Abstract
Purpose
PIK3CA gene mutations have been detected in many malignancies, but the frequency of different mutations and their role in the carcinogenesis of lung adenocarcinoma are still unclear. The purpose of this study was to explore the clinical pathological impact and prognostic implications of PIK3CA mutations in lung adenocarcinoma.
Methods
Five common PIK3CA mutations (E542K, E545K, and E545D mutation in exon 9, H1047R and H1047L mutation in exon 20) were detected by amplification refractory mutation system (ARMS) allele-specific polymerase chain reaction (PCR), in 122 patients with lung adenocarcinoma. The relationships were studied between these mutations and various clinicopathologic variables (age, lymph node status, distant metastasis, clinicopathologic stage, smoking status, and progression-free survival).
Results
In total, 25 mutations were identified, of which 24 mutations were clustered in exon 20, and one mutation in exon 9. The most common mutations were H1047R (18 out of the 122 patients, 14.8%) in exon 20. PIK3CA-mutated tumors were more frequently found in patients with lymph node positive metastasis status (P < 0.05). There was no significant association between PIK3CA mutations and age, distant metastasis, smoking status, or clinicopathologic stage. However, mutations were found less frequently in the early clinicopathologic stage patients (six in 50 cases, 12%) than in advanced stage (19 in 72 cases, 26.4%). Higher frequency of H1047R mutations was associated with poor prognosis, and this association reached statistical significance (P < 0.05).
Conclusion
Our data indicate that the PIK3CA mutations H1047R and H1047L are significant genetic alterations in lung adenocarcinoma. Among lung adenocarcinoma patients who underwent curative resection, PIK3CA mutations were associated with shorter progression-free survival. Our findings demonstrated a significant role of PIK3CA in lung adenocarcinoma.
Keywords: Phosphatidylinositol-3-kinase catalytic subunit (PIK3CA), H1047R mutation, Cancer, Lung neoplasms
Introduction
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer mortality worldwide. Lung adenocarcinoma is the main type of NSCLC.1 Owing to its complex tumorigenesis, lung adenocarcinoma is exceedingly difficult to treat and prognosticate. Recently, in order to find therapeutic targets and prognostic biomarkers, research on lung adenocarcinoma has focused on gene mutations.
The activation of the phosphatidylinositol 3-kinase (PI3K)/AKT pathway is thought to play a critical role in the development of lung cancer.2 As in most cancers, during the development of lung adenocarcinoma, genetic defects accumulate, resulting in the altered activity of the PI3K/AKT signaling pathway. The PIK3CA gene encodes the catalytic p110-α subunit of PI3K.3 It has been suggested that PIK3CA is somatically mutated in lung cancers.4 Most of the previous studies investigated the PIK3CA mutations using gene sequencing technology, and the positive mutation rate was less than 10% in lung cancer.5,6 Recently, the research of Chaft et al detected four types of PIK3CA mutations (E545K, E542K, H1047R, and H1047L) in lung adenocarcinoma; however, the rate of mutation was only 2% by mass spectrometry–based nucleic acid assay technology.7 It was unclear about the relation between PIK3CA mutations and patient prognosis. We need more research to understand the impact of PIK3CA mutations in lung adenocarcinoma. Therefore, we attempted to detect the frequency of PIK3CA mutations using a new method – amplification refractory mutation system (ARMS) allele-specific polymerase chain reaction (PCR) – and also assessed the clinicopathologic significance and prognosis of PIK3CA mutation status, in a large series of lung adenocarcinoma.8
The aim of this study was to identify significantly mutated loci not previously associated with lung adenocarcinoma and to describe the relationships between PIK3CA mutations and clinical features. Moreover, our integration of ARMS-PCR and mutation data provides a novel view of PIK3CA alterations in lung adenocarcinoma. These findings further our understanding of lung adenocarcinoma and provide clues to prognosis and new therapeutic targets.
Materials and methods
Patients and tissue samples
The analysis was conducted in 122 patients with histologically confirmed primary lung adenocarcinoma who underwent surgical resection between April 2008 and September 2010 at the Beijing Chest Hospital, Capital Medical University, Beijing, People’s Republic of China. All patients gave informed consent according to a study protocol that was approved by Beijing Chest Hospital Human Tissue Committee and Research Ethics Board (REB). All tissue samples had been flash frozen in liquid nitrogen within 30 minutes of surgery and stored at −80°C. The age of the studied patients ranged from 36 to 83 years with a median of 61.5 years. The histological classification was based on the 2004 World Health Organization criteria. Tumors were staged according to the 2009 Union for International Cancer Control guidelines. The detailed clinical characteristics of the 122 primary lung adenocarcinoma patients are listed in Table 1.
Table 1.
Clinical characteristics of 122 patients
| Total | Sex
|
P | ||
|---|---|---|---|---|
| Male | Female | |||
| Number of patients | 122 | 72 | 50 | |
| Age (years) | ||||
| 0–60 | 60 | 31 | 29 | 0.10 |
| >60 | 62 | 41 | 21 | |
| Lymph node status | ||||
| Ln (−) | 50 | 34 | 18 | 0.22 |
| Ln (+) | 72 | 38 | 32 | |
| Distant metastasis | ||||
| M (−) | 111 | 64 | 47 | 0.52 |
| M (+) | 11 | 8 | 3 | |
| Clinicopathologic stage | ||||
| I–II | 50 | 30 | 20 | 0.85 |
| III–IV | 72 | 42 | 30 | |
| Smoking status | ||||
| Smoker | 59 | 55 | 4 | 0.000a |
| Nonsmoker | 60 | 16 | 44 | |
| Missing | 3 | 1 | 2 | |
Note:
Smoking status is significantly associated with gender.
Abbreviations: Ln, lymph node; M, metastasis.
DNA extraction
The DNA was extracted from the frozen tissues using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, People’s Republic of China), according to the manufacturer’s protocol. The DNA was qualitatively assessed by agarose gel electrophoresis and was quantified spectrophotometrically, to confirm that A260/A230 value is greater than 2.0 and A260/A280 value between 1.8 and 2.0.
Detection of PIK3CA mutations
For the ARMS-PCR analysis of the five common PIK3CA mutations (E542K, E545K, and E545D mutation in exon 9, H1047R and H1047L mutation in exon 20), samples were loaded, in triplicate, into 96-well reaction plates, using the AmoyDx™ PIK3CA Five Mutation Detection Kit (Amoy Diagnostics Co, Ltd, Xiamen, People’s Republic of China), according to the manufacturer’s instructions, for each sample. The mutation assay for each sample and control assay (PIK3CA mixed standard and no-template controls) were analyzed in the same PCR run, to avoid run-to-run variations in threshold settings. The assays were performed with the Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). Predenaturing was carried out at 95°C for 5 minutes, followed by 15 cycles of denaturing at 95°C for 25 seconds, 64°C for 20 seconds, and 72°C for 20 seconds, in order to reach a full plateau for all samples, and was followed by the detection stage with 20 cycles at 93°C for 25 seconds, 60°C for 35 seconds, and 72°C for 20 seconds. The analysis of the target mutations in the samples was accomplished by measuring the fractional cycle number at which the degree of expression reached a fixed cycle threshold (CT). Based on different mutant CT values, the detection results were categorized as either positive or negative for mutation.
Statistical analysis
Chi-square and the Fisher’s exact test were used to analyze the association of mutations with clinical characteristics. The Kaplan–Meier method was used to estimate survival and progress free survival, and the exact log rank test was used to compare differences in survival the distributions among groups. All data were analyzed by statistical software SPSS 16.0 (SPSS Inc, Chicago, IL, USA). A P-value less than 0.05 was considered to be statistically significant.
Results
PIK3CA mutations were found in lung adenocarcinoma
Among the 122 patients who underwent curative resection of stage I to IV lung adenocarcinoma, we detected PIK3CA mutations in exons 9 and 20, using ARMS-PCR technology. In all, 24 patients (19.7%) harbored at least one mutation in the regions analyzed. A total of 25 point mutations were found in these 24 patients: one mutation occurred in the helical domain (E542K, exon 9), and 24 mutations were in the kinase domain (18 of these were mutations of H1047R and six were of H1047L, exon 20). One patient carried two mutations in exon 20 (H1047R and H1047L). The “hot spot” was H1047R, located in the kinase domain (exon 20). None of the patients showed mutations in both exon 9 and exon 20. The frequency and distribution of the mutations are shown in Table 2.
Table 2.
Frequency of each PIK3CA mutation type in lung adenocarcinoma
| Exon | Total (%) | Exon 20
|
Exon 9
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H1047R
|
H1047L
|
E542K
|
E545K
|
E545D
|
|||||||
| Male (%) | Female (%) | Male (%) | Female (%) | Male (%) | Female (%) | Male (%) | Female (%) | Male (%) | Female (%) | ||
| Number of mutations | 25 (20.5%)a | 10 (8.2%) | 8 (6.6%) | 3 (2.5%) | 3 (2.5%) | 0 | 0 | 0 | 1 (0.8%) | 0 | 0 |
| Age (years) | 0 | 0 | 0 | 1 (0.8%) | 0 | 0 | |||||
| 0–60 | 15 (12.3%) | 5 (4.1%) | 6 (4.9%) | 3 (2.5%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| >60 | 10 (8.2%) | 5 (4.1%) | 2 (1.6%) | 0 | 3 (2.5%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Lymph node status | |||||||||||
| Ln (−) | 5 (4.1%) | 1 (0.8%) | 3 (2.5%) | 1 (0.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ln (+) | 20 (16.4%)b | 9 (7.4%) | 5 (4.1%) | 2 (1.6%) | 3 (2.5%) | 0 | 0 | 0 | 1 (0.8%) | 0 | 0 |
| Distant metastasis | |||||||||||
| M (−) | 22 (18%) | 9 (7.4%) | 7 (5.7%) | 2 (1.6%) | 3 (2.5%) | 0 | 0 | 0 | 1 (0.8%) | 0 | 0 |
| M (+) | 3 (2.5%) | 1 (0.8%) | 1 (0.8%) | 1 (0.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Clinicopathologic stage | |||||||||||
| I–II | 6 (4.9%) | 2 (1.6%) | 3 (2.5%) | 1 (0.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| III–IV | 19 (15.6%) | 8 (6.6%) | 5 (4.1%) | 2 (1.6%) | 3 (2.5%) | 0 | 0 | 0 | 1 (0.8%) | 0 | 0 |
| Smoking status | |||||||||||
| Smoker | 13 (10.7%) | 9 (7.4%) | 0 | 3 (2.5%) | 1 (0.8%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Nonsmoker | 11 (9.0%) | 0 | 8 (6.6%) | 0 | 2 (1.6%) | 0 | 0 | 0 | 1 (0.8%) | 0 | 0 |
| Missing | 1 (0.8%) | 1 (0.8%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Notes:
One sample had both mutations of H1047R and H1047L;
P < 0.05.
Abbreviations: LN, lymph node; M, metastasis.
PIK3CA mutations correlated with lymph node (Ln) status and tumor stage
We analyzed the clinical characteristics of the lung adenocarcinoma cases according to PIK3CA mutation status (Tables 2 and 3). Compared with PIK3CA wild-type tumors, PIK3CA mutations were found more frequently in tumors with LN metastasis (LN [+]), 20 point mutations in LN (+), and five point mutations in LN patients without LN metastasis (LN [−]) (P = 0.029). PIK3CA mutations occurred in 12% (6/50) of patients at early clinicopathologic stage (I–II) and in 26.4% (19/72) of patients at advanced stage (III–IV) disease, but the association was not statistically significant (P = 0.122). PIK3CA mutations were not associated with smoking status in the total group of patients (P = 0.78). No correlation was identified between the PIK3CA mutation rate and distant metastasis, age, or gender.
Table 3.
Clinical characteristics according to PIK3CA mutation status in lung adenocarcinoma
| Total (n = 122) N (%) |
PIK3CA mutation status
|
Pa | ||
|---|---|---|---|---|
| Wild-type (n = 98) N (%) | Mutant (n = 24) N (%) | |||
| Age (years) | ||||
| Mean (range) | 60 (36–83) | 61 (37–83) | 58 (36–73) | |
| 0–60 | 60 (49%) | 46 (47%) | 14 (58%) | 0.32 |
| >60 | 62 (51%) | 52 (53%) | 10 (42%) | |
| Lymph node status | ||||
| Ln (−) | 52 (43%) | 47 (48%) | 5 (21%) | 0.029b |
| Ln (+) | 70 (57%) | 51 (52%) | 19 (79%) | |
| Distant metastasis | ||||
| M (−) | 111 (91%) | 89 (91%) | 22 (92%) | 0.79 |
| M (+) | 11 (9%) | 9 (9%) | 2 (8%) | |
| Clinicopathologic stage | ||||
| I–II | 50 (41%) | 44 (45%) | 6 (25%) | 0.122 |
| III–IV | 72 (59%) | 54 (55%) | 18 (75%) | |
| Smoking status | ||||
| Smoker | 59 (48%) | 47 (48%) | 12 (50%) | 0.78 |
| Nonsmoker | 60 (49%) | 49 (50%) | 11 (46%) | |
| Missing | 3 (2%) | 2 (2%) | 1 (4%) | |
Notes:
χ2 test used for binary/categorical variables;
P < 0.05.
Abbreviations: Ln, lymph node; M, metastasis.
PIK3CA mutations and patient progression-free survival (PFS)
We assessed the influence of PIK3CA mutation on patient PFS in resectable lung adenocarcinoma. The median follow up for the cohort was 19 months (0–47 months). Patients harboring PIK3CA-mutated lung adenocarcinoma showed a marginally significant shorter median PFS of 12 months (0–42 months) compared with patients with PIK3CA wildtype tumors, for whom median PFS was about 20 months (1–47 months). The patients with PIK3CA-mutated tumors experienced an increase in cancer-specific mortality, although statistical significance was not reached (P = 0.385) (Figure 1A). We also assessed the five PIK3CA mutation types separately. In our study, the H1047R mutant is the most common mutation. H1047R mutations associated with survival, and reached statistical significance (P = 0.032) (Figure 1B); the median PFS of H1047R (+) patients was 9 months (95% confidence interval [CI] 7.2–10.8), far under the median PFS of 26 months (95% CI 17.2–36.8) for H1047R (−) patients.
Figure 1.
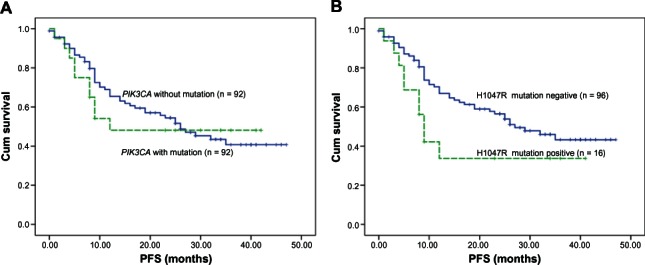
Kaplan–Meier curves for PFS in lung adenocarcinoma with five PIK3CA mutations (A) and for the H1047R mutation type (B).
Notes: In (A), lung adenocarcinoma patients harboring PIK3CA with mutation (n = 20) showed a marginally significant shorter median PFS of 12 months (0–42 months) compared with 20 months (1–47 months) in patients without mutation (n = 92). The patients with PIK3CA-mutated tumors experienced an increase in cancer-specific mortality, although statistical significance was not reached (P = 0.385). In (B), the median PFS of H1047R mutation-positive patients (n = 16) was 9 months (95% CI 7.2–10.8), far under the median PFS of 26 months (95% CI 17.2–36.8) for H1047R mutation-negative patients (n = 96). H1047R mutations associated with survival, and reached statistical significance (P = 0.032). When the mutation of H1047R was included, the association reached statistical significance (P = 0.032).
Abbreviations: CI, confidence interval; cum, cumulative; PFS, progression-free survival.
Discussion
Most of the reported PIK3CA mutations were localized at hotspots in exon 9 and exon 20 of the gene. Exon 20 encodes the catalytic domain of p110α, and mutations in this domain may constitutively activate its enzymatic activity. Exon 9 encodes the helical domain of p110α, and those mutations depress an inhibitory interaction between the N-terminal Src homology 2 (SH2) domain of p85 and the p110α catalytic subunit. Expression of the PIK3CA mutations leads to increased oncogenic potential in vitro and in vivo. Direct gene sequencing has been commonly used to analyze mutations of the PIK3CA gene. However, this method was unable to detect the presence of mutant PIK3CA when present at <30% of the total.8 The ARMS-PCR assay, a new detection method, identified more mutations in the clinical samples than did direct sequencing. The ARMS–PCR assays were able to detect the presence of mutations at 1% of the total. These assays were more sensitive than sequencing and could detect five copies of mutant DNA in proportions as low as 0.1% of the total DNA.8 In our study, we tested the five “hotspot” mutations (E542K, E545K, E545D mutation in exon 9, H1047R and H1047L mutation in exon 20) in 122 lung adenocarcinomas using ARMS–PCR; we found that the positive rate (19.7%) was much higher than in early reports studies.7,9,10
The PIK3CA mutations found in human cancers primarily occur at two “hot spots:” E545K in the helical domain and H1047R in the catalytic domain.7,11,12 These mutations are known to promote the catalytic activity of p110α, thereby leading to constitutive activation of the PI3K signaling pathway.13 Research by Engelman et al indicated that transgenic mice with lung-specific induction of the kinase-domain mutant p110α H1047R (in exon 20) developed lung adenocarcinoma.14 Yamaguchi et al determined that E545K and H1047R mutations in p110α enhanced invadopodia-mediated extracellular matrix degradation and invasion.15 This finding provides mechanistic insight into the role of p110α mutations in cancer invasion, and Yamaguchi suggested that PI3K signaling, via p110α, regulates invadopodia-mediated invasion and migration of breast cancer cells.15 In this study, we analyzed the clinicopathologic significance of five “hotspot” PIK3CA mutations among 122 lung adenocarcinoma patients. The data showed that PIK3CA mutations were associated with LN metastasis. Lung adenocarcinoma patients with LN (+) had many more mutations (20 in 70 cases, 28.6%) than did patients with LN (−) (five in 52 cases, 9.6%) (P = 0.029). Moreover, there was no significant association between PIK3CA mutations and age (P = 0.32), distant metastasis (P = 0.79), smoking status (P = 0.78), or clinicopathologic stage (P = 0.122), by Chi-square test. However, mutations were found less frequently in the early clinicopathologic stage patients (six in 50 cases, 12%) than in advanced stage (19 in 72 cases, 26.4%).
PIK3CA mutations activate AKT through phosphorylation, and pAKT expression has been reported to be associated with poor prognosis.15 Some studies have shown that PIK3CA mutation predicts shorter survival in colorectal and breast cancers.16–21 Because the effect of PIK3CA mutations on lung adenocarcinoma patient prognosis has rarely been studied, we investigated the prognostic significance of PIK3CA mutations in the present study. In our research, we examined on prognostic significance of five hot spot PIK3CA mutations in 122 lung adenocarcinoma patients, focusing on PFS. In our study, the H1047R mutant is the most common mutation. H1047R mutations associated with survival, and reached statistical significance (P = 0.032) (Figure 1B). The median PFS of H1047R (+) patients was 9 months (95% CI 7.2–10.8), which was far under that of H1047R (−) patients, who had a mean PFS of 26 months (95% CI 17.2–36.8). In our cohorts, the data on cancer treatment were limited. Nonetheless, it is unlikely that chemotherapy use differed according to tumor PIK3CA-mutated status because such data were not available to patients or treating physicians.
In summary, our study suggests that PIK3CA mutation is associated with poor prognosis in resectable lung adenocarcinoma. This finding may have considerable clinical implications. Considerable effort has been focused on identifying therapeutic inhibitors of the PI3K/AKT pathway, in lung adenocarcinoma as well as in other common malignancies. Future studies are needed to confirm the association between PIK3CA mutations and the PI3K/AKT pathway and as well, to elucidate the exact mechanisms by which PIK3CA mutations affect tumor behavior. Thus, advances in the understanding of the molecular mechanisms of lung adenocarcinoma will lead to the development of novel and individualized anticancer therapeutic strategies.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez M, Roman E, Santos ES, Raez LE. New targets for non-small-cell lung cancer therapy. Expert Rev Anticancer Ther. 2007;7(10):1423–1437. doi: 10.1586/14737140.7.10.1423. [DOI] [PubMed] [Google Scholar]
- 3.Hiles ID, Otsu M, Volinia S, et al. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell. 1992;70(3):419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 4.Catalogue of Somatic Mutation in Cancer.[homepage on the Internet] England: Sanger Institute; [updated 2012 Dec 06]. Available from: http://www.cancer.sanger.ac.uk/cosmic/gene/analysis?ln=PIK3CAAccessed March 29, 2013 [Google Scholar]
- 5.Okudela K, Suzuki M, Kageyama S, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int. 2007;57(10):664–6671. doi: 10.1111/j.1440-1827.2007.02155.x. [DOI] [PubMed] [Google Scholar]
- 6.An SJ, Chen ZH, Su J, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7(6):e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012;11(2):485–491. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Board RE, Thelwell NJ, Ravetto PF, et al. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54(4):757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- 9.Spoerke JM, O’Brien C, Huw L, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res. 2012;18(24):6771–6783. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- 10.Lu HY, Su D, Pan XD, Jiang H, Ma SL. Mutation and expression of multiple treatment response-related genes in a population with locally advanced non-small cell lung cancer. Oncol Lett. 2012;3(2):415–420. doi: 10.3892/ol.2011.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18(1):77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27(41):5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102(3):802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi H, Yoshida S, Muroi E, et al. Phosphoinositide 3-kinase signaling pathway mediated by p110α regulates invadopodia formation. J Cell Biol. 2011;193(7):1275–1288. doi: 10.1083/jcb.201009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba Y, Nosho K, Shima K, et al. Phosphorylated AKT expression is associated with PIK3CA mutation, low stage, and favorable outcome in 717 colorectal cancers. Cancer. 2011;177(7):1399–1408. doi: 10.1002/cncr.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27(9):1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangone FR, Bobrovnitchaia IG, Salaorni S, Manuli E, Nagai MA. PIK3CA exon 20 mutations are associated with poor prognosis in breast cancer patients. Clinics (Sao Paulo) 2012;67(11):1285–1290. doi: 10.6061/clinics/2012(11)11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fariña Sarasqueta A, Zeestraten EC, van Wezel T, et al. PIK3CA kinase domain mutation identifies a subgroup of stage III colon cancer patients with poor prognosis. Cell Oncol (Dordr) 2011;34(6):523–531. doi: 10.1007/s13402-011-0054-4. [DOI] [PubMed] [Google Scholar]


