SUMMARY
Tail-anchored (TA) proteins are posttranslationally inserted into either the endoplasmic reticulum (ER) or the mitochondrial outer membrane. The C-terminal transmembrane domains (TMDs) of TA proteins enable their many essential cellular functions by specifying the membrane target, but how cells process these targeting signals is poorly understood. Here, we reveal the composition of a conserved multiprotein TMD recognition complex (TRC) and show that distinct TRC subunits recognize the two types of TMD signals. By engineering mutations in a mitochondrial TMD, we switch over its TRC subunit recognition, thus leading to its misinsertion into the ER. Biochemical reconstitution with purified components demonstrates that TRC tethers and enzymatically activates Get3 to selectively hand off ER-bound TA proteins to Get3. Thus, ER-bound TA proteins are sorted at the top of a TMD chaperone cascade that culminates with the formation of Get3-TA protein complexes, which are recruited to the ER membrane for insertion.
INTRODUCTION
Eukaryotic cells employ a variety of sophisticated targeting pathways to efficiently and accurately deliver newly synthesized proteins to organelles (Cross et al., 2009). Membrane proteins are particularly difficult to target, because their hydrophobic membrane-spanning regions are prone to aggregation and interactions with noncognate targeting factors that lead to their mislocalization in the cell. The signal recognition particle (SRP) pathway solves these problems by transferring membrane-spanning regions from the ribosome directly into the Sec61 protein translocation channel that releases them into the endoplasmic reticulum (ER) lipid bilayer (Rapoport, 2007).
Cotranslational insertion by the SRP pathway, however, is not an option for membrane proteins that have their ER-targeting information contained in a single transmembrane domain (TMD) near the C terminus (Kutay et al., 1995; Yabal et al., 2003). TMDs of these tail-anchored (TA) proteins emerge out of the ribosome exit tunnel only after protein synthesis is completed and are then posttranslationally inserted into the ER with their N-terminal ends facing the cytosol. TA proteins that reside in other compartments of the secretory system are delivered there by vesicular trafficking (Kutay et al., 1995; Bulbarelli et al., 2002). On the other hand, mitochondrial TA proteins have TMD-targeting signals that lead to their direct insertion into the mitochondrial outer membrane (Borgese et al., 2001). A few biologically prominent examples of TA proteins include many of the SNAREs, which enable vesicle fusion; components of the Sec61 and mitochondrial outer membrane translocon; regulators of mitochondrial morphology; and members of the Bcl2 family of apoptosis regulators (Wattenberg and Lithgow, 2001; Beilharz et al., 2003; Rehling et al., 2004; Borgese et al., 2007). Despite their critical roles in many important cellular processes, we still know very little about how TMD signals are processed by cells to accurately and efficiently direct them to the appropriate membrane for insertion.
This question of membrane target specificity has been further complicated by the observation that some TA proteins are able to insert spontaneously into protein-free lipid bilayers (Borgese et al., 2003; Brambillasca et al., 2005). Thus, TA proteins are a uniquely problematic targeting substrate because of their potential to spontaneously insert into biological membranes. The discovery that cholesterol blocks spontaneous insertion in vitro (Brambillasca et al., 2005) suggests an appealing membrane avoidance mechanism that would block TA protein misinsertion into cholesterol-rich membranes in vivo. Additional targeting mechanisms are needed, however, to explain how TA proteins are sorted for insertion into either the ER or the outer mitochondrial membrane, as both of these membranes are cholesterol poor (van Meer, 2005; Colbeau et al., 1971). Recently, several complementary in vivo and in vitro studies have revealed the existence of a conserved guided entry of TA proteins (GET) pathway that targets TA proteins to the ER (Stefanovic and Hegde, 2007; Favaloro et al., 2008; Schuldiner et al., 2008). A key step in this pathway is the formation of membrane-targeting complexes between Get3, a cytosolic ATPase, and newly synthesized TA proteins. Get3-TA protein complexes are subsequently recruited for insertion by the Get1/2 receptor in the ER membrane (Schuldiner et al., 2008). Deletion of any one of the GET genes causes ER-bound TA proteins to misinsert into mitochondria or aggregate in the cytosol (Schuldiner et al., 2008). Similar mutant phenotypes have been observed for two additional cytosolic proteins, Get4 and Get5, but their precise role in the GET pathway remains unclear (Jonikas et al., 2009). The current conceptual framework of the GET pathway explains its strong membrane target specificity for the ER but leaves unanswered the central question of how Get3-TA protein complexes are formed in the first place. Unlike SRP, Get3 does not associate detectably with ribosomes (Stefanovic and Hegde, 2007). Thus, it remains unclear whether Get3 surveys all newly synthesized TMD signals and commits only the ER-bound ones into targeting complexes or whether TMD signals are already sorted at an earlier step in the GET pathway before Get3 recognition takes place.
In the present study, we reveal the composition of a conserved multiprotein TMD recognition complex (TRC) that stands in the GET pathway between TA protein synthesis by ribosomes and Get3 recognition. We show that distinct components of TRC recognize the targeting information of ER-bound versus mitochondrial TMDs. Moreover, engineered mutations in a mitochondrial TMD that switch its recognition between these TRC components result in its misinsertion into the ER membrane. Finally, by reconstituting TA protein handoff from TRC to Get3 with purified components, we demonstrate that TRC acts as an active tethering device that brings Get3 into proximity with its substrates and enzymatically activates Get3 for TMD recognition.
RESULTS
Get4 and Get5 Escort TA Proteins to Get3
A recent global analysis of genes required for protein folding in the ER predicted a role for Get4 and Get5 in the biogenesis of ER-bound TA proteins (Jonikas et al., 2009). This prediction was based on the observation that Δget4 and Δget5 have a pattern of genetic interactions that closely resembles the Δget3 pattern. Additional in vivo and in vitro experiments supported this prediction, but the precise function of Get4 and Get5 remained unclear. As a starting point for this study, we postulated that the reason why Get4 and Get5 are bound to only a small fraction of cytosolic Get3 (Jonikas et al., 2009) is because their function is to deliver TA proteins to Get3. Furthermore, we reasoned based on the absence of Get4 and Get5 from the membrane-bound Get1-Get2-Get3 complex (Jonikas et al., 2009) that following Get4/5-mediated substrate delivery, Get3-TA protein complexes would become free for recruitment to the ER membrane. To test this hypothesis, we in vitro translated (IVT) in the presence of 35S-labeled methionine Sec22, an early secretory pathway TA protein, in GET3FLAG extracts and immunoprecipitated any Get3-Sec22 with anti-FLAG resin. We observed that Get3 bound to Sec22, but not Sec22 lacking the hydrophobic TMD (Figure 1A). Furthermore, fusion of Sec22’s TMD to Sumo (SumoTMD), an otherwise soluble cytosolic protein, resulted in the formation of Get3-SumoTMD (Figure S1A). By comparison, Δget4 and Δget5 extracts had much less Get3-Sec22 (Figure 1B). Next, we extended this immunoprecipitation (IP) approach to monitor Get4 and Get5 coimmunoprecipitation (coIP) with Sec22 and detected TMD-dependent formation of Get4-Sec22 and Get5-Sec22 (Figures 1C, S1A, and S1B). Importantly, both associations persisted in Δget3 extracts (Figure 1C). In parallel experiments (see below), we have shown that under our IP conditions, Get4 and Get5 are in a multiprotein complex with each other and several other proteins. To test if this complex can deliver Sec22 to Get3, we incubated recombinant Get3 (Figure S1C) with Get4FLAG-Sec22 bound to anti-FLAG resin. Strikingly, addition of Get3 resulted in Sec22 elution (Figures 1D and S1D). Moreover, Sec22 elution was abolished by three point mutations in Get3 (Get3LFI: L187E, F190E, and I212E) (Figure S1C) that we engineered in the predicted TMD-binding site (Mateja et al., 2009; Bozkurt et al., 2009; Yamagata et al., 2010) (Figure S1E) Finally, we asked if the eluted Sec22 is in a membrane-targeting complex with Get3 by monitoring insertion into ER-derived membranes (microsomes). To facilitate detection of insertion, we used Sec22 with an engineered N-glycan acceptor site after the TMD that upon insertion gains access to the glycosylation machinery, which is restricted to the ER lumen (Schuldiner et al., 2008). We found that eluted Sec22 was efficiently inserted into microsomes in a Get1/2-dependent manner (Figure 1E). Taken together, these data demonstrate that Get4 and Get5 escort newly synthesized TA proteins to Get3.
Figure 1. Get4 and Get5 Escort TA Proteins to Get3.
(A) In vitro translation (IVT) of Sec22 or Sec22 lacking the transmembrane domain (Sec22Delta;TMD) in the presence of 35S-labeled methionine in GET3 or GET3FLAG extracts. Following anti-FLAG immunoprecipitation (IP) and 3× FLAG peptide elution as shown in the accompanying schematic, 3% of total (T) and 30% of elution (E) were resolved by SDS-PAGE and analyzed by autoradiography. Dashed lines in all the figures signify that the data came from the same gel.
(B and C) In vitro translation (IVT) of Sec22 in the indicated extracts was followed by anti-FLAG immunoprecipitation (IP) and 3× FLAG peptide elution. Samples were prepared and analyzed by autoradiography and immunoblotting (IB) as in (A).
(D) In vitro translation (IVT) of Sec22 in GET4FLAG Δget3 extract was followed by anti-FLAG immunoprecipitation (IP) and elution with recombinant Get3 (0.4 μg/μl) for 20 min at room temperature or mock treatment, as shown in the accompanying schematic. Following centrifugation, the eluted material was resolved by SDS-PAGE and analyzed by autoradiography. Elution efficiency is defined as the percentage of immobilized Sec22 input (determined by elution with SDS-PAGE loading buffer) that was eluted by Get3.
(E) Get3 elution from (D) was split and incubated with wild-type (WT) or Δget1/2 microsomes for 30 min at room temperature or mock treated. Samples were resolved by SDS-PAGE and analyzed by autoradiography. The positions of Sec22 and glycosylated Sec22 (gSec22) are indicated. Sec22 contains a C-terminal Opsin tag with an N-glycan acceptor site that facilitates detection of correctly inserted Sec22, as shown in the accompanying schematic. Insertion efficiency is defined as the percentage of total Sec22 that is glycosylated. Note that some Sec22 may be correctly inserted but not glycosylated.
Sgt2 Is a Scaffold Protein that Brings Get4 and Get5 into Proximity with TA Proteins
Previous studies have shown that Get4 and Get5 bind to each other and to several other proteins (Ito et al., 2001; Krogan et al., 2006; Liou and Wang, 2005; Liou et al., 2007; Chang et al., 2010). This raises the possibility that one or more proteins tightly bound to Get4/5 enable their coIP with Sec22. We will now present a systematic biochemical analysis, which established that both Get4-Sec22 and Get5-Sec22 formation depends on Sgt2 (small glutamine-rich tetratricopeptide repeat-containing protein 2), a scaffold protein with the following topology: an N-terminal domain that binds Get4 and Get5, a central tetratrico-peptide repeat (TPR) domain that binds heat shock proteins (Hsps), and a C-terminal domain that binds TA proteins.
Affinity purification followed by mass spectrometry showed that Get4 and Get5 exist in a complex with each other, as well as Hsp104, Hsp70s, Sgt2, and Ybr137w (Figure S2A). We detected relatively few Get3-derived peptides in our preparations, suggesting that Get3 is not an integral component of this complex. To exclude the possibility that Get3 detection by mass spectrometry was obscured for technical reasons, we repeated the purifications in a Δget3 genetic background, but we observed no apparent change in the molecular composition of the complex (Figure 2A). Several extensions of this approach enabled us to define the subunit organization of the complex. First, in the absence of Sgt2, Get5 remained bound only to Get4 (Figure 2A). Second, in the absence of Get5, Get4 was not bound to any of the other complex components (Figure 2A). Third, in the absence of Get4, Get5’s apparent association with Sgt2 (and consequently Hsps and Ybr137w; see below) was diminished (Figures 2A and S2B) due to partial Get5 proteolysis under these conditions (Figures S2B and S2C). Fourth, Hsp and Ybr137w binding to the rest of the complex was abolished by two point mutations (R171A and R175A) in the TPR domain of Sgt2 (Figures S2D and S3A) that, on the basis of structural homology modeling (Chivian and Baker, 2006) (Figure S2E), we expected would disrupt critical electrostatic interactions with the acidic C-terminal tails of Hsps (Scheufler et al., 2000) (Figures S3B). Lastly, in the absence of Ybr137w, Get5 remained bound to all the other complex components (Figure 2A). In summary, Get4 and Get5 are part of a multi-protein complex in which Get5 mediates the association between Get4 and Sgt2, while the TPR domain of Sgt2 mediates the association of Hsp104, Hsp70s, and Ybr137w with the rest of the complex (Figure 2B).
Figure 2. Sgt2 Is a Scaffold Protein that Binds Get4/5, Heat Shock Proteins, and TA Proteins.
(A) Cytosolic lysates from the indicated FLAG-tagged strains were immunoprecipitated (IP) with anti-FLAG resin. Following washing, the resin was eluted with 3× FLAG peptide. Eluted proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. Protein identities were established by mass spectrometry analysis (Figure S2A). Note that the Get5FLAG band from the Δget4 preparation is difficult to see because of partial proteolysis (Figure S2B).
(B) Schematic showing the subunit organization of the affinity-purified Get4/5FLAG cytosolic complex. The presence of multiple acidic residues in the C-terminal tail of Ybr137w suggests that like Hsp104 and Hsp70, it interacts directly with the TPR domain of Sgt2. The schematic is not intended to specify the copy number of each component in the complex.
(C and D) In vitro translation (IVT) of Sec22 or Sec22ΔTMD in the indicated extracts was followed by anti-FLAG immunoprecipitation (IP) and 3× FLAG peptide elution. Samples were prepared and analyzed by autoradiography and immunoblotting (IB) as in Figure 1.
(E) Cytosolic lysates from the indicated FLAG-tagged strains were immunoprecipitated (IP) with anti-FLAG resin. Following washing, the resin was eluted with 3× FLAG peptide. Eluted proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. Protein identities were established by mass spectrometry analysis. * indicates the position of a protein fragment derived from Sgt2.
Next, we asked if Sec22 can bind to Get4 and Get5 when they are no longer bound to the rest of the complex, but we observed no detectable Get5-Sec22 formation in Dsgt2 extracts (Figure 2C). As most of the cellular Get4/5 is in a complex with Sgt2 (Figure S3C), we postulated that Sgt2 is required for Get5-Sec22 formation, because Sec22 was in fact bound to one of the other components of the complex in our original Get4 and Get5 immunoprecipitates (see Figure 1C). To test this, we first established that we could detect formation of Sgt2-Sec22 in a TMD-dependent manner (Figure 2D). Importantly, Sgt2-Sec22 persisted in Δget3, Δget5, and Δget3/5 extracts (Figure 2D). To determine if Sgt2 binding to Hsps and Ybr137w is required for Sec22 coIP with Sgt2, we monitored Sgt2-Sec22 formation in the SGT2R171A, R175AFLAG Δget5 extract. Surprisingly, even though Sgt2 was no longer bound to Get4, Get5, Hsps, and Ybr137w under these conditions (Figure 2E), Sgt2-Sec22 still persisted (Figure 2D).
If Hsps are not required for Sgt2-Sec22 formation, how then does Sgt2 recognize the hydrophobic TMD of Sec22? Sequence alignment of distant Sgt2 homologs revealed that the central TPR domain is flanked by less conserved N- and C-terminal domains (Dutta and Tan, 2008). Intriguingly, we noticed that the C-terminal domains of Sgt2 homologs are rich in methionine residues (Figure 3A), a feature they share with the domain of SRP that recognizes hydrophobic signal sequences (Keenan et al., 1998; Clérico et al., 2009; Janda et al., 2010). Therefore, we asked if the C-terminal domain of Sgt2 is required for Sec22 binding by monitoring Sgt2-Sec22 formation in extracts expressing truncations of Sgt2 (Figure S3D). We observed that deletion of the C-terminal domain (Sgt2ΔC) abolished Sec22 binding, while deletion of the first 58 amino acids (Sgt2ΔN), although critical for Get4/5 binding (see next section), were not required for Sgt2-Sec22 formation (Figure 3B). Lastly, to establish if Sgt2 recognition of Sec22 can occur in the absence of any other yeast proteins, we coexpressed in bacteria Sgt2 with SumoTMD. This fusion strategy is commonly used to enhance the solubility of proteins overexpressed in bacteria (Butt et al., 2005) and didn’t interfere with the insertion of the TMD into microsomes (Figure S3E). In full agreement with our yeast coIP studies, Sgt2 and Sgt2ΔN formed a complex with SumoTMD in bacteria, whereas Sgt2ΔC did not (Figure 3C).
Figure 3. The Methionine-Rich C-Terminal Domain of Sgt2 Binds to the Hydrophobic Transmembrane Domains of TA Proteins.
(A) Schematic of the three protein domains of Sgt2. The bracketed regions indicate the span of the indicated Sgt2 truncations. Shown below is the Clustal W2 amino acid sequence alignment of the C-terminal domains of Sgt2 homologs from Saccharomyces cerevisiae (NP_014649.1), Homo sapiens (NP_003012.1), Caenorhabditis elegans (NP_494893.1), and Drosophila melanogaster (NP_609842.1). ESPript 2.0 was used to highlight identical (white in color, boxed with red), well-conserved (red in color, boxed in white), and methionine residues (shaded blue). Note that the percent of methionines in the C-terminal domain of S. cerevisiae Sgt2 (residues 208 through 346) is 7.2%, compared to an average of 7.7% for the other three Sgt2 C-terminal domains. The average percent of methionines in the M domain of SRP homologs (see Figure 5 in Keenan et al., 1998) is 2.3%.
(B) In vitro translation (IVT) of Sec22 in the indicated extracts followed by anti-FLAG immunoprecipitation (IP) and 3× FLAG peptide elution. Ten percent of the IP input (T) and 20% of 3× FLAG peptide elution (E) were resolved by SDS-PAGE and analyzed by autoradiography and immunoblotting (IB).
(C) Cytosolic lysates (T) from the indicated E. coli strains were immunoprecipitated (IP) with Ni-NTA resin. Following washing, the resin was eluted with imidazole (E). Eluted proteins were resolved by SDS-PAGE and visualized by immunoblotting (IB), as indicated.
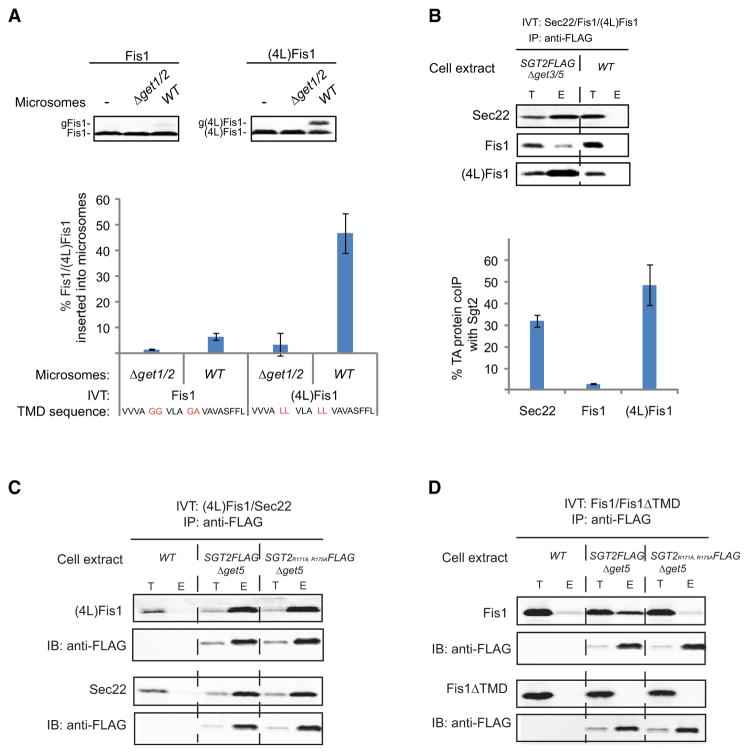
To test if Sgt2 can distinguish between ER and mitochondrial TMD targeting determinants, we took advantage of the observation that substitution of four leucines (4L) at specific positions in the TMD of Fis1, a mitochondrial TA protein, causes it to mislocalize Fis1 to the ER in vivo (Beilharz et al., 2003). Consistent with this, (4L)Fis1 inserted into microsomes in a Get1/2-dependent manner ten times more efficiently than Fis1 (Figure 4A), thus making its insertion efficiency comparable to that of Sec22. This change in Fis1 membrane targeting was mirrored by a 10-fold increase in the efficiency of Fis1 coIP with Sgt2, thus making it comparably efficient to Sgt2-Sec22 formation (Figure 4B). As expected, Sgt2-(4L)Fis1 persisted in SGT2FLAG Δget5 and SGT2R171A, R175AFLAG Δget5 extracts (Figure 4C), arguing that the (4L)Fis1 TMD is being recognized by Sgt2. Unexpectedly, TPR mutations completely abolished Sgt2-Fis1 formation (Figure 4D), which suggests that Fis1 TMD is indirectly associating with Sgt2 by binding to Hsps. Importantly, Sgt2-Fis1 formation was still TMD dependent under these conditions (Figure 4D). Collectively, our findings establish that the methionine-rich C-terminal domain of Sgt2 selectively recognizes the hydrophobic targeting determinants of the subset of TA proteins that are bound for the ER.
Figure 4. Sgt2 Selectively Recognizes the Targeting Determinants of TA Proteins Destined for the Secretory Pathway.
(A) In vitro translation (IVT) of Fis1 or (4L)Fis1 in WT extract was followed by incubation with either WT or Δget1/2 microsomes for 30 min at room temperature. A C-terminal Opsin tag with an N-glycan acceptor site was used to monitor insertion (see Figure 1E). Samples were resolved by SDS-PAGE and analyzed by autoradiography (shown is a representative analysis). The positions of Fis1/(4L)Fis1 and glycosylated Fis1/(4L)Fis1 (gFis1/g(4L)Fis1) are indicated. Shown graphically are the average and standard deviation (three independent experiments) of the percentage of Fis1 and (4L)Fis1 inserted. Also shown highlighted in red are the four leucine substitutions in the (4L)Fis1 transmembrane domain (TMD) sequence.
(B) The indicated TA proteins were in vitro translated to comparable final concentrations in SGT2FLAG Δget3/5 extract (data not shown). Following anti-FLAG immunoprecipitation and 3× FLAG peptide elution, total (T) and eluted (E) samples were resolved by SDS-PAGE and analyzed by autoradiography (shown is a representative analysis) and immunoblotting with anti-FLAG (data not shown). Shown graphically are the average and standard deviation (three independent experiments) of each TA protein’s coimmunoprecipitation (coIP) efficiency with Sgt2.
(C and D) In vitro translation (IVT) of the indicated TA proteins in the indicated extracts was followed by anti-FLAG immunoprecipitation (IP) and 3× FLAG peptide elution. Samples were prepared and analyzed by autoradiography and immunoblotting (IB) as in Figure 1.
Reconstitution of TA Protein Handoff from Sgt2 to Get3 with Purified Components
To determine the precise role of Sgt2 in TA protein biogenesis, we first established that Dsgt2 extracts have a defect in Sec22 insertion into microsomes, a hallmark of get deletion extracts (Figure S4A). In contrast, Sec22 insertion was unaffected in Dybr137w (Figure S5B) and SGT2R171A, R175A extracts (data not shown). Next, we incubated Sgt2-Sec22 purified from Δget3/5 extract with microsomes but observed no detectable Sec22 insertion (Figure 5A). Under the same conditions, addition of recombinant Get3 resulted in only trace amounts of inserted Sec22 (Figure 5A). Strikingly, incubation of microsomes with Sgt2-Sec22, preformed recombinant Get4-Get5 complexes (Figure S1C), and Get3 resulted in efficient, Get1/2-dependent Sec22 insertion (Figure 5A). Next, we tested if formation of Get3-Sec22 was occurring by an active Sec22 transfer mechanism from Sgt2 to Get3. Thus, we incubated Sgt2FLAG-Sec22 bound to anti-FLAG resin with Get3, Get4, and Get5, either individually, in pairwise combinations, or all three combined, and monitored Sec22 elution. Remarkably, we observed nearly quantitative elution of Sec22 from Sgt2 in the combined presence of Get3, Get4, and Get5 (Figure 5B). All the other incubations containing Get3 resulted in only marginal Sec22 elution (Figure 5B), which is in agreement with the low, but above background, level of Sec22 insertion into microsomes incubated with Sgt2-Sec22 and Get3. The remaining elutions, which contained Get4 and/or Get5 without Get3, were indistinguishable from background (Figure 5B). Three additional lines of evidence strongly argue that Sec22 eluted with Get3/4/5 is in a membrane-targeting complex with Get3. First, Sec22 quantitatively immunoprecipitated with Get3 following elution (Figure S4C) and could then be efficiently inserted into microsomes in a Get1/2-dependent manner (Figure S4D). Second, Sec22 elution was abolished by three point mutations (Get3LFI: L187E, F190E, and I212E) in Get3’s predicted TMD-binding site (Mateja et al., 2009; Bozkurt et al., 2009; Yamagata et al., 2010) (Figure 5C). Third, (4L)Fis1 was eluted with an efficiency comparable to that observed for Sec22, while Fis1 was not appreciably eluted under the same conditions (Figure 5D). Lastly, we asked if Sgt2-associated Hsps and Ybr137w are required for Get3-Sec22 formation under these conditions but observed no apparent decrease in Sec22 elution from Sgt2R171A, R175A (Figure S4E). These data clearly demonstrate that our reconstituted in vitro system is an excellent tool for dissecting the mechanism of Get4/5-facilitated TA protein handoff from Sgt2 to Get3.
Figure 5. In Vitro Reconstitution of TA Protein Handoff from Sgt2 to Get3.
(A) In vitro translation (IVT) of Sec22 in SGT2FLAG Δget3/5 extract followed by anti-FLAG immunoprecipitation (IP) and 3× FLAG peptide elution. Elution was split and incubated for 20 min at room temperature with Get proteins (Get3 at 35 ng/μl and Get4-Get5 at 70 ng/μl) or mock treated. This was followed by incubation for 20 min at room temperature with GET1/2 or Δget1/2 microsomes, as indicated. To minimize any potential microsome contamination by cytosolic Sgt2, Get3, and Get5, GET1/2 microsomes were prepared from the Δsgt2&; Delta get3/5 strain. The positions of Sec22 and glycosylated Sec22 (gSec22) are indicated (see Figure 1E).
(B) In vitro translation (IVT) of Sec22 in SGT2FLAG Δget3/5 extract was followed by anti-FLAG immunoprecipitation (IP) and elution with the indicated Get proteins (all at 64 ng/μl) for 20 min at room temperature or mock treatment, as shown in the accompanying schematic. Following centrifugation, elutions were collected, the resin washed, and eluted again, but this time with gel loading buffer (resin after elution). The elutions were resolved by SDS-PAGE and analyzed by autoradiography. The data are separately boxed on the left and right to signify that they came from experiments performed on different days.
(C and D) In vitro translation (IVT) of the indicated TA proteins in SGT2FLAG Δget3/5 extract was followed by anti-FLAG immunoprecipitation (IP) and elution with the indicated Get proteins for 20 min at room temperature. Samples were prepared and analyzed by autoradiography as in (B).
Get4 and Get5 Employ a Dual Mechanism to Facilitate TA Protein Handoff
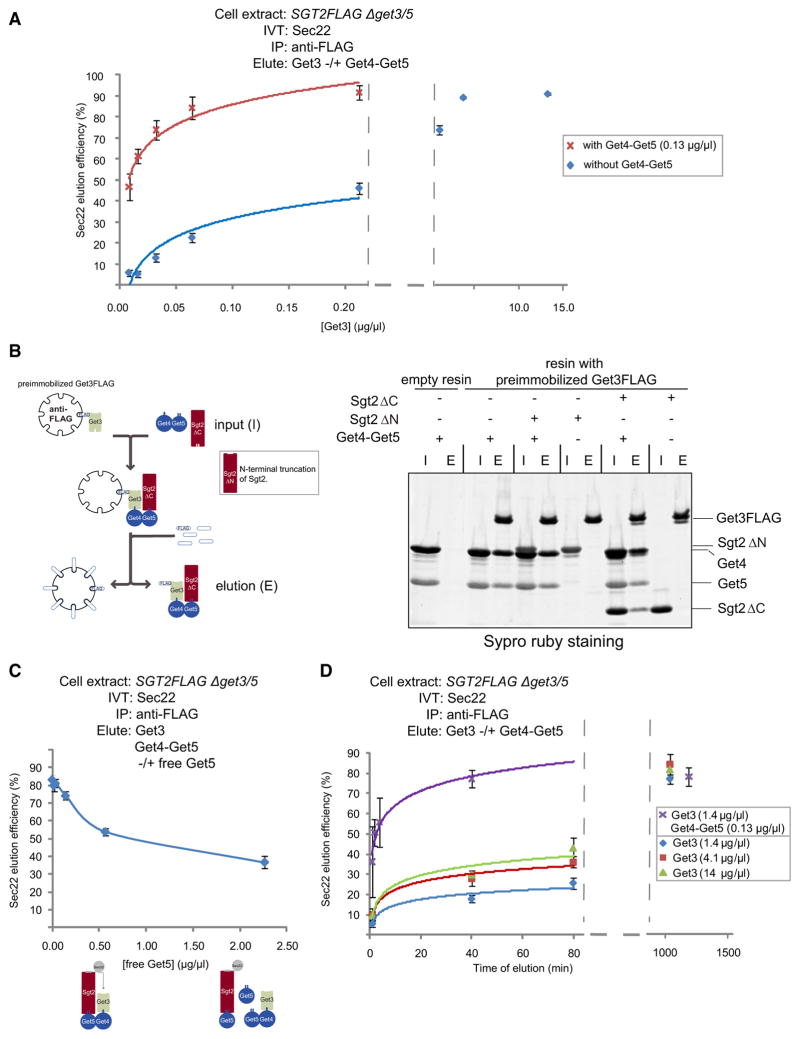
To elucidate the mechanism by which Get4 and Get5 facilitate Get3-Sec22 formation, we first asked if higher concentrations of Get3 alone could drive Sec22 release from Sgt2. Indeed, increasing concentrations of Get3 stimulated Sec22 elution from immobilized Sgt2FLAG until very little Sec22 remained on the resin (Figure 6A). Even at these relatively high Get3 concentrations, Sec22 elution was still dependent on a functional Get3 TMD-binding site (Figure S5A). Next, we monitored the Get3 concentration dependence of Sec22 elution in the presence of Get4-Get5 and observed that half-maximal elution was achieved at a Get3 concentration approximately 25 times lower (Figure 6A), suggesting a close cooperation between these components. A plausible explanation for how Get4 and Get5 sensitize Sec22 elution under these conditions is that they tether Get3 to Sgt2, thus increasing the local concentration of Get3 near its substrate. To test this idea, we monitored Get3 binding to Sgt2 in vitro. First, we expressed and purified recombinant Sgt2ΔC and Sgt2ΔN but were unsuccessful in obtaining full-length Sgt2 of similarly high quality (data not shown). Nonetheless, our gel filtration experiments established that Get4-Get5 forms a stable complex with Sgt2ΔC but not Sgt2ΔN (Figure-S5B), consistent with the previous finding that Get5 interacts with the N terminus of Sgt2 (Liou et al., 2007). Next, we incubated Sgt2ΔC or Sgt2ΔN with recombinant Get3FLAG immobilized on anti-FLAG resin but detected no appreciable binding (Figure 6B). Importantly, Get4-Get5 bound to Get3 under identical conditions (Figure 6B). In support of the idea that Get4-Get5 increases the local concentration of Get3 near Sgt2 during Get3-Sec22 formation, Sgt2ΔC but not Sgt2ΔN formed a complex with Get3-Get4-Get5 (Figure 6B). To test the functional relevance of this tethering interaction, we reasoned that excess Get5 should inhibit Get3-Sec22 formation by sequestering Get3 and Sgt2 into separate complexes. In agreement with this prediction, we observed that increasing concentrations of Get5 progressively reduced the efficiency of Sec22 elution (Figure 6C). Finally, we reasoned that if Get3 were being passively tethered to Sgt2 during substrate handoff, Get4-Get5 should only stimulate the rate of Get3-Sec22 formation under conditions when Get3 is limiting. In contrast, we observed that even at saturating concentrations of Get3, the rate of Sec22 elution was further increased by the addition of Get4-Get5 (Figure 6D). In summary, our data strongly argue that Get4 and Get5 use a dual mechanism to facilitate the biogenesis of ER-bound TA proteins. First, they tether Get3 into proximity with TA proteins held by Sgt2. Second, they catalytically hasten the assembly of Get3 membrane-targeting complexes from tethered Get3 and Sgt2-held TA proteins.
Figure 6. Get4 and Get5 Facilitate TA Protein Handoff by a Dual Mechanism.
(A) In vitro translation (IVT) of Sec22 in SGT2FLAG Δget3/5 extract was followed by anti-FLAG immunoprecipitation (IP) and elution at different concentrations of Get3 in either the presence or the absence of Get4-Get5 for 20 min at room temperature. Shown are the average and standard deviation (two independent experiments) of percentage Sec22 eluted at different Get3 concentrations (starting at 8 ng/μl). Note that the data for Get3 concentrations after the axis break were not used in the curve fitting and that the background elution following mock treatment is ~4%. A separate hyperbolic fitting (which included the background elution and elution data at all the Get3 concentrations; see Experimental Procedures) yielded a half-maximal Get3 concentration of 0.23 μg/μl (without Get4/5) and 0.0092 μg/μl (with Get4/5).
(B) Anti-FLAG resin preimmobilized with Get3FLAG or empty resin was incubated with the indicated recombinant proteins and eluted with 3× FLAG peptide, as shown in the accompanying schematic. Input (I) and elutions (E) were resolved by SDS-PAGE and visualized by SYPRO Ruby staining.
(C) In vitro translation (IVT) of Sec22 in SGT2FLAG Δget3/5 extract was followed by anti-FLAG immunoprecipitation (IP) and elution at different concentrations of free Get5 in the presence of Get3 and preformed Get4-Get5 complex. Samples were prepared and analyzed as in Figure 5B. Shown are the average and standard deviation (two independent experiments) of percentage Sec22 eluted at different concentrations of free Get5. Also shown is a schematic of the sequestration of Sgt2 and Get3 into separate complexes in the presence of excess Get5.
(D) In vitro translation (IVT) of Sec22 in SGT2FLAG Δget3/5 extract was followed by anti-FLAG immunoprecipitation (IP) and elution with the indicated Get proteins. Notably, elution was carried out at 4°C to slow down the kinetics of elution (data not shown) and allow enough time for sample processing. Samples were prepared and analyzed as in Figure 5B. Shown are the average and standard deviation (two independent experiments) of the percentage Sec22 eluted at different times following the addition of Get proteins to resin-immobilized Sgt2. Note that data at time points after the axis break were not used in the curve fitting.
DISCUSSION
The fact that certain TA proteins can insert into protein-free lipid bilayers has led to the evocative proposal that they are a vestige of a spontaneous membrane insertion world (Borgese et al., 2007). Along the same lines, it has been argued that TA proteins enabled the evolutionary elaboration of intracellular membrane-bound compartments, as evidenced by their presence in the modern protein translocation machineries of the ER and mitochondrial outer membrane (Borgese et al., 2007). Moreover, the very identity of membrane compartments along the secretory/endocytic system is dependent on SNAREs, most of which are TA proteins. At the same time, the existence of numerous organelles presents eukaryotic cells with the challenge of preventing TA proteins from inserting into membrane off-targets. While the relatively high cholesterol content of certain organelles may help prevent TA protein misinsertion into them (Brambillasca et al., 2005), additional mechanisms are required to sort TA proteins for insertion into the inherently permissive lipid bilayers of the ER and mitochondrial outer membrane (Borgese et al., 2007).
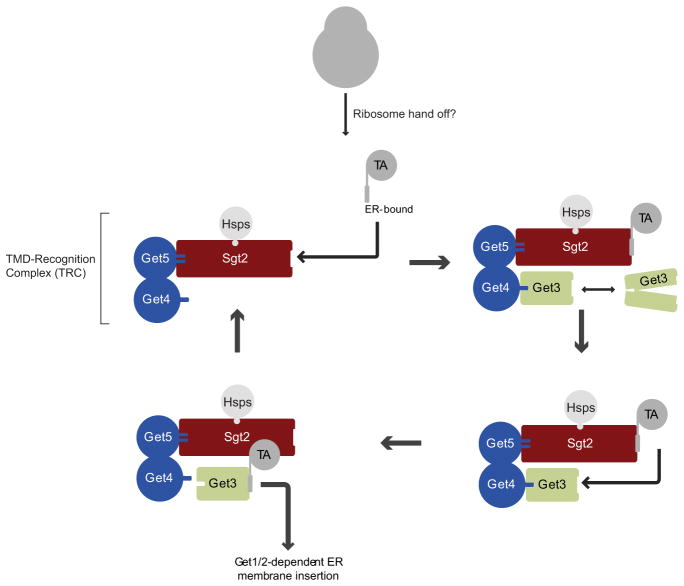
In the present study, we have uncovered a mechanism that efficiently and accurately sorts TA proteins for insertion into the ER (Figure 7). Specifically, we have shown that a multiprotein TRC (according to the nomenclature of Stefanovic and Hegde, 2007) escorts newly synthesized TA proteins to Get3. Sgt2 is the component of TRC that recognizes the targeting determinants of ER-bound TMDs with its C-terminal domain. We speculate, by analogy with SRP (Keenan et al., 1998; Janda et al., 2010), that the high frequency of hydrophobic, flexible methionine side chains in the C-terminal domain of Sgt2 enables the structural plasticity necessary for the recognition of the diverse TMD sequences of ER-bound TA proteins. TRC also associated with mitochondrial TA proteins, but in this case TMD recognition was mediated by a distinct TRC component that is bound to the TPR domain of Sgt2 and very likely corresponds to Hsp70. While Sgt2 is not absolutely required for TA protein targeting to mitochondria (see below), the role of Hsp70s in this process should be explored further. In an effort to understand how TRC segregates recognition of ER-bound from mitochondrial TA proteins, we have shown that 4L substitutions in a mitochondrial TMD were sufficient to switch over its recognition to Sgt2 and in doing so reprogram it for sorting to the ER. Further supporting the idea that Sgt2 TMD recognition is a key decision point in TA protein sorting is a recent large-scale analysis of the genetic landscape of the cell (Costanzo et al., 2010), which found that Pex15, a TA protein client of the GET pathway (Schuldiner et al., 2008; Jonikas et al., 2009), is mislocalized to mitochondria in Dsgt2 cells. Future structural and biophysical studies should help reveal the precise molecular basis of Sgt2 TMD selectivity.
Figure 7. Model of the TMD Chaperone Cascade for Sorting TA Proteins.
See Discussion for details.
Hsp104/70/40 are TRC components associated with Sgt2’s TPR domain. In several other contexts, these chaperones form a disaggregase system capable of harnessing the energy of ATP hydrolysis into a mechanical force that unfolds aggregated substrates by threading them through the narrow central pore of Hsp104’s hexameric ring (Haslberger et al., 2010). Intriguingly, earlier studies have shown that TA proteins form cytosolic aggregates in get mutants (Schuldiner et al., 2008; Jonikas et al., 2009). Moreover, cellular perturbations that partially inactivate Get3, such as redox stress (Favaloro et al., 2008), are likely to make newly synthesized TA proteins more aggregation prone. TRC has all the components necessary to buffer such insults on TA protein homeostasis by salvaging aggregated TA proteins for insertion. Recent technical advances in monitoring Hsp104 activity in vivo (Tessarz et al., 2008) now enable testing of this idea.
Following TRC recognition, ER-bound but not mitochondrial TA proteins are efficiently handed off to Get3. Two additional TRC components, Get4 and Get5, facilitate this process by tethering Get3 to Sgt2 and, additionally, enhancing the rate of Get3-TA protein complex formation. Uncovering the precise mechanistic basis of this latter enzymatic effect is an important future goal. While we didn’t observe a stable association between TMD and Get4/5, it is still possible that Get4/5 form an active TMD conduit between Sgt2 and Get3 during handoff. Another, not mutually exclusive, possibility is that Get4/5 convert Get3 into a more receptive state for TMD binding. Interestingly, several recent crystallography studies have captured the Get3 dimer in several different conformations (Mateja et al., 2009; Suloway et al., 2009; Hu et al., 2009; Bozkurt et al., 2009; Yamagata et al., 2010). In the closed conformation, the Get3 subunits are juxtaposed to form a large hydrophobic groove. This groove is disrupted, however, in other, open crystallographic forms of Get3. Here, we have shown that point mutations in this groove abolished TA protein handoff from TRC to Get3. Thus, Get4 and Get5 may facilitate Get3-TA protein complex formation by stabilizing Get3 in its closed conformation.
The TA protein sorting mechanism described here in yeast is very likely to be conserved in metazoans. Indeed, the biochemical identification of the mammalian Get3 homolog, Asna1, as the ER-targeting factor for Sec61β, an ER TA protein, pointed to the existence of additional proteins associated with Asna1 (Stefanovic and Hegde, 2007). While the identities of these proteins and their precise role in TA protein targeting remain to be determined, the human homologs of Get4 (C7orf20), Get5 (Ubl4A), and Sgt2 (α and βSGT) are now a list of possible candidates for further testing.
A previous study has shown that bacterial coexpression of Get3 with a TA protein results in the formation of Get3-TA protein targeting complexes (Bozkurt et al., 2009). Given that bacteria lack apparent homologs of Get4, Get5, and Sgt2, why do eukaryotic cells rely on TRC for the delivery of TA proteins to Get3? Many mitochondrial TMDs are very similar to ER-bound TMDs on the basis of hydrophobicity and flanking charge (Borgese et al., 2007). While the precise enzymological framework of the TA protein handoff from TRC to Get3 remains to be established, double checking of the targeting information by Sgt2 and Get3 has the potential to amplify the fidelity of TA protein sorting beyond the theoretical thermodynamic limit of Get3 recognition (Fersht, 1999). Another possibility is that ER-bound TA protein misinsertion into mitochondria and aggregation in the cytosol are such kinetically fierce off-pathways that they necessitate an elaborate TMD chaperone cascade extending from the ribosome to the ER lipid bilayer. Intriguingly, a mass spectrometry study reported that Get5 is ribosome associated (Fleischer et al., 2006). Our study now paves the way for biochemical efforts aimed at defining the path of TA protein travel from the ribosome to TRC.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Sec22 and Fis1 in vitro transcription plasmids were described previously (Schuldiner et al., 2008). The construction of the other plasmids used in this study is described in the Supplemental Experimental Procedures.
Recombinant Protein Expression and Purification
Unless otherwise indicated, pET-based bacterial expression was done by IPTG induction in BL21 DE3 E. coli cells as detailed in the Supplemental Experimental Procedures along with the description of the ensuing purification steps.
Saccharomyces cerevisiae Strain Construction
The construction and genotypes of all the yeast strains used in this study are described in the Supplemental Experimental Procedures and listed in Table S1 therein.
Large-Scale Native FLAG IP
Yeast cells grown to late-log phase (OD600 ~1.6) were lysed, frozen by ball milling (Retch PM 100), and subjected to anti-FLAG affinity purification as described previously (Jonikas et al., 2009).
Get3 FLAG Pull-Down
Recombinant Get3FLAGHis (6.5 μg) was incubated with 35 μl anti-FLAG-M2 affinity gel slurry (Sigma) pre-equilibrated in binding buffer (50 mM HEPES-KOH [pH 6.8], 10 mM Mg[OAc]2, 150 mM NaCl, 2 mM DTT, 10% glycerol, 3 mM ATP) for 30 min at 4°C with agitation. The resin was then washed three times with 200 μl ice-cold binding buffer before incubating it with 4× molar excess of input proteins, indicated in Figure 6B (75 μl final volume in binding buffer), for 30 min at 4°C. Following three binding buffer washes, bound proteins were eluted with 1 mg/ml 3× FLAG peptide (Sigma), resolved by SDS-PAGE, and visualized by SYPRO Ruby (Invitrogen) staining using a Typhoon imaging system with ImageQuant TL (GE) software.
Immunoblotting
Following semidry transfer (Bio-Rad; Hercules, CA) onto nitrocellulose membranes and blocking (LI-COR; Lincoln, NE), immunoblots were incubated with primary antibodies in TBST (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) with 5% milk as follows: anti-FLAG mouse M2 monoclonal (Sigma) at 1:2000 dilution; anti-His mouse monoclonal (QIAGEN), 1:500; anti-Opsin mouse monoclonal (Sigma), 1:1000; rabbit anti-Hexokinase (US Biologicals; Swampscott, MA), 1:20,000. Following washing in TBST, immunoblots were further incubated with fluorescently labeled secondary antibodies in TBST with 5% milk as follows: Cy5 goat anti-mouse IgG (Invitrogen), 1:10,000; Alexa Fluor goat anti-rabbit IgG (Invitrogen), 1:10,000. After extensive TBST washing and a PBS rinse, western detection was carried out on a Typhoon imaging system with ImageQuant TL (GE) software.
In Vitro Transcription, Translation, and Microsome Insertion
Capped mRNAs were in vitro transcribed from the appropriate PCR products using the mMESSAGE mMACHINE T7 kit (Ambion; Austin, TX) as described previously (Schuldiner et al., 2008).
Yeast in vitro translation (IVT) extracts and translocation-competent, ER-derived membrane (microsomes) were prepared as described before (Schuldiner et al., 2008). One notable exception was that after the low-speed spin, lysates for IVT were centrifuged in a Sw55 Ti rotor for 30 min at 49,000 rpm.
IVT and insertion were performed as described before (Schuldiner et al., 2008). Insertion efficiency was determined by phosphorimager analysis using a Typhoon imaging system with ImageQuant TL software (GE).
Native FLAG IP and Elution Following In Vitro Translation
Anti-FLAG M2 affinity gel slurry (5 μl) equilibrated in IP buffer (20 mM HEPES-KOH [pH 7.4], 2 mM Mg[OAc]2, 100 mM KOAc, 2 mM DTT, 14% glycerol, 1× Roche complete protease inhibitors cocktail, 3 mM ATP) was incubated with 15 μl IVTs at 4°C for 30 min with agitation. Following washing with ice-cold IP buffer, the resin was eluted by one of two methods: FLAG peptide elution, 15 μl 0.75 mg/ml 3× FLAG peptide in IP buffer for 30 min at 4°C with agitation; Get protein elution, 15 μl final volume in IP buffer with recombinant Get protein concentrations, incubation times, and elution temperatures as described in the figure legends. Half-maximal Get3 concentrations in Figures S1D and 6A were calculated by fitting the data to a nonlinear regression hyperbola equation, y = y0 + a * x/(b + x) (SigmaPlot 10.0), to determine the b value.
Supplementary Material
Acknowledgments
We thank W. Lane and the Harvard Microsequencing Facility for mass spectrometry analysis; M. Tung for generating the Get3 TMD-binding mutant; Z. Newby for experimental assistance on recombinant Get4 and Get5 expression; E. O’Shea for allowing us the extensive use of her equipment; M. Rust, J. Markson, A. Rizvi, N. Bradshaw, A. Whynot, B. Burton, A. Drummond, L. Colwell, and K. Foster for useful discussions; B. Stern, E. O’Shea, and members of the Denic lab for critical reading of the manuscript; and B. Toyama for generous help with graphics. V.D. wishes to thank J. Weissman and A. Murray for encouragement and stimulating discussions. Harvard University supported this work.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, Supplemental References, five figures, and one table and can be found with this article online at doi:10.1016/j.molcel.2010.08.038.
References
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Borgese N, Gazzoni I, Barberi M, Colombo S, Pedrazzini E. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol Biol Cell. 2001;12:2482–2496. doi: 10.1091/mbc.12.8.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, Wild K, Bange G, Favaloro V, Rippe K, Hurt E, Dobberstein B, Sinning I. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA. 2009;106:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, Hegde RS, Borgese N. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulbarelli A, Sprocati T, Barberi M, Pedrazzini E, Borgese N. Trafficking of tail-anchored proteins: transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells. J Cell Sci. 2002;115:1689–1702. doi: 10.1242/jcs.115.8.1689. [DOI] [PubMed] [Google Scholar]
- Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Chuang YC, Ho YC, Cheng MY, Sun YJ, Hsiao CD, Wang C. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 2006;34:e112. doi: 10.1093/nar/gkl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clérico EM, Szymanska A, Gierasch LM. Exploring the interactions between signal sequences and E. coli SRP by two distinct and complementary crosslinking methods. Biopolymers. 2009;92:201–211. doi: 10.1002/bip.21181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeau A, Nachbaur J, Vignais PM. Enzymic characterization and lipid composition of rat liver subcellular membranes. Biochim Biophys Acta. 1971;249:462–492. doi: 10.1016/0005-2736(71)90123-4. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Dutta S, Tan YJ. Structural and functional characterization of human SGT and its interaction with Vpu of the human immunodeficiency virus type 1. Biochemistry. 2008;47:10123–10131. doi: 10.1021/bi800758a. [DOI] [PubMed] [Google Scholar]
- Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W.H. Freeman; 1999. pp. 377–400. [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslberger T, Bukau B, Mogk A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol. 2010;88:63–75. doi: 10.1139/o09-118. [DOI] [PubMed] [Google Scholar]
- Hu J, Li J, Qian X, Denic V, Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS ONE. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Li J, Oubridge C, Hernández H, Robinson CV, Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou ST, Wang C. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Arch Biochem Biophys. 2005;435:253–263. doi: 10.1016/j.abb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Liou ST, Cheng MY, Wang C. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones. 2007;12:59–70. doi: 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N. Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Suloway CJ, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenberg B, Lithgow T. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic. 2001;2:66–71. doi: 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- Yabal M, Brambillasca S, Soffientini P, Pedrazzini E, Borgese N, Makarow M. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J Biol Chem. 2003;278:3489–3496. doi: 10.1074/jbc.M210253200. [DOI] [PubMed] [Google Scholar]
- Yamagata A, Mimura H, Sato Y, Yamashita M, Yoshikawa A, Fukai S. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.