SUMMARY
The innate immune system detects viral infection predominantly by sensing viral nucleic acids. We report the identification of a viral sensor, consisting of RNA helicases DDX1, DDX21, and DHX36, and the adaptor molecule TRIF, by isolation and sequencing of poly I:C-binding proteins in myeloid dendritic cells (mDCs). Knockdown of each helicase or TRIF by shRNA blocked the ability of mDCs to mount type I interferon (IFN) and cytokine responses to poly I:C, influenza A virus, and reovirus. Although DDX1 bound poly I:C via its Helicase A domain, DHX36 and DDX21 bound the TIR domain of TRIF via their HA2-DUF and PRK domains, respectively. This sensor was localized within the cytosol, independent of the endosomes. Thus, the DDX1-DDX21-DHX36 complex represents a dsRNA sensor that uses the TRIF pathway to activate type I IFN responses in the cytosol of mDCs.
INTRODUCTION
Type I interferon (IFN) response represents the most important and powerful innate immune response against viral infection (García-Sastre and Biron, 2006). The innate immune system detects viral infection predominantly by sensing viral nucleic acids to produce type I IFN (Theofilopoulos et al., 2005; Gilliet et al., 2008.). During the past decade, major efforts with genomic and genetic approaches have identified three major classes of innate immune receptors for sensing microbial nucleic acids including Toll-like receptors (TLR; TLR3, 7, 8, 9) (Iwasaki and Medzhitov, 2004; Takeuchi and Akira, 2007), retinoic acid-inducible gene I-like helicases (RLH: RIG-I, LGP2, MDA-5) (Kato et al., 2006; Myong et al., 2009; Pippig et al., 2009; Takeuchi and Akira, 2009), and nucleotide-binding domain and leucine-rich repeat containing (NLR) proteins (Martinon and Tschopp, 2005; Kufer et al., 2005). Poly I:C is a synthetic form of RNA that mimics double-stranded viral RNA. Many cell types, such as epithelial cells, fibroblasts, and myeloid cells, sense poly I:C to produce type I IFN via at least three different sensor systems, including: (1) the TLR3-TRIF endosomal pathway (Akira and Takeda, 2004; Alexopoulou et al., 2001; Yamamoto et al., 2002; Yamamoto et al., 2003); (2) the RIG-I or MDA5-IPS-1 mitochondria pathway (Kato et al., 2005; Yoneyama et al., 2005; Meylan et al., 2005; Seth et al., 2005; Matsui et al., 2006; Yoneyama and Fujita, 2007); and (3) the PKR cytosolic pathway (Williams, 2001; García et al., 2007). Several studies suggested that different cell types may use different receptors to sense poly I:C or viral dsRNA. Although TLR3 was reported to play a key role in sensing poly I:C by epithelial cells (Guillot et al., 2005; Rudd et al., 2006; Matsukura et al., 2007), it only played a moderate or minor role in sensing poly I:C in macrophages or conventional DCs (Alexopoulou et al., 2001; Yamamoto et al., 2003, López et al., 2004). By contrast, RIG-I and MDA5 were found to play a more important role than TLR3 in sensing poly I:C in fibroblasts and DCs (Kato et al., 2005, Kato et al., 2006; Kato et al., 2008). Although MDA5-IPS-1 was found to preferentially sense long-form poly I:C (over 1000 bp), RIG-I-IPS-1 was found to preferentially sense short form poly I:C (>300 bp and < 1000 bp) or dsRNA with 5′ triphosphate (Kato et al., 2006; Kato et al., 2008; Schlee et al., 2009a; Schlee et al., 2009b).
The genomic and genetic approaches have left a major gap in our understanding of how these receptors bind nucleic acids and whether additional receptors or coreceptors exist. For example, although it is known that TLR3 uses TRIF as an adaptor molecule for signal transduction when sensing poly I:C, cells derived from TRIF-deficient mice displayed more reduced IFN-β and NF-κB responses than those of cells derived from TLR3-deficient mice in response to poly I:C (Yamamoto et al., 2003), suggesting the presence of a TLR3-independent, TRIF-dependent poly I:C sensor. We investigated this issue by isolating and characterizing poly I:C-binding proteins in mDCs by using biotinylated poly I:C and protein pull-down experiments, followed by protein sequencing with liquid chromatography (LC)-mass spectrometry. We found that poly I:C pulled down two known dsRNA sensors, PKR and LGP2, as well as three new members of the DExD/H-box helicase family, DDX1, DDX21, and DHX36 (Fuller-Pace, 2006; Linder, 2006). We demonstrated here that DDX1, DDX21, DHX36 represent a dsRNA sensor that uses the TRIF pathway to activate type I IFN responses.
RESULTS
Isolation of Poly I:C-Binding Proteins in mDCs
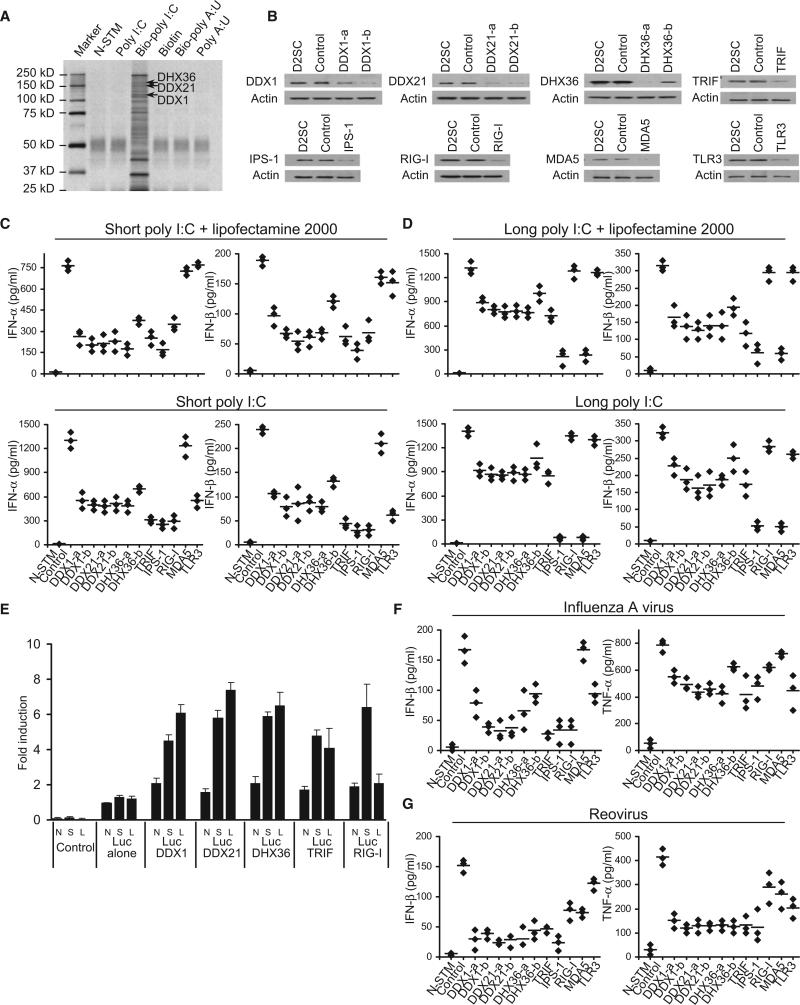
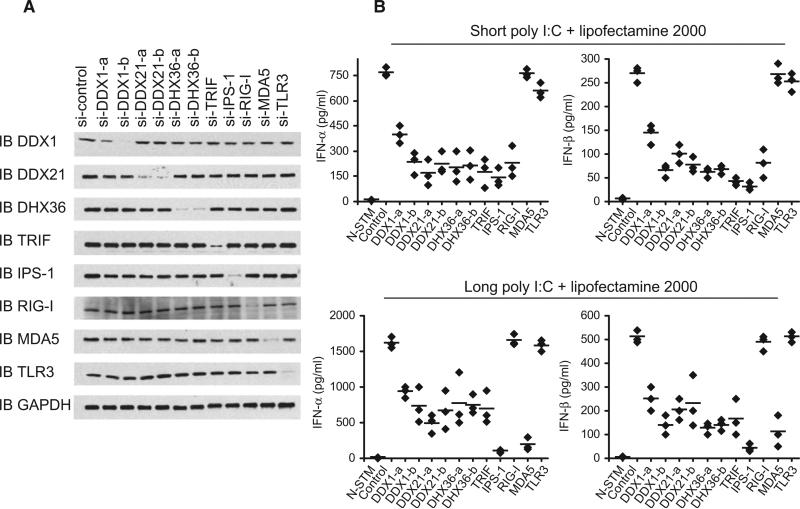
We generated biotinylated poly I:C that was 0.2–1 kb in length; poly I:C is frequently used to induce type I IFN responses. Biotinylated poly A:U (bio-poly A:U) was also generated to use as a control (Table S1 and Figure S1 available online). To purify poly I:C-bound protein complexes, we initially incubated D2SC cells with culture medium, poly I:C, bio-poly I:C, biotin, or biopoly A:U for 8 hr. Whole-cell lysates from the treated D2SC cells were prepared and subjected to purification with NeutrAvidin beads (NA beads). The proteins bound to bio-poly I:C were separated by gel electrophoresis. As shown in Figure 1A, we identified several protein bands that were unique to the bio-poly I:C, yet absent from the controls. The protein bands within 250 kD to 25 kD from bio-poly I:C and poly I:C pull-downs were excised from the gel and analyzed by LC-mass spectrometry. We obtained ~50 unique sequences with five or more hits. We found two known dsRNA sensors, PKR and LGP2, which validated our method for isolating dsRNA-binding proteins in mDCs. However, we did not find the two other known dsRNA sensors, RIG-I and MDA5. We also identified three members of the DExD/H helicase family, DDX1, DDX21, and DHX36. Because members of this family, including RIG-I (DDX58) (Yoneyama et al., 2004), LGP2 (DHX58) (Rothenfusser et al., 2005), MDA5 (IFIH1) (Kato et al., 2006), and Dicer (Deddouche et al., 2008), have been shown to play key roles in sensing dsRNA and viral infection, we investigated the function of DDX1, DDX21, and DHX36. We established stable D2SC cell lines, which are derived from BALB/c mice spleen primary cells and have functional attributes of immature DCs, expressing small heteroduplex RNA (shRNA) to knockdown expression of DDX1, DDX21, DHX36, RIG-I, MDA5, IPS-1, or TLR3. Two different clones of shRNA for DDX1, DDX21, and DHX36 were selected after screening the shRNA sets from Open Biosystems. A scrambled shRNA served as the control. Efficient knockdown of protein expression was confirmed, as shown in Figure 1B. The cells were then stimulated by a short poly I:C (0.2-1 kb) or long poly I:C (1.5–8 kb) delivered with or without Lipofectamine 2000. The production of type I IFN (IFN-α and IFN-β), TNF-α, and IL-6 by the cultured cells was measured by ELISA.
Figure 1. Characterization of poly I:C-Binding Proteins in mDCs.
(A) Silver staining of poly I:C-biotin-associated proteins purified with NA-beads from D2SC cells without stimulation (N-STM) or stimulated with poly I:C, biotin-poly I:C, biotin, biotin-poly A:U, or poly A:U. Proteins identified by LC-mass spectrometry are indicated.
(B) Immunoblot analysis showing the knockdown efficiency of indicated proteins in D2SC cells without treatment (D2SC, first lane), treated with scrambled shRNA (control, second lane), or treated with shRNA for targeting protein (third lane and fourth lane). β-Actin blots are shown as loading controls (lower panel).
(C and D) ELISA of type I IFN production by D2SC with the indicated shRNA after 16 hr stimulation with short poly I:C delivered by Lipofectamine 2000 or short poly I:C directly (C), or long poly I:C delivered by Lipofectamine 2000 or long poly I:C directly (D). N-STM, D2SC treated with scrambled shRNA without stimulation. Individual diamond represents the value from each independent experiment. Error bars represent the average value from at least three independent experiments.
(E) L929 cells were transfected with 100 ng of the IFN-β promoter luciferase reporter vector (Luc) together with indicated expression vectors to a total of 500 ng of various expression vectors or empty vector. Renilla-luciferase reporter gene (2 ng) was transfected simultaneously for the internal control. Cells were no-stimulated (N), stimulated with 1 μg/ml of short poly I:C (S), or long poly I:C (L) delivered with Lipofectamine 2000. Data represent the mean ± SD of triplicate or quadruplicate measurements. The data shown represent at least three independent experiments.
(F and G) ELISA of IFN-β and TNF-α production by DCs with the indicated shRNA upon 16 hr stimulation with flu A (F) or reovirus (G) at a multiplicity of infection (MOI) = 10. N-STM, D2SC treated with scrambled shRNA without stimulation. Individual diamond represents the value from each independent experiment. Error bars represent the average value from at least three independent experiments.
Sensing Short Poly I:C
D2SC mDCs treated with scrambled shRNA produced high amounts of type I IFN, TNF-α, and IL-6 after stimulation with a short poly I:C delivered with Lipofectamine 2000. This cytokine response was strongly attenuated (~80%) in D2SC mDCS expressing shRNA targeting DDX1, DDX21, DHX36, TRIF, or IPS-1, partially attenuated (~50%) in D2SC mDCs expressing shRNA targeting RIG-I and was not affected in D2SC mDCs expressing shRNA targeting TLR3 or MDA5 (Figure 1C and Figure S2).
Because Lipofectamine 2000 may deliver the majority of poly I:C into the cytosol, we determined the cytokines produced by D2SC cells in response to poly I:C without Lipofectamine 2000, in order to measure the endosomal poly I:C sensing by TLR3 (Figure 1C). DDX1, DDX21, and DHX36 knockdown led to a 60% reduction in type I IFN production, whereas RIG-I, IPS-1, and TRIF knockdown led to 80% reduction in type I IFN. Again, MDA5 knockdown had no effect on type I IFN production by D2SC mDCs in response to short poly I:C with or without Lipofectamine 2000, confirming a previous report showing that MDA5 only plays an important role in sensing long poly I:C (Kato et al., 2006). Although TLR3 knockdown had no marked effect on the cytokine responses of D2SC cells to poly I:C with Lipofectamine 2000, it led to a 60% and 70% reduction in IFN-α and IFN-β production, respectively, by D2SC cells in response to poly I:C without Lipofectamine 2000, confirming a previous study showing that TLR3 only senses poly I:C in the endosomes (Diebold et al., 2003). D2SC cells treated with TRIF shRNA displayed more reduced cytokine responses than the D2SC cells treated with TLR3 shRNA in response to poly I:C, suggesting the presence of TLR3-independent, TRIF-dependent poly I:C sensors. A similar effect of the above molecules on TNF-α and IL-6 production by D2SC in response to poly I:C without Lipofectamine 2000 was observed (Figure S2). These data indicate that DDX1, DDX21, DHX36, TRIF, RIG-I, and IPS-1 all play important roles in sensing short poly I:C, MDA-5 plays no role in sensing short poly I:C, and TLR3 only senses short poly I:C in the endosomes.
Sensing Long Poly I:C
We next investigated whether DDX1, DDX21, and DHX36 also play a role in sensing long poly I:C (1.5–8 kb) in D2SC cells. DDX1, DDX21, DHX36, and TRIF knockdown resulted in about a 40%–50% reduction in IFN-β production by D2SC cells in response to long poly I:C with or without Lipofectamine delivery. Whereas MDA5 and IPS-1 knockdown led to a 90% of reduction in type I IFN, RIG-I and TLR3 knockdown had little effect on type I IFN production by D2SC cells in response to long poly I:C (Figure 1D). Our data suggest that DDX1, DDX21, DHX36, and TRIF all play important roles in sensing both short (0.2–1 kb) and long (1.5–8 kb) poly I:C in D2SC cells. Our study also confirms a previous study showing that MDA5-IPS-1 preferentially sense long poly I:Cs and RIG-I-IPS-1 preferentially sense short poly I:Cs in DCs (Kato et al., 2008). In addition, TLR3 only plays an important role in sensing short poly I:C without Lipofectamine delivery.
To further confirm the role of DDX1, DDX21, and DHX36 in sensing poly I:C, we overexpressed DDX1, DDX21, DHX36, TRIF, or RIG-I in L929 cells, a mouse fibroblast cell line that was used previously for RIG-I overexpression experiments (Yoneyama et al., 2004). We found that overexpression of DDX1, DDX21, DHX36, or TRIF led to increased IFN-β promoter activation in L929 cells after stimulation with short or long poly I:C (Figure 1E). Overexpression of RIG-I led to increased IFN-β promoter activation in L929 cells after stimulation with short poly I:C, but not with long poly I:C.
Sensing Other Forms of Nucleic Acids and Viral Infection
To determine whether DDX1, DDX21, and DHX36 sense other nucleic acids, we stimulated D2SC cells treated with shRNA with 5′ triphosphate RNA (RIG-I ligand) (Myong et al., 2009) or poly dA-dT. As shown in Figure S2, DDX1, DDX21, and DHX36 knockdown, as well as MDA5 knockdown, had little effect on the production of type I IFN by the mDCs in response to poly dA-dT or 5′ triphosphate RNA, whereas IPS-1 and RIG-I knockdown led to a 90% reduction in type I IFN levels in response to RIG-I ligand 5′ triphosphate RNA. The size of RNA ligands used is shown in Figure S2.
To determine the function of DDX1, DDX21, and DHX36 in sensing viral infection, we cultured D2SC cells treated with shRNA with influenza A virus. DDX1, DDX21, and DHX36 knockdown resulted in about a 60%–70% reduction in IFN-β production and a 40%–50% reduction in TNF-α production by D2SC cells in response to influenza A virus (Figure 1F). We confirmed a previous study showing that TRIF, RIG-I, and IPS-1 play critical roles, TLR3 plays a moderate role, and MDA5 plays no role in sensing influenza A viral infection (Guillot et al., 2005; Loo et al., 2008; Kato et al., 2008). To further determine whether DDX1/DDX21/DHX36 sense other RNA viruses, we cultured D2SC cells treated with shRNA with reovirus. DDX1, DDX21, and DHX36 knockdown all resulted in about a 70%–80% reduction in IFN-β production and a 60%–70% reduction in TNF-α production by D2SC cells in response to reovirus (Figure 1G).
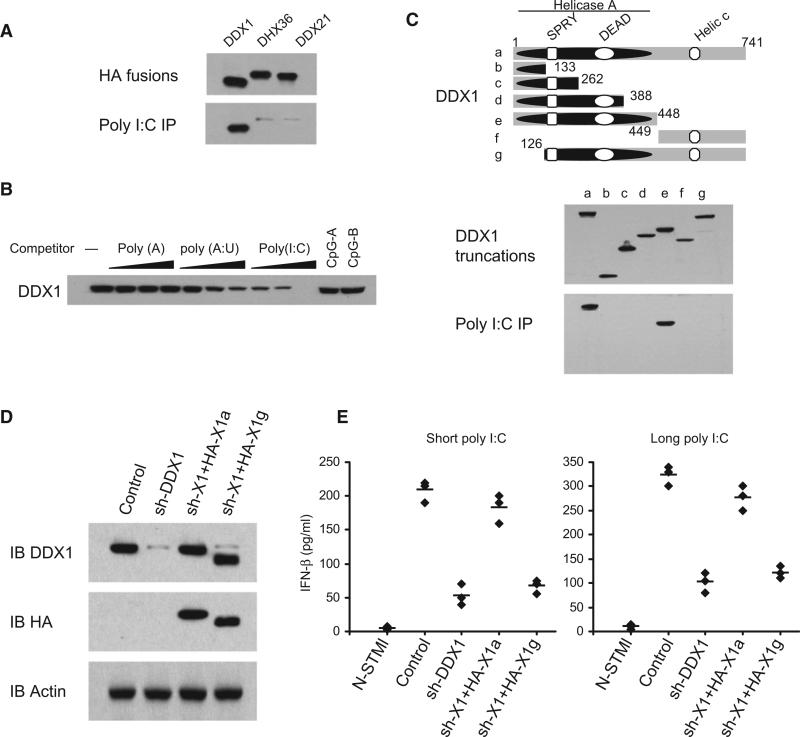
The Helicase A Domain of DDX1 Binds poly I:C
To determine whether the three helicases directly bind poly I:C, we prepared recombinant HA-tagged helicases by transfecting HEK293T cells with plasmids encoding the recombinant proteins and then purifying them with anti-HA beads. Each purified helicase was then incubated with bio-poly I:C. Only DDX1, but not DDX21 or DHX36, was found to bind poly I:C (Figure 2A). By performing competition experiments using increasing amounts of unlabeled poly I:C, poly A:U, CpG, and poly A, we found that only unlabeled poly I:C could block the binding of bio-poly I:C to DDX1 (Figure 2B). To map the poly I:C-binding site of DDX1, we prepared truncated versions of DDX1. Bio-poly I:C pull-down experiments indicated that the Helicase A domain of DDX1 binds poly I:C (Figure 2C). To determine whether recombinant DDX1 could rescue the DDX1 shRNA-induced defect, HA-DDX1a (full size DDX1) or HA-DDX1g (deletion of poly I:C binding site) was expressed in the DDX1 shRNA cells (Figure 2D). This shRNA selectively targets the 3′ UTR of DDX1 mRNA so that only the expression of endogenous DDX1 was knocked down. As shown in Figure 2E, the full size DDX1 could rescue the IFN-β responses to short and long poly I:C, whereas The DDX1 with deletion of the poly I:C-binding domain failed to rescue. These data indicate that DDX1 dsRNA binding activity is necessary for eliciting the type I IFN response to poly I:C.
Figure 2. The Helicase A Domain of DDX1 Binds Poly I:C.
(A) Pull-down assays were performed by incubating purified HA-DDX1, HA-DHX36, or HA-DDX21 with poly I:C-biotin-NA beads. Bound proteins were analyzed by immunoblotting with anti-HA.
(B) The mixture of HA-DDX1 and poly I:C-biotin-NA beads were incubated without polynucleotides or with polynucleotides at 0.5, 5, or 50 μg/ml concentration. Bound proteins were analyzed by immunoblotting with anti-HA.
(C) Schematic representations of DDX1 and its serial deletion mutants: (a) DDX1 full size, (b) DDX1 dSPRY, (c) DDX1 dDEAD, (d) DDX1 nDEAD, (e) DDX1 nHELICa, (f) DDX1 cHELICc, and (g) DDX1 SPY. Helicase A, Helicase ATP-binding domain; DEAD, Asp-Glu-Ala-Asp box motif; SPRY, SPla and the RYanodine domain; and Helic C, helicase C-terminal domain. Numbers denote amino acid residues. Truncated HA fusions were purified (middle panel) and individually incubated with poly I:C-biotin along with NA beads. Bound proteins were analyzed by immunoblotting with anti-HA (lower panel).
(D) Immunoblot of endogenous DDX1, recombinant HA-DDX1a and HA-DDX1g in D2SC with the indicated antibodies. D2SC treated with scrambled shRNA (control) or with shRNA targeting DDX1 (sh-DDX1). The sh-DDX1 cells were then transfected with HA-DDX1a (sh-X1+HA-X1a) or HA-DDX1g (sh-X1+HA-X1g) expression plasmids.
(E) ELISA of IFN-β production by indicated cells stimulated with short or long poly I:C delivered with Lipofectamine 2000 for 16 hr. Individual diamond represents the value from each independent experiment. Error bars represent the average value from at least three independent experiments.
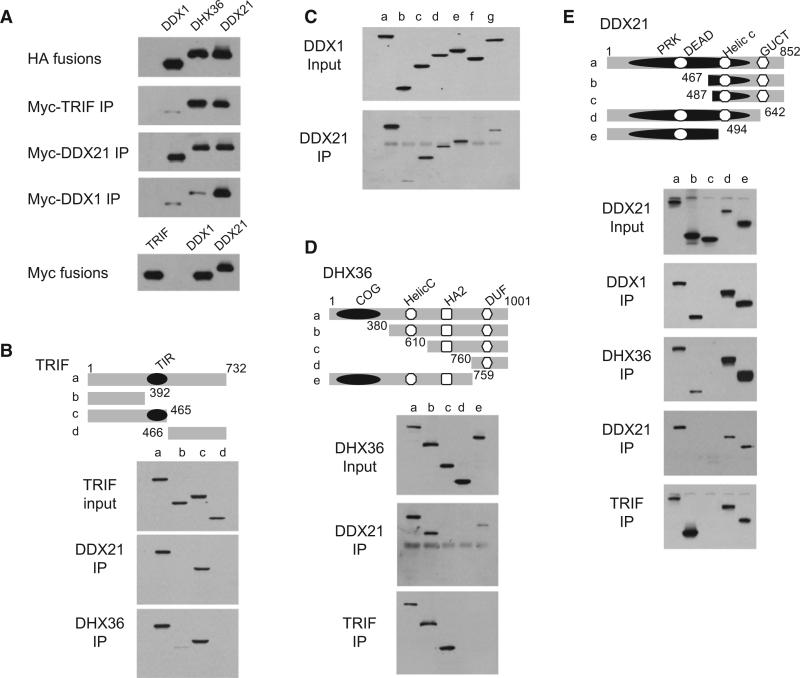
DDX21 and DHX36 Bind the TIR Domain of TRIF
Because DDX1, DDX21, DHX36, and TRIF knockdown displayed similar effects on the cytokine responses of D2SC cells to short and long poly I:C, we hypothesized that DDX1, DDX21, and DHX36 may use TRIF as an adaptor molecule for signal transduction. A Myc-tagged version of TRIF was incubated with HA-tagged versions of DDX1, DHX36, or DDX21. Anti-Myc pull-down experiments showed that TRIF binds DDX21 and DHX36, but not the poly I:C binding DDX1 (Figure 3A). To map the binding site of TRIF with DDX21 and DHX36, we prepared truncated versions of TRIF and conducted pull-down assays with DDX21 or DHX36. As indicated in Figure 3B, we observed that the TIR domain of TRIF interacts with DDX21 and DHX36. To determine which domains of DDX21 and DHX36 mediate interaction with TRIF, we incubated Myc-TRIF with truncated versions of DDX21 or DHX36. As indicated in Figures 3D and 3E, DDX21 and DHX36 bind TRIF via their PRK and HA2-DUF domains, respectively.
Figure 3. Interactions among DDX1, DDX21, DHX36, and TRIF.
(A) HA-DDX1, HA-DHX36, or HA-DDX21 protein was incubated with Myc-DDX1, Myc-DDX21 or Myc-TRIF, then incubated with anti-Myc beads. Bound proteins were analyzed by immunoblotting with anti-HA.
(B) Schematic representations of TRIF and its serial truncations (top panel): (a) TRIF full size, (b) TRIF N terminal, (c) TRIF N terminal plus TIR domain, and (d) TRIF C-terminal TIR domain: the Toll and interleukin-1 receptor homology domain. Numbers denote amino acid residues. HEK293T cells were transfected with HA-tagged TRIF full size or truncation expression plasmids. Proteins were purified with anti-HA beads. TRIF truncations were individually incubated with full-size Myc-DDX21 or Myc-DHX36. After incubation with anti-Myc beads, the bound proteins were analyzed by immunoblotting with anti-HA.
(C) DDX1 truncations, as illustrated in Figure 2C, were individually incubated with Myc-DDX21. Bound proteins were analyzed by immunoblotting with anti-HA (lower panel).
(D) Schematic representations of DHX36 and its serial truncations (top panel): (a) DHX36 full size, (b) DHX36 dCOG, (c) DHX36 dHELICc, (d) DHX36 dHA2, and (e) DHX36 dDUF. PRK, ATP-dependent RNA helicase RhlB; GUCT, GUCT domain; COG, CRISPR-associated helicase Cas3; HA2, Helicase-associated domain; and DUF, domain of unknown function. DHX36 truncations were individually incubated with the full-size Myc-DDX21 or Myc-TRIF. After incubation with anti-Myc beads, the bound proteins were analyzed by immunoblotting with anti-HA.
(E) Schematic representations of DDX21 and its serial truncations (top panel): (a) DDX21 full size, (b) DDX21 dDEAD, (c) DDX21 cHELICc, (d) DDX21 dGUCT, and (e) DDX21 dHELICc. DDX21 truncations were individually incubated with full-size Myc-DDX1, DHX36, DDX21 itself, or TRIF. After incubation with anti-Myc beads, the bound proteins were analyzed by immunoblotting with anti-HA.
DDX21 Bridges DDX1 and DHX36
Because DDX1, DDX21, and DHX36 appear to play similar roles in sensing poly I:C in D2SC cells, we investigated whether the three helicases have the ability to form a complex. Myc-tagged versions of DDX1 or DDX21 were incubated with HA-tagged versions of DDX1, DHX36, or DDX21. Anti-Myc pull-down experiments showed that DDX1 binds DDX21, DDX21 binds DHX36, and DDX21 can bind itself, whereas there is no direct interaction between DDX1 and DHX36 (Figure 3A). The domains involved in DDX1, DDX21, and DHX36 interactions were further determined by mutagenesis experiments. HA-tagged versions of DDX1, DDX21, and DHX36 were prepared and incubated with Myc-tagged versions of DDX1, DDX21, or DHX36. Pull-down experiments indicated that the SPRY domain of DDX1 binds the PRK domain of DDX21 (Figures 3C and 3E). The PRK domain of DDX21 binds the Helicase C-HA2-DUF domains of DHX36 (Figures 3D and 3E). The N terminus of DDX21 can bind together (Figure 3E).
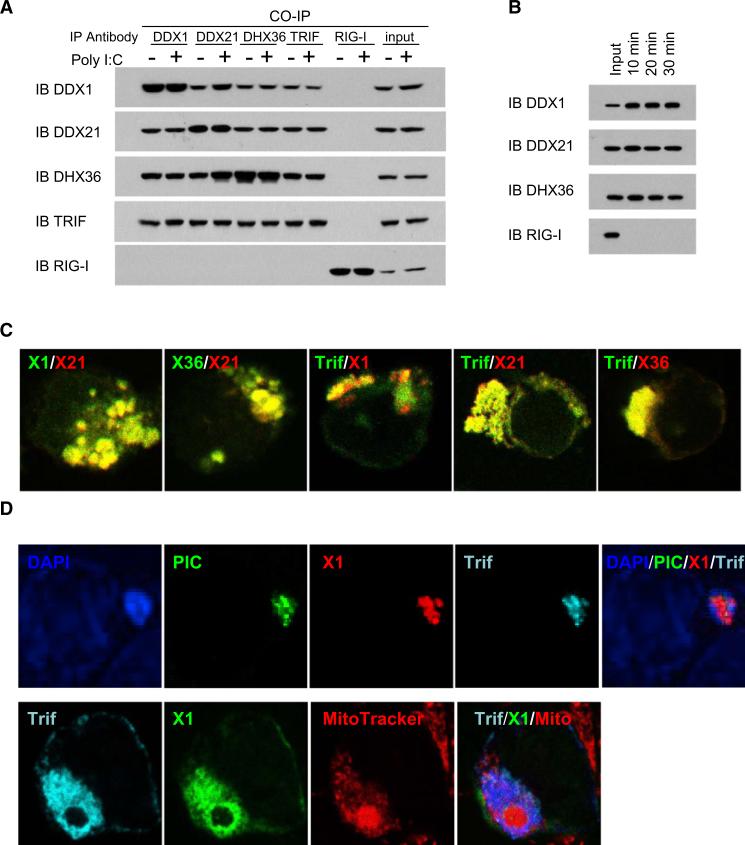
Identification of the Endogenous DDX1-DDX21-DHX36-TRIF Complex
To further determine whether endogenous DDX1, DDX21, and DHX36 exist as a complex in D2SC cells, we cultured the cells with medium or poly I:C for 16 hr and then immunoprecipitated them by using antibody to DDX1, DDX21, DHX36, TRIF, or RIG-I. DDX1 antibody precipitated DDX21 and DDX36, DDX21 antibody precipitated DDX1 and DDX36, and the DDX36 antibody precipitated DDX1 and DDX21 (Figure 4A). Interestingly, the antibody against TRIF (Figure 4A), but not RIG-I or MDA5 (data not shown), precipitated all three helicases. These data showed that endogenous DDX1, DDX21, DHX36, and TRIF proteins exist as a complex in D2SC cells, with or without poly I:C stimulation. To determine whether poly I:C stimulation modified the expression of the endogenous DDX1, DDX21, DHX36, and TRIF protein complex, D2SC cells were incubated with bio-poly I:C for 10, 20, and 30 min. Whole-cell lysates from the treated D2SC cells were prepared and subjected to precipitation with avidin-conjugated beads. The proteins bound to bio-poly I:C were detected by antibodies against DDX1, DDX21, DHX36, and RIG-I. DDX1, DDX21, and DHX36, but not RIG-I, could be detected after stimulation for 10, 20, and 30 min (Figure 4B). These data indicate that the DDX1, DDX21, DHX36, and TRIF complex exists in resting cells and the complex formation does not require stimulation.
Figure 4. Identification of the DDX1-DDX21-DHX36-TRIF Complex in Cells.
(A) Whole-cell lysates from D2SC cells, with or without poly I:C stimulation, were incubated with the indicated antibodies and protein G beads. Bound proteins were analyzed by immunoblotting with the indicated antibodies.
(B) D2SC cells were incubated with bio-poly I:C for 10, 20, or 30 min. Whole-cell lysates from the treated D2SC cells were prepared and subjected to purification with NA-beads. The proteins bound to bio-poly I:C were detected with indicated antibodies.
(C) HEK293T cells were cotransfected with indicated expression vectors. Cells were stained with Myc or HA antibodies.
(D) HEK293T cells were cotransfected with indicated expression vectors. Twenty-four hours later, cells were stimulated with poly I:C or Alexafluor 488 labeled poly I:C for 4 hr, then stained with Myc antibody or HA antibody. MitoTracker was used to probe the mitochondrion. DAP1 served as the nuclei marker.
Localization of the DDX1-DDX21-DHX36-TRIF Complex in the Cytosol
Because antibodies to DDX1, DDX21, and DHX36 for cell staining are not available, we decided to express HA-tagged together with Myc-tagged versions of DDX1, DDX21, DHX36, and TRIF in HEK293T cells to obtain information about their subcellular localization. As shown in Figure 4C, DDX1 (green) was colocalized with DDX21 (red); DHX36 (green) was colocalized with DDX21 (red); and TRIF (green) was colocalized with DDX1 (red), DDX21 (red), and DHX36 (red). We found that DDX1 (I panel), DDX21 (II panel), DHX36 (III panel), and TRIF (IV panel) were not colocalized with nuclear staining (DAPI), early endosome staining (transferin receptor [TfR]), or late endosome staining (LAMP1) (Figure S3). These data suggest that the DDX1-DDX21-DHX36-TRIF complex is localized in the cytosol. To determine whether poly I:C interacts with DDX1-DDX21-DHX36-TRIF complex in the living cells, we cotransfected DDX1 and TRIF with labeled poly I:C into HEK293T cells. Confocal imaging analysis showed the colocalization of poly I:C (green) with DDX1 (red) and TRIF (blue) (Figure 4D, upper panel). Furthermore, there was increased expression of TRIF and DDX1 in the mitochondria after poly I:C activation (Figure 4D, lower panel, and Figure S3), indicating that the DDX1-DDX21-DHX36-TRIF complex translocates to the mitochondria upon poly I:C stimulation.
Poly I:C Triggers NF-κB and IRF3 Signaling through TRIF
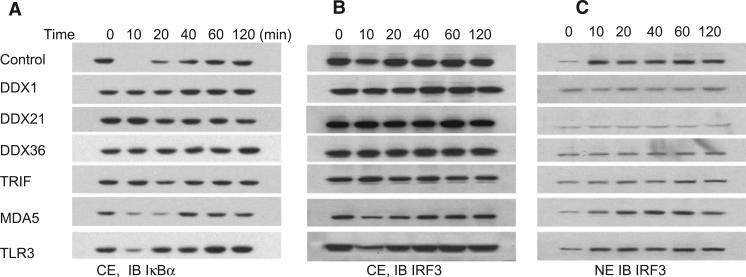
It was previously shown that poly I:C-induced cytokine and type I IFN production in fibroblasts involves the activation of NF-κB and IRF3 (Lee and Kim, 2007). We investigated whether poly I:C-induced helicase and TRIF downstream signaling involved IkB activation. Scrambled shRNA (control) or TLR3-, RIG-1-, TRIF-, DDX1-, DDX21-, and DHX36-knockdown D2SC cells were stimulated with short poly I:C delivered by Lipofectamine 2000, and cell cytoplasmic extracts were prepared and analyzed by SDS-PAGE. In scrambled shRNA and MDA5-and TLR3-knockdown D2SC cells, degradation of IκBα was detected at 10 min and sustained for up to 120 min. In contrast, degradation of IκBα was barely detectable in helicase- and TRIF-knockdown D2SC cells (Figure 5A). We then analyzed the nuclear translocation of IRF3. D2SC cells were activated by short poly I:C delivered by Lipofectamine 2000. Nuclear and cytoplasmic proteins extracts were prepared and then analyzed by immunoblotting. In scrambled shRNA and TLR3- and MDA5-knockdown D2SC cells, we observed an increase in IRF3 nuclear translocation at 10 min. By contrast, helicaseand TRIF-knockdown D2SC cells had no increase in IRF3 nuclear translocation (Figures 5B and 5C). β-actin and HDAC1 were used as controls to confirm the purity of nuclear and cytosolic fractions (Figure S4). These data suggest that DDX1, DDX21, DHX36, and TRIF are required for activating NF-κB and IFN-response signal transduction pathways upon dsRNA stimulation.
Figure 5. DDX1, DDX21, DHX36, or TRIF Knockdown Prevents IκBα Degradation and IRF3 Translocation.
Immunoblot (IB) of IκBα (A) and IRF3 (B and C) from protein extracts of D2SC cells treated with indicated shRNA upon stimulation with short poly I:C delivered with Lipofectamine 2000 for indicated time. Scrambled shRNA-treated D2SC cells served as the control. CE, cytoplasmic protein extracts; NE, nuclear protein extracts.
Function of DDX1, DDX21, DHX36, and TRIF in Primary Bone Marrow-Derived DCs
To determine whether the helicases and TRIF complex sense poly I:C in primary cells, we prepared GM-CSF-derived mDCs and treated them with siRNA to knock down expression of DDX1, DDX21, DHX36, TRIF, MDA5, or TLR3. Two different siRNAs for each helicase were selected from siRNA sets available through Dharmacom and Santa Cruz Biotechnology. Figure 6A shows the specificity and efficiency of siRNA knockdown of DDX1, DDX21, DHX36, TRIF, IPS-1, RIG-I, MDA5, and TLR3 protein expression. Compared to mDCs treated with scrambled siRNA, mDCs treated with siRNA for DDX1, DDX21, DHX36, or TRIF produced much reduced type I IFN in response to short or long poly I:C (Figure 6B). The mDCs treated with siRNA for IPS-1 produced much reduced amounts of type I IFN in response to short or long poly I:C. The mDCs treated with siRNA for RIG-I produced reduced amounts of type I IFN in response to short poly I:C and normal amounts of type I IFN in response to long poly I:C. In contrast, mDCs treated with siRNA for MDA5 produced normal amounts of type I IFN in response to short poly I:C, but much reduced levels of type I IFN in response to long poly I:C. These data indicate that the DDX1, DDX21, DHX36 and TRIF complex plays a critical role in sensing both short and long poly I:C in BM-derived DCs.
Figure 6. DDX1, DDX21, or DHX36 Knockdown in Bone Marrow Derived DCs Abolishes their Cytokine Responses to poly I:C.
(A) Immunoblot (IB) showing the knockdown efficiency of siRNA targeting the indicated genes in BMDCs. Non-targeting siRNA served as a control (first left lane). GAPDH blots are shown as loading controls (lower panel).
(B) ELISA of type I IFN production by DCs with the indicated siRNA after 16 hr stimulation with short or long poly I:C delivered by Lipofectamine 2000. N-STM, DCs treated with scrambled siRNA without stimulation. Individual diamond represents the value from each independent experiment. Error bars represent the average value from at least three independent experiments.
The Relationship of the DDX1-DDX21-DHX36-TRIF Pathway to TLR3, MyD88, and IPS-1 Pathways
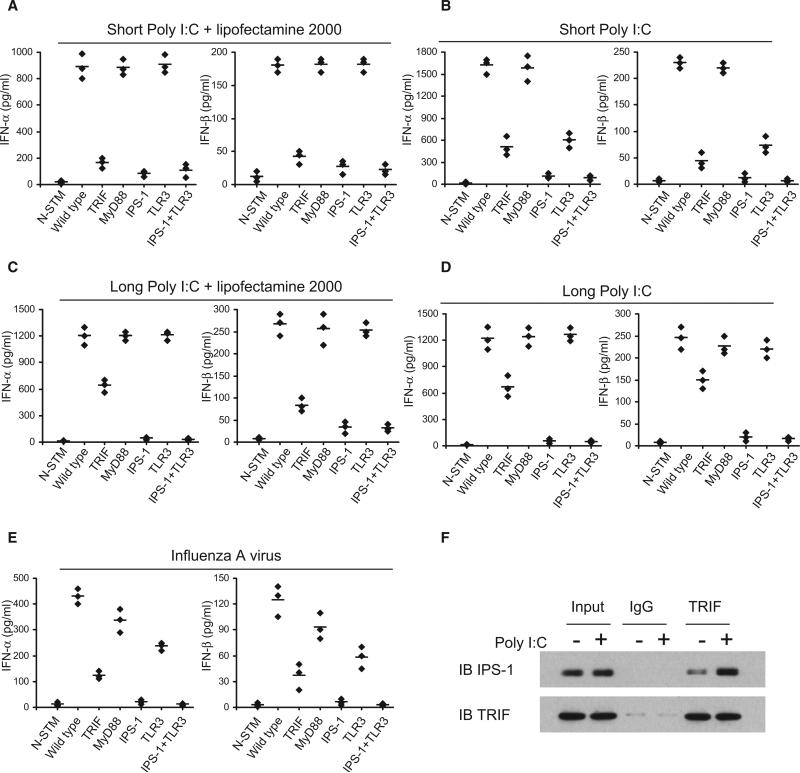
Our biochemical and functional data with siRNA and shRNA indicate that the DDX1-DDX21-DHX36 complex represents a TLR3-independent, TRIF-dependent poly I:C sensor in mDCs. To provide further supporting evidence at the genetic level, we analyzed the type I IFN responses to poly I:C by mDCs derived from mice that are deficient for TRIF, MyD88, IPS-1, TLR3, or TLR3 plus IPS-1. Bone marrow-derived GM-CSF DCs were prepared from those mutant mice. The cells were stimulated with short or long poly I:C, with or without Lipofectamine 2000, and production of type I IFN was measured by ELISA. As shown in Figures 7A–7D, in response to short poly I:C delivered by Lipofectamine 2000, mDCs from TRIF-, IPS-1-, and TLR3- plus IPS-1-deficient mice made little type I IFN; mDCs from both MyD88- and TLR3-deficient mice made normal amounts of type I IFN. No difference was observed between IPS-1- and TLR3 plus IPS-1-deficient mice. These data suggest that, similar to macrophages (Diebold et al., 2003), there are TLR3-independent, TRIF-dependent and IPS-1-dependent poly I:C sensors in mDCs. These data also support a previous report showing that TRIF and IPS-1 share a common signaling pathway and that IPS-1 functions downstream of TRIF and upstream of IRF3 (Xu et al., 2005). In response to short poly I:C without Lipofectamine 2000, mDCs from the IPS-1-deficient mice displayed over a 90% reduction in type I IFN responses; mDCs from TRIF- and TLR3-deficient mice showed over 70% reduction in type I IFN responses; and mDCs from MyD88-deficient mice showed strong type I IFN responses, similar to that of wild-type mice. In response to long poly I:C, with or without Lipofectamine 2000, mDCs from IPS-1-deficient mice made little type I IFN responses; mDCs from TRIF-deficient mice displayed over a 50% reduction in type I IFN responses; and mDCs from MyD88- and TLR3-deficient mice or wild-type mice made normal type I IFN responses.
Figure 7. The Relationship of DDX1-DDX21-DHX36-TRIF Pathway to TLR3, MyD88, and IPS-1 Pathways.
(A–E) BMDCs were prepared from wild-type or indicated knockout mice. ELISA of type I IFN production by DCs stimulated with short poly I:C delivered by Lipofectamine 2000 (A), short poly I:C directly (B), long poly I:C delivered by Lipofectamine 2000 (C), long poly I:C directly (D), or Flu A at a MOI of 10 (E). N-STM, DCs without stimulation. Individual diamond represents the value from each independent experiment. Error bars represent the average value from at least three independent experiments.
(F) Whole-cell lysates from D2SC cells, with or without poly I:C stimulation, were incubated with anti-TRIF and Protein G beads. Bound proteins were analyzed by immunoblotting with the indicated antibodies.
To determine the role of TLR3, TRIF, and IPS-1 in sensing viral infection, we infected GM-CSF mDCs prepared from mutant mice with influenza A virus. The mDCs from IPS-1- and TLR3-IPS-1-deficient mice made little type I IFN responses; mDCs from TRIF-deficient mice showed a 70% reduction in type I IFN responses; mDCs from TLR3-deficient mice showed a 50% reduction in type I IFN responses; and mDCs from MyD88-deficient or wild-type mice made normal type I IFN responses (Figure 7E). These data suggest that IPS-1 and TRIF play critical roles and TLR3 plays a moderate role in sensing influenza A viral infection.
To further determine whether the DDX1-DDX21-DHX36-TRIF signaling is independent of MDA5 or RIG-I signaling, we performed experiments with mouse embryonic fibroblasts (MEFs) derived from MDA5- and RIG-I-deficient mice. There was a residual response to poly I:C in MDA5- and RIG-I-deficient MEFs, in agreement with a previous report (Kato et al., 2006). These residual responses were decreased substantially after knockdown of DDX1, DDX21, DHX36, or TRIF by shRNA with 90% efficiency (Figure S5), suggesting that DDX1-DDX21-DHX36-TRIF is independent of RIG-I and MDA5 to sense poly I:C.
TRIF Interacts with IPS-1
A previous study showed that IPS-1, also known as VISA, interacts with TRIF and functions downstream of TRIF and upstream of IRF-3 in 293T cells during poly I:C-induced type I IFN responses (Xu et al., 2005). To investigate the potential interaction between TRIF and IPS-1 in mDCs, we performed anti-TRIF co-IP experiments in resting and poly I:C activated mDCs. As shown in Figure 7F, TRIF could pull down IPS-1 at the endogenous level in both resting and poly I:C stimulated mDCs. Interestingly, poly I:C stimulation upregulated levels of the TRIF-IPS-1 complex in mDCs. These data support the idea that TRIF and IPS-1 share a common signaling pathway in sensing dsRNA.
DISCUSSION
Although different poly I:C sensors including PKR, TLR3, RIG-1, MDA-5, and LGP2 have been reported, the relative importance of these sensors in response to dsRNA has been confusing. The first sensor for poly I:C was identified as PKR (Maran et al., 1994). However, mutant mice data indicated that PKR was not directly essential in response to poly I:C (Yang et al., 1995; Chu et al., 1999; Maggi et al., 2000). Later, several studies suggested that RIG-I and TLR-3 were the major sensors (Alexopoulou et al., 2001; Yamamoto et al., 2003; Yoneyama et al., 2004, Schulz et al., 2005). More recent studies suggested that MDA5, but not RIG-I or TLR3, was more important to sense poly I:C (Diebold et al., 2003; Kato et al., 2006; Kato et al., 2008). One potential explanation is that different dsRNA or poly I:C sensors may recognize dsRNA with different subcellular localization, length, or structure. By using a direct biochemical approach, we identified a multi-helicase-TRIF complex in myeloid DCs that directly binds poly I:C and triggers type I IFN and proinflammatory cytokine responses. This complex senses both short and long forms of poly I:C in the cytosol. This complex contains DDX1, which directly binds poly I:C, and DDX21 and DHX36, which serve as bridges to TRIF. By gene knockdown experiments, we further demonstrated that each of the four components within the complex is essential for cytokine responses of mDCs to poly I:C. By contrast, we found that MDA5 or TLR3 contributes little to the mDC's response to short poly I:C in the cytosol, confirming previous reports (Diebold et al., 2003; Kato et al., 2006). We therefore have identified a third molecular sensor of poly I:C in the cytosol of mDCs.
In eukaryotes, there are total of 59 DExD/H helicases, which have been grouped into the RIG-1-like, DEAH/RHA, DEAD-Box, and Ski2-like subfamilies (Linder, 2006). Our study suggests that the role played by DExD/H helicases in antiviral innate immune responses may be broader than previously thought. Indeed, a recent study demonstrated that Dicer, another RIG-I-like DExD/H helicase, was found to sense viral nucleic acids in the Drosophila innate immune system (Deddouche et al., 2008). The potential roles of other DExD/H helicase family members in sensing microbial nucleic acids remain to be explored.
It is well established that different classes of pattern recognition receptors preferentially use different adaptor molecules for signal transduction (Lee and Kim, 2007). Whereas members of RLR use IPS-1 (Kumar et al., 2006), NLR uses ASC (apoptosis-associated spek-like protein containing a card) and Caspase-1 (Ichinohe et al., 2009; Allen et al., 2009; Thomas et al., 2009), TLR3 uses TRIF, and the majority of the other TLRs use MyD88 (O'Neill and Bowie, 2007). At least four different receptors have been demonstrated to sense poly I:C, including PKR, TLR3, RIG-1 and MDA-5 (Alexopoulou et al., 2001; Kato et al., 2006; Balachandran et al., 2000; Yamamoto et al., 2003; Kawai et al., 2005; Kato et al., 2008). Although TLR3 has been demonstrated to use TRIF for signal transduction to activate IRF-3-mediated type I IFN responses (Oshiumi et al., 2003), our data from TLR-3 and TRIF knockdown and knockout experiments showed that TLR3 played a marginal role whereas TRIF played a major role in sensing cytosolic poly I:C, suggesting the presence of a TLR3-independent, TRIF-dependent poly I:C sensor. Our study suggests that the DDX1-DDX21-DHX36 complex represents this missing poly I:C sensor, which uses DDX1 to bind poly I:C and uses DDX21 and DXH36 to bind TRIF. A previous study suggested that RIG-I and MDA-5, but not TLR3, are type I IFN-induced genes in response to poly I:C (Ueta et al., 2011). Because the DDX1-DDX21-DHX36-TRIF complex is expressed constitutively in mDCs and not regulated by type I IFNs, this complex may represent an early sensor of poly I:C that triggers the initial IFN production. This initial IFN production will upregulate RIG-1 and MDA-5, which then further amplifies the IFN responses to poly I:C. This may explain the overlapping functions of DDX1-DDX21-DHX36-TIF, RIG-I, and MDA5. Taken together, we demonstrated here that DDX1-DDX21-DHX36 represents a dsRNA sensor that uses the adaptor molecule TRIF to activate the NF-κB pathway and type I IFN responses in dendritic cells.
EXPERIMENTAL PROCEDURES
Reagents
Poly I:C and 5′ triphosphate RNA were purchased from Invivogen. Poly dA-dT was purchased from Sigma. The silver staining kit and Lipofectamine 2000 were purchased from Invitrogen. The following antibodies were used for immunoprecipitation and/or immunoblotting: anti-DDX1, DDX21, TLR3, TRIF (Novus Biologicals), anti-DHX36 (abcam), anti-MDA5, RIG-I, IPS-1, IκBα, GAPDH, HDAC1, Actin, Alexa 555-Myc, Alexa 488-HA (Cell Signaling), anti-IRF3, TfR (SantCruz), anti-HA-HRP, and Myc-HRP (Sigma). Anti-HA or Myc beads were purchased from Sigma. NA-beads were purchased from Pierce.
In Vitro Culture of Primary mDC and siRNA
Single-cell suspensions of bone marrow cells were cultured in RPMI 1640 medium containing 10% FCS, 1 mM sodium pyruvate, HEPES, penicillin, streptomycin, and b-mercaptoethanol supplemented with murine GM-CSF (50 ng/ml) (R&D Systems). Fresh GM-CSF was provided on days 3 and 5. Cells were recovered on 6 days for siRNA knockdown. Transfection of primary mDCs was performed with Mouse Dendritic Cell Nucleofector Kit (Amaxa) with 0.5 million of cells and 0.4 nmole of siRNAs. Otherwise, cells were recovered on day 7 for stimulation.
D2SC Cell Culture and Lentiviral Infection
The murine splenic DC cell line D2SC was maintained in Iscove's modified Dulbecco's medium containing 5% heat-inactivated fetal calf serum and 1% penicillin-streptomycin (Invitrogen-GIBCO). D2SC cells were infected with the lentiviral vector carrying a helicase shRNA sequence or a scrambled shRNA. After 1–2 days of growth, cells were selected by adding puromycin (2 ng/ml) in the medium. Cells were stimulated with poly I:C (3 μg/ml), poly dA-dT (10 μg/ml), 5′ triphosphate RNA (1 μg/ml) plus lipofectamine 2000, or poly I:C (40 μg/ml) directly with or influenza A virus, as indicated in the figure legends.
MEF Cell Culture and Lentiviral Infection
The MEF cells were maintained in Dulbecco's Modified Eagle Medium containing 15% heat-inactivated fetal calf serum and 1% penicillin-streptomycin (Invitrogen-GIBCO). MEF cells were infected with the lentiviral vector carrying a helicase shRNA sequence or a scrambled shRNA. After 1–2 days of growth, cells were selected by adding puromycin (1 ng/ml) in the medium. Cells were stimulated with poly I:C (5 μg/ml) plus Lipofectamine 2000. The knockdown efficiency was detected with real time RT-PCR.
Measurement of Cytokine Production from Mouse Dendritic
Mouse dendritic cells transfected with siRNAs or infected with shRNA were cultured with poly I:C (3 μg/ml) plus Lipofectamine 2000, poly I:C (40 μg/ml) directly, or influenza A virus, as indicated in the figure legends. The concentrations of type I IFN, IL-6 and TNF-α in the culture supernatants were measured by ELISA. IFN-α and IFN-β ELISA kits were purchased from PBL Interferon-Source. IL-6 and TNF-α ELISA kits were purchased from R&D Systems, Inc.
Purification of poly I:C-Binding Protein Complexes from D2SC Cells
Poly I:C was labeled with biotin with a biotin labeling kit (Ambion). One hundred million D2SC cells were treated with 3 μg/ml of biotin-labeled polyI:C plus Lipofectamine 2000 and cultured for 8 hr. Cells were lysed in NP40 lysis buffer (50 mM Tris-Cl [pH7.5], 1 mM EDTA, 150 mM NaCl, 1.0% NP40, and 10% Glycerol), and subjected to ultracentrifugation. Cleared lysate was incubated with NA bead overnight. Beads were washed extensively with lysis buffer, separated on a 4%–20% gradient polyacrylamide gel, and stained with silver. All bands were analyzed by LC-mass spectrometry.
In Vitro Pull-Down and Immunoblotting Assay
Lysates from HEK293T cells transfected with the indicated expression plasmids were incubated with anti-HA beads. Proteins were eluted from beads after six washings with PBS. Purified proteins were incubated for 4 hr with biotin-poly I:C in the absence or presence of poly I:C or other competitor. Centrifugation was followed by incubation with NA beads and beads were analyzed by immunoblotting with anti-HA-HRP.
Confocal Microscopy
Poly I:C was labeled with ULYSIS Alexafluor 488 nucleic acid labeling kit (Invitrogen). HEK293T cells were cotransfected with HA- or Myc- empty vector or tagged DDX1, DDX21, DHX36, or TRIF expression plasmids. After 24 hr, cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% saponin. Cells were then blocked with 10% goat serum for 30 min, incubated with Alexa 555-Myc and Alexa 488-HA overnight at 4° C, and examined with confocal microscopy.
IFN-β Luciferase Reporter Assay
L929 cells seeded on 48-well plates (1 × 105 cells/well) were transfected with 100 ng of the luciferase reporter vector controlled by the IFN-β promoter together with a total of 500 ng of various expression vectors or empty control vector. Twenty-four hours later, cells were stimulated with 1 μg/ml of poly I:C delivered with Lipofectimine 2000. Cells were harvest after 6 hr stimulation. The luciferase activity in the total cell lysate was detected with the Dual-luciferase Reporter Assay System (Promega). A total of 2 ng of Renilla-luciferase reporter gene was transfected simultaneously for the internal control.
Recombinant DDX1 Rescues the DDX1 shRNA-Induced Defect
D2SC cells were infected with the lentiviral vector carrying a scrambled shRNA or the DDX1 shRNA sequence targeting the 3′ UTR. After 1–2 days of growth, cells were selected by adding puromycin (2 ng/ml) in the medium and transfected with HA-DDX1 or HA-DDX1 mutant expression plasmid. Twenty-four hours after transfection, cells were stimulated with short or long poly I:C. The concentrations of IFN-β in the culture supernatants were measured by ELISA.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Wentz for critical reading. We thank all colleagues in our laboratory.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Data include Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at doi:10.1016/j.immuni.2011.03.027.
REFERENCES
- Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL. The DExD/ H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat. Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: A lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: Sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J. Biol. Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Fritz JH, Philpott DJ. NACHT-LRR proteins (NLRs) in bacterial infection and immunity. Trends Microbiol. 2005;13:381–388. doi: 10.1016/j.tim.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: A family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, García-Sastre A, Katze MG, Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CB, Moltedo B, Alexopoulou L, Bonifaz L, Flavell RA, Moran TM. TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J. Immunol. 2004;173:6882–6889. doi: 10.4049/jimmunol.173.11.6882. [DOI] [PubMed] [Google Scholar]
- Maggi LB, Jr., Heitmeier MR, Scheuner D, Kaufman RJ, Buller RM, Corbett JA. Potential role of PKR in double-stranded RNA-induced macrophage activation. EMBO J. 2000;19:3630–3638. doi: 10.1093/emboj/19.14.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maran A, Maitra RK, Kumar A, Dong B, Xiao W, Li G, Williams BR, Torrence PF, Silverman RH. Blockage of NF-kappa B signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Matsui K, Kumagai Y, Kato H, Sato S, Kawagoe T, Uematsu S, Takeuchi O, Akira S. Cutting edge: Role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 2006;177:5785–5789. doi: 10.4049/jimmunol.177.9.5785. [DOI] [PubMed] [Google Scholar]
- Matsukura S, Kokubu F, Kurokawa M, Kawaguchi M, Ieki K, Kuga H, Odaka M, Suzuki S, Watanabe S, Homma T, et al. Role of RIG-I, MDA-5, and PKR on the expression of inflammatory chemokines induced by synthetic dsRNA in airway epithelial cells. Int. Arch. Allergy Immunol. 2007;143(Suppl 1):80–83. doi: 10.1159/000101411. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Rudd BD, Smit JJ, Flavell RA, Alexopoulou L, Schaller MA, Gruber A, Berlin AA, Lukacs NW. Deletion of TLR3 alters the pulmonary immune environment and mucus production during respiratory syncytial virus infection. J. Immunol. 2006;176:1937–1942. doi: 10.4049/jimmunol.176.3.1937. [DOI] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009a;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G. Approaching the RNA ligand for RIG-I? Immunol. Rev. 2009b;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Schulz O, Diebold SS, Chen M, Näslund TI, Nolte MA, Alexopoulou L, Azuma YT, Flavell RA, Liljeström P, Reis e Sousa C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol. Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Thomas PG, Dash P, Aldridge JR, Jr., Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M, Kawai T, Yokoi N, Akira S, Kinoshita S. Contribution of IPS-1 to polyI:C-induced cytokine production in conjunctival epithelial cells. Biochem. Biophys. Res. Commun. 2011;404:419–423. doi: 10.1016/j.bbrc.2010.11.136. [DOI] [PubMed] [Google Scholar]
- Williams BR. Signal integration via PKR. Sci. STKE. 2001;2001:re2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 2007;282:15315–15318. doi: 10.1074/jbc.R700007200. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr., Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.