Chem Theory Comput. 2010;6:3713–3720. [Google Scholar]
- 4.Schlitter J, Klahn M. A New Concise Expression for the Free Energy of a Reaction Coordinate. J Chem Phys. 2003;118:2057–2060. [Google Scholar]
- 5.Zwanzig RW. High-Temperature Equation of State by a Perturbation Method. I. Nonpolar Gases. J Chem Phys. 1954;22:1420–1426. [Google Scholar]
- 6.Kong X, Brooks CL. Lambda-Dynamics: A New Approach to Free Energy Calculations. J Chem Phys. 1996;105:2414–2423. [Google Scholar]
- 7.Knight JL, Brooks CL. Lambda-Dynamics Free Energy Simulation Methods. J Comput Chem. 2009;30:1692–1700. doi: 10.1002/jcc.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Z, Brooks CL, Kong X. Efficient and Flexible Algorithm for Free Energy Calculations Using the λ-Dynamics Approach. J Phys Chem B. 1998;102:2032–2036. [Google Scholar]
- 9.Abrams JB, Rosso L, Tuckerman ME. Efficient and Precise Solvation Free Energies via Alchemical Adiabatic Molecular Dynamics. J Chem Phys. 2006;125:074115. doi: 10.1063/1.2232082. [DOI] [PubMed] [Google Scholar]
- 10.Laio A, Parrinello M. Escaping Free-Energy Minima. Proc Natl Acad Sci U S A. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Micheletti C, Laio A, Parrinello M. Reconstructing the Density of States by History-Dependent Metadynamics. Phys Rev Lett. 2004;92:170601. doi: 10.1103/PhysRevLett.92.170601. [DOI] [PubMed] [Google Scholar]
- 12.Ensing B, De Vivo M, Liu Z, Moore P, Klein ML. Metadynamics as a Tool for Exploring Free Energy Landscapes of Chemical Reactions. Acc Chem Res. 2005;39:73–81. doi: 10.1021/ar040198i. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Min D, Liu Y, Yang W. Essential Energy Space Random Walk via Energy Space Metadynamics Method to Accelerate Molecular Dynamics Simulations. J Chem Phys. 2007;127(9):094101. doi: 10.1063/1.2769356. [DOI] [PubMed] [Google Scholar]
- 14.Min D, Chen M, Zheng L, Jin Y, Schwartz MA, Sang QXA, Yang W. Enhancing QM/MM Molecular Dynamics Sampling in Explicit Environments via an Orthogonal-Space-Random-Walk-Based Strategy. J Phys Chem B. 2011;115(14):3924–3935. doi: 10.1021/jp109454q. [DOI] [PubMed] [Google Scholar]
- 15.Laio A, Rodriguez-Fortea A, Gervasio FL, Ceccarelli M, Parrinello M. Assessing the Accuracy of Metadynamics. J Phys Chem B. 2005;109:6714–6721. doi: 10.1021/jp045424k. [DOI] [PubMed] [Google Scholar]
- 16.Laio A, Gervasio FL. Metadynamics: a Method to Simulate Rare Events and Reconstruct the Free Energy in Biophysics, Chemistry and Material Science. Rep Prog Phys. 2008;71:126601. [Google Scholar]
- 17.Jorgensen WL. The Many Roles of Computation in Drug Discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 18.Yaffe D, Cohen Y, Espinosa G, Arenas A, Giralt F. A Fuzzy ARTMAP-Based Quantitative Structure–Property Relationship (QSPR) for the Henry's Law Constant of Organic Compounds. J Chem Inf Comput Sci. 2002;43:85–112. doi: 10.1021/ci025561j. [DOI] [PubMed] [Google Scholar]
- 19.Katritzky AR, Oliferenko AA, Oliferenko PV, Petrukhin R, Tatham DB, Maran U, Lomaka A, Acree WE. A General Treatment of Solubility. 1. The QSPR Correlation of Solvation Free Energies of Single Solutes in Series of Solvents. J Chem Inf Comput Sci. 2003;43:1794–1805. doi: 10.1021/ci034120c. [DOI] [PubMed] [Google Scholar]
- 20.Katritzky AR, Oliferenko AA, Oliferenko PV, Petrukhin R, Tatham DB, Maran U, Lomaka A, Acree WE. A General Treatment of Solubility. 2. QSPR Prediction of Free Energies of Solvation of Specified Solutes in Ranges of Solvents. J Chem Inf Comput Sci. 2003;43:1806–1814. doi: 10.1021/ci034122x. [DOI] [PubMed] [Google Scholar]
- 21.Simonson T, Bruenger AT. Solvation Free Energies Estimated from Macroscopic Continuum Theory: An Accuracy Assessment. J Chem Phys. 1994;98:4683–4694. [Google Scholar]
- 22.Roux B, Simonson T. Implicit Solvent Models. Biophys Chem. 1999;78:1–20. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- 23.Bashford D, Case DA. Generalized Born Models of Macromolecular Solvation Effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 24.Feig M, Onufriev A, Lee MS, Im W, Case DA, Brooks CL. Performance Comparison of Generalized Born and Poisson Methods in the Calculation of Electrostatic Solvation Energies for Protein Structures. J Comput Chem. 2004;25:265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 25.Chamberlin AC, Cramer CJ, Truhlar DG. Performance of SM8 on a Test To Predict Small-Molecule Solvation Free Energies. J Phys Chem B. 2008;112:8651–8655. doi: 10.1021/jp8028038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS. Predicting Small-Molecule Solvation Free Energies: An Informal Blind Test for Computational Chemistry. J Med Chem. 2008;51:769–779. doi: 10.1021/jm070549+. [DOI] [PubMed] [Google Scholar]
- 27.Ribeiro R, Marenich A, Cramer C, Truhlar DG. Prediction of SAMPL2 Aqueous Solvation Free Energies and Tautomeric Ratios Using the SM8, SM8AD, and SMD Solvation Models. J Comput Aided Mol Des. 2010;24:317–333. doi: 10.1007/s10822-010-9333-9. [DOI] [PubMed] [Google Scholar]
- 28.Goncalves PFB, Stassen H. Calculation of the Free Energy of Solvation from Molecular Dynamics Simulations. Pure Appl Chem. 2004;76:231–240. [Google Scholar]
- 29.Mobley DL, Bayly CI, Cooper MD, Shirts MR, Dill KA. Small Molecule Hydration Free Energies in Explicit Solvent: An Extensive Test of Fixed-Charge Atomistic Simulations. J Chem Theory Comput. 2009;5:350–358. doi: 10.1021/ct800409d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shivakumar D, Deng Y, Roux B. Computations of Absolute Solvation Free Energies of Small Molecules Using Explicit and Implicit Solvent Model. J Chem Theory Comput. 2009;5:919–930. doi: 10.1021/ct800445x. [DOI] [PubMed] [Google Scholar]
- 31.Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J Chem Theory Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 32.Kamerlin SCL, Haranczyk M, Warshel A. Progress in Ab Initio QM/MM Free-Energy Simulations of Electrostatic Energies in Proteins: Accelerated QM/MM Studies of pKa, Redox Reactions and Solvation Free Energies. J Phys Chem B. 2008;113:1253–1272. doi: 10.1021/jp8071712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy M, Erion M. Relative Solvation Free Energies Calculated Using an Ab Initio QM/MM-Based Free Energy Perturbation Method: Dependence of Results on Simulation Length. J Comput Aided Mol Des. 2009;23:837–843. doi: 10.1007/s10822-009-9300-5. [DOI] [PubMed] [Google Scholar]
- 34.Min D, Zheng L, Harris W, Chen M, Lv C, Yang W. Practically Efficient QM/MM Alchemical Free Energy Simulations: The Orthogonal Space Random Walk Strategy. J Chem Theory Comput. 2010;6:2253–2266. doi: 10.1021/ct100033s. [DOI] [PubMed] [Google Scholar]
- 35.Hu H, Yang WT. Free Energies of Chemical Reactions in Solution and in Enzymes with Ab Initio Quantum Mechanics/Molecular Mechanics Methods. Annu Rev Phys Chem. 2008;59(1):573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beutler TC, Mark AE, van Schaik RC, Gerber PR, van Gunsteren WF. Avoiding Singularities and Numerical Instabilities in Free Energy Calculations Based on Molecular Simulations. Chem Phys Lett. 1994;222:529–539. [Google Scholar]
- 37.Banba S, Guo Z, Brooks CL. Efficient Sampling of Ligand Orientations and Conformations in Free Energy Calculations Using the λ-Dynamics Method. J Phys Chem B. 2000;104:6903–6910. [Google Scholar]
- 38.Guo Z, Brooks CL. Rapid Screening of Binding Affinities: Application of the λ-Dynamics Method to a Trypsin-Inhibitor System. J Am Chem Soc. 1998;120:1920–1921. [Google Scholar]
- 39.Guo Z, Durkin J, Fischmann T, Ingram R, Prongay A, Zhang R, Madison V. Application of the λ-Dynamics Method To Evaluate the Relative Binding Free Energies of Inhibitors to HCV Protease. J Med Chem. 2003;46:5360–5364. doi: 10.1021/jm030040o. [DOI] [PubMed] [Google Scholar]
- 40.One in-house developed software for QM/MM MD simulations (http://www.qm4d.info/).
- 41.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 42.Berendsen HJC, Postma JPM, Gunsteren WFV, DiNola A, Haak JR. Molecular Dynamics with Coupling to an External Bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 43.Martyna GJ, Klein ML, Tuckerman M. Nose-Hoover Chains: The Canonical Ensemble via Continuous Dynamics. J Chem Phys. 1992;97:2635–2643. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.