Abstract
Background
The transforming-growth-factor-β-signaling pathway has been identified as being involved in colorectal cancer.
Objective
To determine how diet and lifestyle factors in combination with genetic variation in the transforming-growth-factor-β-signaling pathway alters colorectal cancer risk.
Design
We use data from two population-based case-control studies.
Patients
Participants included colon cancer cases (n=1574) and controls (1970) and rectal cancer cases (n=791) and controls (999).
Main Outcome Measures
Newly diagnosed cases of colon or rectal cancer.
Results
Colon and rectal cancer risk increased with the number of at-risk genotypes within the transforming-growth-factor-β-signaling pathway (odds ratio 3.68 95% confidence interval 2.74,4.94 for colon cancer and odds ratio 3.89, 95% confidence interval 2.66,5.69 for rectal cancer). A high at-risk lifestyle score also resulted in significant increased risk with number of at-risk lifestyle factors (odds ratio 2.99 95% confidence interval 2.32,3.85 for colon cancer and odds ratio 3.37 95% confidence interval 2.24,5.07 for rectal cancer). The combination of high-risk genotype and high-risk lifestyle results in the greatest increase in risk (odds ratio 7.89 95% confidence interval 4.45,13.96 for colon cancer; odds ratio 8.75 95% confidence interval 3.66,20.89 for rectal cancer).
Limitations
Results need validation in other large studies of colon and rectal cancer.
Conclusions
In summary, our data suggest that there is increased colon and rectal cancer risk with increasing number of at-risk genotypes and at-risk lifestyle factors. While the integrity of the pathway can be diminished by number of high-risk genotypes, this risk can be in part offset by maintaining a healthy lifestyle.
Keywords: TGF-β, NSAIDs, estrogen, cigarette smoke, colorectal cancer, Smad, BMP, RUNX
Introduction
The TGF-β-signaling pathway is involved in all aspects of tumorigenesis, including cell growth regulation, avoidance of apoptosis, promotion of inflammation and angiogenesis, immune response, and stimulation of tumor invasion and metastasis.1 A functional TGF-β-signaling pathway can restrict cell growth by inhibiting cell proliferation, mediating differentiation, and inducing apoptosis; a dysfunctional TGF-β-signaling pathway is associated with cancer development, progression, and metastasis.2 The TGF-β cytokine family includes bone morphogenetic proteins (BMP), which have been independently associated with colon and rectal cancer.3 Regulation of TGF-β signaling involves interaction with Smad proteins and runt-related transcription factors (RUNX), including RUNX1, RUNX2, and RUNX3.4–6 TGF-β signaling also entails non-Smad pathways that include mitogen-activated protein kinase 1 (MAPK1), also known as extracellular signal-regulated kinase 2 (ERK2), eukaryotic translation initiation factor 4E (eIF4E)7, NFκB, and the Akt pathway including PTEN and mTOR.8–10
The importance of the TGF-β-signaling pathway in colon and rectal cancer has been supported in numerous studies. Recent gene-set analysis of GWAS colon cancer data have identified it as a key pathway associated with colon cancer.11 Our previous work further demonstrates the risk associated with genetic variation in components of this pathway with colon and rectal cancer.3, 12–15 Given the importance of this pathway in colon and rectal cancer, it is reasonable to determine if lifestyle factors impact the susceptibility associated with genetic risk. Given the biological function of the pathway it is possible that lifestyle factors that influence inflammation, insulin or energy-related factors may importantly modify genetic risk. Factors previously identified with colon and rectal cancer risk that may influence this pathway are use of aspirin/NSAIDs, estrogen status, use of cigarettes, body size, physical activity, and dietary intake.16–23
Our goal in this study is to evaluate the association of the composite genetic variation in the TGF-β-signaling pathway with risk of colon and rectal cancer. To do this we have developed a genotype risk score that summarizes individual risk across genotypes in the pathway that were associated individually with colon and rectal cancer risk. Similarly, we have summarized individual lifestyle risk through a composite score of those lifestyle variables that independently influenced colon and rectal cancer risk. We evaluate the composite genotype and lifestyle scores as they relate to colon and rectal cancer and to each other to more comprehensively describe the association between the TGF-β-signaling pathway and colon and rectal cancer.
Methods
Two study populations are included in these analyses. The first study, a population-based case-control study of colon cancer, included cases (n=1,558) and controls (n=1,958) identified between October 1, 1991 and September 30, 1994 living in the Twin Cities Metropolitan Area of Minnesota, Kaiser Permanente Medical Care Program of Northern California (KPMCP) and a seven county area of Utah and who had complete genotype data The second study, with identical data collection methods, included cases with cancer of the rectosigmoid junction or rectum (n=756) and controls (n=962) who were identified between May 1997 and May 2001 in Utah and KPMCP21 with complete genotype data. Eligible cases were between 30 and 79 years old at time of diagnosis, English speaking, mentally competent to complete the interview, had no previous history of CRC, and no known (as indicated on the pathology report) familial adenomatous polyposis, ulcerative colitis, or Cohn’s disease. Approximately 91% of the colon cancer study and 88% of the rectal cancer study was non-Hispanic white.
TagSNP Selection and Genotyping
DNA for genotyping was available from buffy coats (California and Minnesota) and immortalized cell lines (Utah) for both cases and controls. TagSNPs were selected using the following parameters: r2=0.8 defined LD blocks using a Caucasian LD map, minor allele frequency or MAF>0.1, range= −1500 bps from the initiation codon to +1500 bps from the termination codon, and 1 SNP/LD bin. All markers were genotyped using a multiplexed bead array assay format based on GoldenGate chemistry (Illumina, San Diego, California). A genotyping call rate of 99.85% was attained. Blinded internal replicates represented 4.4% of the sample set. The duplicate concordance rate was 100.00%
For TGFβ1, representative markers rs4803455 and rs1800469 were chosen based on a prevalent minor allele frequency and previous findings described in the literature24. They were genotyped using a TaqMan assay from Applied Biosystems (Foster City, California). Each 5ul PCR reaction contained 20 ng of genomic DNA, primers, probes, and TaqMan Universal PCR Master Mix (containing AmpErase UNG, AmpliTaq Gold enzyme, dNTPs, and reaction buffer). PCR was carried out under the following conditions: 50°C for 2 minutes to activate UNG, 95°C for 10 min, followed by 40 cycles of 92°C for 15 sec, and 60°C for 1 minute using 384 well duel block ABI 9700. Fluorescent endpoints of the TaqMan reactions were measured using a 7900HT sequence detection instrument.
Environmental data
Trained and certified interviewers collected diet and lifestyle data as previously outlined.25, 26 The referent year for the study was the calendar year approximately two years prior to date of diagnosis (cases) or selection (controls). Information was collected on demographic factors such as age, sex, and study center; diet, physical activity, regular use of aspirin and non-steroidal drug use, measured and self-reported height and weight, and other lifestyle factors including medical, family history of first-degree relatives with colorectal cancer, and reproductive history were assessed. Physical active data included information on amount, frequency, and intensity of activities for the referent year as well as 10 and 20 years prior to the referent year27. Dietary data were based on the CARDIA diet history questionnaire26, 28 Cigarette smoking patterns were determined through questions that focused on age first having smoked cigarettes, age at which cigarette smoking stopped, and the number of cigarettes usually smoked. All data were collected in a rigorous manner using quality control procedures that included review of audiotapes of 10% of interviews as well as any interview where the interview reported that the respondent provided questionable data.
We focused the analysis on those variables that have been previously associated with colon and rectal cancer. Recent use of aspirin/NSAID, long-term physical activity, cigarette smoking, and Western Dietary Pattern have been reported as associated with both colon and rectal cancer.19, 21 For colon cancer we included Prudent Diet Pattern and BMI, taking into account estrogen status of women.23 For rectal cancer we included dietary fiber and calcium rather than the Prudent dietary pattern because these factors appeared to be more strongly associated with rectal cancer risk than the Prudent dietary pattern. Dietary factors including Western and Prudent Diet, calcium, and fiber were categorized into three levels: low, intermediate (two mid-quartiles), and high.
Statistical Methods
All statistical analyses were performed using SAS version 9.2 (SAS Institute, Cary, NC). SNP selection was based on previously reported significant associations between genes in the pathway and risk of colon and rectal cancer. Genes included were TGFβ1 (2 SNPs), TGFβR1 (3 SNPs), Smad1 (5 SNPs), Smad2 (4 SNPs), Smad3 (37 SNPs), Smad4 (2 SNPs), Smad7 (11 SNPs), BMP1 (11 SNPs), BMP2 (5 SNPs), BMP4 (3 SNPs), BMPR1A (9 SNPs), BMPR1B (21 SNPs), BMPR2 (11 SNPs), GDF10 (7 SNPs), RUNX1 (40 SNPs), RUNX2 (19 SNPs), RUNX3 (9 SNPs), eiF4E (3 SNPs), eiF4EBP2 (2 SNPs), eiF4EBP3 (2 SNPs), and MAPK1 (6 SNPs), NFκB1 (13 SNPs); PTEN (5 SNPs), FRAP1 (3SNPs), and AKT1 (2 SNPs).
To summarize risk associated with multiple variants across the pathway we created a summary polygenic score29 that was based on all at-risk genotypes identified from previous analyses for colon and rectal cancer. The score for each SNP was based on the inheritance model and its associated risk. All genotypes were evaluated so that they were coded with the low-risk genotype as 0. For the co-dominant or additive model a score of zero, one, or two was assigned directly correlated to the number of high-risk alleles, while scores of zero or two were assigned for the dominant and recessive models. For instance a genotype that was directly associated with risk would be scored 0 for wildtype, 1 for heterozygote, and 2 for homozygote variant in the co-dominant model. If the association were recessive, it would be coded either 0 or 2. In the instance of an inverse association, the wildtype would be coded 2 whereas the homozygote variant would be coded as a 0. After assigning a score for each SNP previously identified as being significant, the scores were summed across SNPs to generate an individual polygenic summary score. Individuals missing SNP data were dropped from the analysis. For the colon cancer study this represented 2.3% of all participants, while for the rectal cancer study 6.2% of participants were dropped because they were missing complete genotype data. The continuous score variable was redefined as a categorical variable based on the frequency distribution within the study population. Cut-points were used that would allow at least 10% of the population in any category for analysis. Only one SNP was used in instances where in high linkage disequilibrium was detected (r2>0.70)
Lifestyle factors were selected based on variables previously identified as contributing to risk in these studies. Summary scores were developed for lifestyle factors in a similar manner as we did for genotype summary scores. Lifestyle factors were assigned values of 0, 1, or 2, depending on their risk. For instance the physical activity variable was assigned a risk of 2 if no lifetime activity was reported (the high-risk lifestyle factor), 1 if they were intermediary, and 0 if they were in the top range of physical activity. For dichotomous variables such as recent NSAID use, a two was assigned for non-users and 0 for users. Cigarette smoking was categorized based on usual amount used for colon cancer and packyears of cigarettes smoked for rectal cancer. Those who smoked the most, 20 or more cigarettes a day or 20 or more packyears were assigned a 2, those who never smoked were assigned 0, those who smoked less than 20 cigarettes a day or 20 packyears were assigned a 1.
Odds ratios (OR) and 95% confidence intervals (CI) were estimated from multiple logistic regression models adjusting for age, sex, center, and race. Categories of exposure for summary scores were based on the distribution of the population, with most of the population being in the middle of the distribution which followed a bell-shaped curve. Our goal was to have close to a 10% sample in all categories to provide stability to risk estimates. Linear trend was based on the improved fit of the model when the score variables were treated as continuous. P values for interaction were calculated for both the continuous and categorical models by comparing a full model that included an interaction term to a reduced model without an interaction term using a likelihood ratio test.
Results
SNPs contributing to colon and rectal cancer risk are shown in Table 1. For colon cancer, 16 genes and 35 SNPs were statistically significantly associated with colon cancer after adjustment for age, sex, center, and race. Fewer genes were associated with rectal cancer. We observed that 11 genes and 18 SNPs significantly altered risk of rectal cancer. For the most part, the SNPs that were most important for colon and rectal cancer differed. BMP2 rs3178250 and RUNX3 rs6672420 were associated with both colon and rectal cancer, although the associations were in the opposite directions for RUNX3 rs6672420.
Table 1.
Summary of SNPs identified as contributing to colon and rectal cancer risk in TGFB-signaling pathway
| Study | Symbol | SNP | MAF | Heterozygote OR | Homozygote Rare OR |
Dom OR |
|---|---|---|---|---|---|---|
| Colon | BMP2 | rs1979855 | T/C | 1.48 (1.25,1.76) | ||
| BMPR1A | rs2168730 | A/G | 1.71 (1.15,2.55) | |||
| rs2883420 | T/C | 0.86 (0.72,1.03) | 0.76 (0.57,1.01) | 0.84 (0.71,1.00) | ||
| BMPR1B | rs2120834 | G/C | 0.78 (0.62,0.99) | |||
| rs7662504 | A/C | 0.87 (0.73,1.04) | 0.79 (0.61,1.01) | 0.85 (0.72,1.01) | ||
| rs9307147 | A/G | 0.87 (0.72,1.04) | 0.64 (0.50,0.81) | |||
| BMPR2 | rs4303700 | G/A | 1.55 (1.11,2.17) | |||
| EIF4E | rs11727086 | A/G | 1.25 (1.06,1.48) | |||
| EIF4EBP3 | rs250425 | C/T | 0.82 (0.70,0.97) | |||
| MAPK1 | rs8136867 | A/G | 0.77 (0.65,0.92) | |||
| NFKB1 | rs4648090 | G/A | 0.51 (0.26,0.98) | |||
| PTEN | rs12572106 | T/C | 0.49 (0.27,0.89) | |||
| RUNX1 | rs2242878 | C/T | 1.35 (1.11,1.64) | |||
| rs2834645 | T/C | 0.80 (0.67,0.95) | 0.70 (0.47,1.03) | 0.78 (0.66,0.93) | ||
| rs2834683 | T/C | 0.78 (0.64,0.96) | ||||
| rs8134380 | A/T | 0.79 (0.66,0.95) | ||||
| rs11702779 | G/A | 0.82 (0.68,0.98) | ||||
| RUNX2 | rs598953 | T/A | 1.27 (1.02,1.59) | |||
| rs2819854 | C/T | 1.34 (1.10,1.62) | ||||
| SMAD2 | rs4940086 | T/C | 1.43 (1.11,1.84) | |||
| SMAD3 | rs1498506 | A/C | 0.83 (0.69,1.00) | 0.73 (0.58,0.93) | ||
| rs4601989 | C/T | 0.62 (0.43,0.89) | ||||
| SMAD7 | rs12953717 | C/T | 1.29 (1.04,1.59) | |||
| TGFB1 | rs1800469 | G/A | 0.72 (0.52,0.99) | |||
| rs4803455 | C/A | 1.22 (0.97,1.53) | 1.34 (1.03,1.74) | |||
| TGFBR1 | rs1571590 | A/G | 1.78 (1.19,2.67) | |||
| Rectal | BMP1 | rs4076873 | A/C | 0.79 (0.62,1.01) | 0.59 (0.35,1.02) | 0.76 (0.60,0.96) |
| BMPR1A | rs2168730 | A/G | 1.92 (1.19,3.09) | |||
| BMPR2 | rs2228545 | G/A | 1.76 (1.09,2.85) | 1.83 (1.14,2.95) | ||
| FRAP1 | rs1057079 | A/G | 1.19 (0.93,1.51) | 1.57 (1.04,2.38) | ||
| GDF10 | rs762454 | A/G | 2.05 (1.39,3.01) | |||
| rs11598444 | G/A | 0.73 (0.54,0.98) | 0.55 (0.19,1.61) | 0.72 (0.54,0.96) | ||
| MAPK1 | rs11913721 | A/C | 0.92 (0.71,1.18) | 0.70 (0.49,0.98) | ||
| NFKB1 | rs230510 | A/T | 0.82 (0.64,1.05) | 0.62 (0.44,0.88) | ||
| PTEN | rs532678 | C/T | 1.36 (1.08,1.71) | |||
| RUNX1 | rs2242878 | C/T | 1.51 (1.15,1.98) | |||
| rs2834645 | T/C | 1.35 (1.05,1.73) | 1.87 (1.04,3.37) | |||
| rs8127225 | T/C | 2.33 (1.13,4.83) | ||||
| rs11701453 | G/C | 1.41 (1.11,1.79) | ||||
| rs11702779 | G/A | 0.47 (0.32,0.69) | ||||
| rs17227161 | C/T | 0.72 (0.55,0.95) | 0.42 (0.17,1.04) | 0.70 (0.54,0.92) | ||
| RUNX2 | rs1200428 | C/A | 2.11 (1.17,3.80) | |||
| rs2819863 | G/C | 1.43 (1.05,1.96) | ||||
| rs6930053 | C/T | 1.23 (0.94,1.61) | 1.70 (1.18,2.44) | |||
| rs7750470 | T/C | 3.01 (1.54,5.90) | ||||
| RUNX3 | rs6672420 | A/T | 1.33 (1.02,1.75) | |||
| rs6688058 | G/A | 2.39 (1.05,5.41) | ||||
| rs9438876 | G/A | 1.34 (1.04,1.74) | ||||
| SMAD2 | rs1792689 | C/T | 0.71 (0.54,0.93) | |||
| SMAD3 | rs1866317 | C/G | 1.36 (1.02,1.81) | 2.40 (0.93,6.19) | 1.41 (1.06,1.87) | |
| rs4776881 | T/C | 1.36 (1.06,1.74) | ||||
| rs11071933 | C/G | 0.58 (0.46,0.75) | ||||
| rs17293443 | T/C | 2.30 (1.29,4.12) |
Minor Allele Frequency (MAF) based on white control population.
Dominant model listed in "Heterozygote OR" column; recessive model listed in "Homozygote Rare OR" column.
A statistically significant linear trend for increasing risk with increasing mutational load was seen for both colon and rectal cancer (p<0.0001) (Table 2). Over a three-fold increase in risk was observed for both diseases at the highest category of high-risk genotypes. Similarly, if one looked at summary of risk associated with lifestyle factors, a three-fold increase in risk was associated with both colon cancer and rectal cancer. Again, significant linear trend was observed for both colon and rectal cancer that showing increasing risk with number of “at-risk” lifestyle factors.
Table 2.
Summary of risk associated with TGF-β-signaling pathway genes and lifestyle for colon and rectal cancer.
| N | N | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Score Intervals | Controls | Cases | OR | (95% CI) | Score Intervals | Controls | Cases | OR | (95% CI) |
| Genotype | Colon | Lifestyle | Colon | ||||||
| (14 – 21) | 345 | 151 | 1.00 | (0 – 3) | 338 | 160 | 1.00 | ||
| (22 – 26) | 1052 | 729 | 1.61 | (1.30,2.00) | (4 – 5) | 734 | 456 | 1.31 | (1.05,1.64) |
| (27 – 29) | 405 | 440 | 2.59 | (2.05,3.28) | (6 – 7) | 625 | 575 | 1.95 | (1.56,2.43) |
| (30 – 36) | 154 | 235 | 3.62 | (2.73,4.79) | (8 – 12) | 259 | 364 | 2.99 | (2.32,3.85) |
| P Trend | <.0001 | <.0001 | |||||||
| Rectal | Rectal | ||||||||
| (10 – 17) | 215 | 94 | 1.00 | (0 – 3) | 164 | 83 | 1.00 | ||
| (18 – 21) | 459 | 305 | 1.51 | (1.14,2.01) | (4 – 6) | 516 | 342 | 1.31 | (0.97,1.76) |
| (22 – 25) | 252 | 293 | 2.65 | (1.97,3.56) | (7 – 8) | 213 | 219 | 2.03 | (1.47,2.82) |
| (26 – 31) | 33 | 62 | 4.26 | (2.61,6.94) | (9 – 12) | 66 | 110 | 3.37 | (2.24,5.07) |
| P Trend | <.0001 | P Trend | <.0001 | ||||||
Adjusted for age, center, race, and sex.
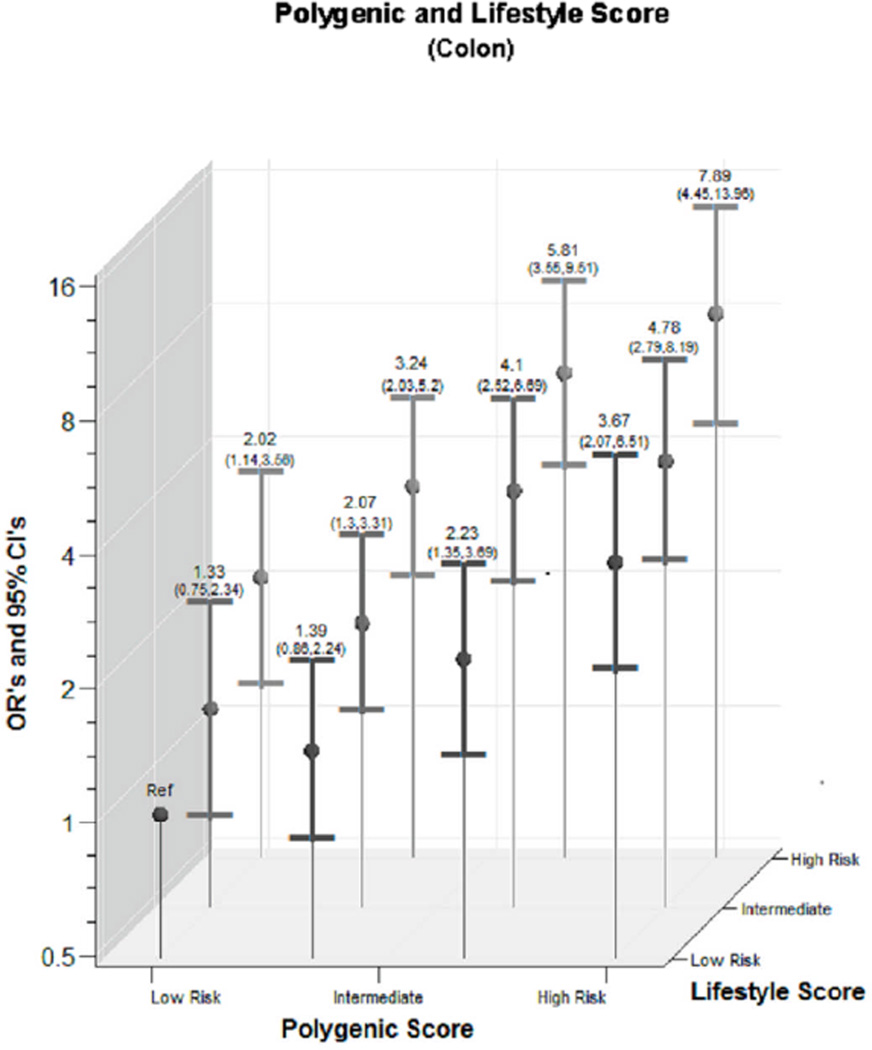
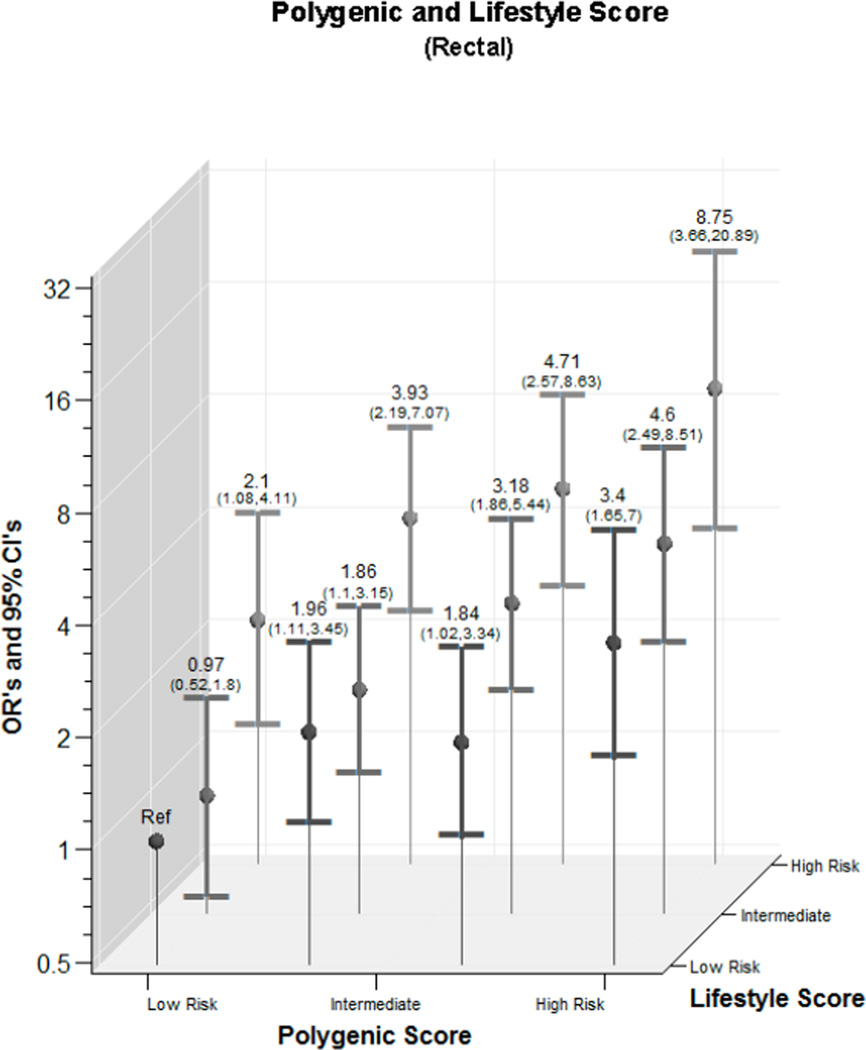
We observed a statistically significant interaction between lifestyle score and genotype score for colon cancer on the categorical interaction (p=<0.001) but not continuous variable interaction (p=0.745); for rectal cancer the p for interaction was 0.157 for categorical and 0.760 for continuous) (Figure 1). The magnitude of the combined risk was similar for both colon and rectal cancer. For colon cancer, those with the highest genotype and lifestyle score had an OR of 7.89 (95% CI 4.45,13.96); for rectal cancer the estimate of association was 8.75 (95% CI 3.66,20.89).
Figure 1.
- Colon Cancer
- Rectal Cancer
We further stratified the data on three lifestyle factors that have been previously shown to interact with TGFβ1 and are associated with both colon and rectal cancer (Table 3), recent use of NSAIDs, exposure to cigarette smoke, and recent exposure to estrogen. Although statistically significant interactions were not observed, clear patterns of associations emerged for all three factors. For those protective factors, NSAIDS and recent estrogen exposure, there is reduced risk of both colon and rectal cancer at low genotype scores where individuals have the fewest “at-risk” genotypes. As the genotype score increases with increasing number of “at-risk” genotypes, the risk increases and becomes significant at the 3rd or 4th category of genotype score, however, the risk is less than for those exposed to NSAID or estrogen than without the exposure. For cigarette smoking exposure, the risk associated with high-risk genotype score is less for non-smokers than for those who smoke cigarettes.
Table 3.
Interaction between NSAID use, cigarette smoking, and estrogen exposure and summary TGF-β-signaling pathway genetic score
| Controls | Cases | Controls | Cases | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Summary Scores | N | N | OR | (95% CI) | N | N | OR | (95% CI) | |
| Genotypes | No Recent Aspirin/NSAID Use | Recent Aspirin NSAID Use | |||||||
| Colon | (14 – 21) | 202 | 108 | 1.00 | 141 | 41 | 0.55 | (0.36,0.84) | |
| (22 – 26) | 616 | 493 | 1.53 | (1.18,2.00) | 427 | 228 | 1.03 | (0.77,1.37) | |
| (27 – 29) | 233 | 290 | 2.45 | (1.83,3.28) | 168 | 146 | 1.70 | (1.23,2.35) | |
| (30 – 36) | 85 | 162 | 3.72 | (2.61,5.30) | 68 | 70 | 2.02 | (1.34,3.04) | |
| Rectal | (10 – 16) | 85 | 36 | 1.00 | 56 | 14 | 0.59 | (0.29,1.20) | |
| (17 – 20) | 239 | 171 | 1.68 | (1.08,2.60) | 186 | 83 | 1.05 | (0.66,1.68) | |
| (21 – 25) | 176 | 231 | 3.09 | (1.99,4.78) | 175 | 152 | 2.05 | (1.31,3.21) | |
| (26 – 31) | 21 | 39 | 4.26 | (2.20,8.24) | 11 | 22 | 4.84 | (2.12,11.03) | |
| Non Smoker/Non Recent Smoker | Recent Smoker | ||||||||
| Colon | (14 – 21) | 287 | 128 | 1.00 | 58 | 23 | 0.87 | (0.52,1.48) | |
| (22 – 26) | 860 | 567 | 1.51 | (1.19,1.91) | 191 | 160 | 1.87 | (1.39,2.52) | |
| (27 – 29) | 333 | 348 | 2.46 | (1.90,3.19) | 72 | 91 | 2.82 | (1.94,4.10) | |
| (30 – 36) | 129 | 191 | 3.46 | (2.54,4.70) | 25 | 44 | 3.98 | (2.33,6.81) | |
| Rectal | (10 – 16) | 119 | 41 | 1.00 | 24 | 9 | 1.06 | (0.46,2.48) | |
| (17 – 20) | 363 | 203 | 1.61 | (1.09,2.39) | 67 | 54 | 2.26 | (1.36,3.75) | |
| (21 – 25) | 297 | 307 | 2.99 | (2.02,4.41) | 56 | 74 | 3.73 | (2.27,6.14) | |
| (26 – 31) | 30 | 51 | 4.88 | (2.75,8.68) | 3 | 11 | 10.20 | (2.70,38.44) | |
| No Recent Estrogen* | Recent Estrogen Exposure* | ||||||||
| Colon | (14 – 21) | 85 | 41 | 1.00 | 61 | 23 | 0.61 | (0.32,1.14) | |
| (22 – 26) | 277 | 228 | 1.70 | (1.12,2.58) | 190 | 93 | 0.83 | (0.52,1.34) | |
| (27 – 29) | 106 | 116 | 2.34 | (1.48,3.71) | 82 | 69 | 1.47 | (0.87,2.46) | |
| (30 – 36) | 55 | 64 | 2.47 | (1.46,4.16) | 28 | 37 | 2.23 | (1.18,4.21) | |
| Rectal | (10 – 16) | 26 | 12 | 1.00 | 34 | 7 | 0.37 | (0.13,1.10) | |
| (17 – 20) | 70 | 46 | 1.40 | (0.64,3.06) | 113 | 60 | 0.95 | (0.44,2.06) | |
| (21 – 25) | 67 | 73 | 2.33 | (1.09,5.00) | 91 | 83 | 1.66 | (0.77,3.56) | |
| (26 – 31) | 6 | 10 | 3.65 | (1.07,12.43) | 11 | 11 | 1.70 | (0.56,5.13) | |
Adjusted for age, center, race, and sex.
Among women only.
Interaction between BMI, physical activity, and dietary factors with polygenic risk is shown in Table 4. For colon cancer the risk from being obese increased with increasing genotype risk. Physical activity and prudent diet had minimal effect on modifying risk from the high-risk genotype. Western diet appeared to have the strongest interaction with the polygenic risk summary score. Eating a diet that strongly followed the Western dietary pattern increased risk of colon and rectal cancer the most as genotype risk increased.
Table 4.
Associations between genetic summary score, BMI, physical activity, and diet and risk of colon and rectal cancer
| Controls | Cases | Controls | Cases | Controls | Cases | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary Scores | N | N | OR | (95% CI) | N | N | OR | (95% CI) | N | N | OR | (95% CI) | |
| Genotypes | BMI | ||||||||||||
| Normal (<25) | Overweight (25–29) | Obese (30+) | |||||||||||
| Colon | (17 – 30) | 105 | 27 | 1.00 | 112 | 63 | 2.18 | (1.29,3.68) | 58 | 27 | 1.77 | (0.94,3.30) | |
| (31 – 38) | 419 | 237 | 2.21 | (1.41,3.48) | 443 | 261 | 2.28 | (1.45,3.58) | 211 | 179 | 3.29 | (2.05,5.25) | |
| (39 – 43) | 165 | 166 | 3.99 | (2.48,6.43) | 169 | 197 | 4.50 | (2.81,7.22) | 106 | 142 | 5.21 | (3.18,8.53) | |
| (44 – 56) | 69 | 74 | 4.24 | (2.48,7.26) | 72 | 109 | 6.01 | (3.57,10.10) | 23 | 68 | 11.38 | (6.02,21.51) | |
| Rectal | (4 – 11) | 76 | 26 | 1.00 | 88 | 35 | 1.12 | (0.62,2.04) | 52 | 23 | 1.25 | (0.64,2.43) | |
| (12 – 15) | 116 | 93 | 2.30 | (1.36,3.88) | 172 | 114 | 1.86 | (1.12,3.09) | 101 | 75 | 2.08 | (1.22,3.57) | |
| (16 – 19) | 94 | 85 | 2.62 | (1.53,4.48) | 118 | 100 | 2.37 | (1.41,4.00) | 58 | 78 | 3.76 | (2.14,6.61) | |
| (20 – 28) | 25 | 39 | 4.49 | (2.29,8.80) | 30 | 53 | 4.91 | (2.60,9.27) | 24 | 28 | 3.21 | (1.58,6.51) | |
| Physical Activity | |||||||||||||
| Low | Intermediate | High | |||||||||||
| Colon | (17 – 30) | 81 | 34 | 1.00 | 104 | 43 | 0.99 | (0.58,1.70) | 91 | 40 | 1.06 | (0.61,1.84) | |
| (31 – 38) | 296 | 228 | 1.91 | (1.23,2.96) | 374 | 233 | 1.50 | (0.97,2.33) | 405 | 217 | 1.28 | (0.82,1.98) | |
| (39 – 43) | 131 | 180 | 3.45 | (2.17,5.47) | 157 | 184 | 2.81 | (1.78,4.43) | 152 | 144 | 2.30 | (1.44,3.66) | |
| (44 – 56) | 43 | 77 | 4.54 | (2.62,7.87) | 60 | 95 | 3.91 | (2.33,6.57) | 62 | 80 | 3.06 | (1.81,5.17) | |
| Rectal | (4 – 11) | 39 | 25 | 1.00 | 77 | 26 | 0.51 | (0.26,0.99) | 100 | 33 | 0.49 | (0.26,0.93) | |
| (12 – 15) | 88 | 72 | 1.28 | (0.71,2.31) | 143 | 97 | 0.99 | (0.56,1.75) | 161 | 113 | 1.03 | (0.58,1.80) | |
| (16 – 19) | 59 | 85 | 2.26 | (1.23,4.14) | 86 | 86 | 1.51 | (0.84,2.72) | 127 | 95 | 1.08 | (0.61,1.92) | |
| (20 – 28) | 15 | 34 | 3.54 | (1.60,7.81) | 31 | 44 | 2.06 | (1.04,4.09) | 33 | 44 | 1.93 | (0.98,3.81) | |
| Western Diet | |||||||||||||
| Low | Intermediate | High | |||||||||||
| Colon | (17 – 30) | 60 | 21 | 1.00 | 143 | 57 | 1.11 | (0.62,2.00) | 73 | 39 | 1.49 | (0.79,2.81) | |
| (31 – 38) | 241 | 117 | 1.35 | (0.79,2.34) | 576 | 363 | 1.79 | (1.07,3.00) | 258 | 198 | 2.18 | (1.27,3.71) | |
| (39 – 43) | 117 | 78 | 1.90 | (1.07,3.39) | 228 | 284 | 3.56 | (2.09,6.03) | 95 | 146 | 4.32 | (2.46,7.59) | |
| (44 – 56) | 35 | 39 | 3.09 | (1.57,6.08) | 96 | 139 | 4.26 | (2.42,7.48) | 34 | 74 | 6.09 | (3.19,11.63) | |
| Rectal | (4 – 11) | 62 | 16 | 1.00 | 94 | 94 | 33 | 1.35 | (0.69,2.67) | 60 | 35 | 2.18 | (1.09,4.37) |
| (12 – 15) | 107 | 68 | 2.43 | (1.30,4.57) | 169 | 114 | 2.58 | (1.41,4.70) | 116 | 100 | 3.22 | (1.73,5.97) | |
| (16 – 19) | 87 | 65 | 2.85 | (1.51,5.40) | 119 | 126 | 4.01 | (2.19,7.36) | 66 | 75 | 4.26 | (2.23,8.14) | |
| (20 – 28) | 25 | 32 | 4.81 | (2.25,10.28) | 31 | 46 | 5.60 | (2.74,11.47) | 23 | 44 | 7.14 | (3.37,15.12) | |
| Prudent Diet | |||||||||||||
| Low | Intermediate | High | |||||||||||
| Colon | (17 – 30) | 63 | 26 | 1.00 | 140 | 66 | 1.07 | (0.62,1.85) | 73 | 25 | 0.78 | (0.41,1.48) | |
| (31 – 38) | 229 | 183 | 1.92 | (1.17,3.17) | 587 | 336 | 1.33 | (0.83,2.15) | 259 | 159 | 1.36 | (0.83,2.25) | |
| (39 – 43) | 114 | 142 | 3.00 | (1.78,5.06) | 225 | 269 | 2.79 | (1.70,4.56) | 101 | 97 | 2.16 | (1.26,3.70) | |
| (44 – 56) | 36 | 57 | 3.91 | (2.10,7.27) | 91 | 138 | 3.56 | (2.09,6.06) | 38 | 57 | 3.37 | (1.81,6.26) | |
| Rectal | (4 – 11) | 68 | 35 | 1.00 | 93 | 28 | 0.56 | (0.31,1.01) | 55 | 21 | 0.70 | (0.37,1.35) | |
| (12 – 15) | 99 | 93 | 1.81 | (1.10,2.98) | 176 | 125 | 1.32 | (0.83,2.11) | 117 | 64 | 0.99 | (0.59,1.65) | |
| (16 – 19) | 96 | 86 | 1.71 | (1.03,2.82) | 121 | 105 | 1.61 | (0.99,2.63) | 55 | 75 | 2.49 | (1.45,4.27) | |
| (20 – 28) | 26 | 40 | 2.92 | (1.54,5.55) | 32 | 50 | 2.88 | (1.57,5.27) | 21 | 32 | 2.72 | (1.36,5.42) | |
| Calcium Intake | |||||||||||||
| Low | Intermediate | High | |||||||||||
| Colon | (17 – 30) | 65 | 25 | 1.00 | 143 | 72 | 1.31 | (0.76,2.25) | 68 | 20 | 0.77 | (0.39,1.53) | |
| (31 – 38) | 265 | 185 | 1.81 | (1.10,2.98) | 537 | 323 | 1.58 | (0.97,2.56) | 273 | 170 | 1.66 | (1.01,2.75) | |
| (39 – 43) | 126 | 144 | 3.02 | (1.79,5.08) | 211 | 259 | 3.25 | (1.98,5.35) | 103 | 105 | 2.67 | (1.56,4.57) | |
| (44 – 56) | 48 | 67 | 3.70 | (2.04,6.70) | 84 | 123 | 3.94 | (2.29,6.77) | 33 | 62 | 4.89 | (2.61,9.18) | |
| Rectal | (4 – 11) | 53 | 14 | 1.00 | 109 | 47 | 1.59 | (0.80,3.15) | 54 | 23 | 1.54 | (0.71,3.32) | |
| (12 – 15) | 83 | 63 | 2.79 | (1.42,5.47) | 211 | 140 | 2.43 | (1.30,4.56) | 98 | 79 | 2.90 | (1.50,5.64) | |
| (16 – 19) | 73 | 79 | 4.01 | (2.05,7.84) | 129 | 130 | 3.70 | (1.95,7.01) | 70 | 57 | 2.92 | (1.46,5.81) | |
| (20 – 28) | 16 | 30 | 6.98 | (2.99,16.29) | 37 | 63 | 6.13 | (2.99,12.56) | 26 | 29 | 3.99 | (1.80,8.84) | |
| Dietary Fiber Intake | |||||||||||||
| Low | Intermediate | High | |||||||||||
| Colon | (17 – 30) | 61 | 34 | 1.00 | 161 | 55 | 0.58 | (0.34,0.98) | 54 | 28 | 0.87 | (0.47,1.63) | |
| (31 – 38) | 278 | 195 | 1.23 | (0.78,1.94) | 553 | 332 | 1.04 | (0.67,1.62) | 244 | 151 | 1.06 | (0.66,1.69) | |
| (39 – 43) | 118 | 138 | 2.06 | (1.26,3.35) | 222 | 260 | 2.04 | (1.29,3.23) | 100 | 110 | 1.89 | (1.14,3.12) | |
| (44 – 56) | 46 | 62 | 2.35 | (1.33,4.16) | 84 | 122 | 2.62 | (1.58,4.34) | 35 | 68 | 3.26 | (1.81,5.87) | |
| Rectal | (4 – 11) | 56 | 26 | 1.00 | 93 | 35 | 0.79 | (0.43,1.46) | 67 | 23 | 0.70 | (0.36,1.37) | |
| (12 – 15) | 74 | 83 | 2.36 | (1.35,4.14) | 191 | 121 | 1.32 | (0.79,2.23) | 127 | 78 | 1.25 | (0.72,2.17) | |
| (16 – 19) | 85 | 71 | 1.77 | (1.01,3.11) | 121 | 132 | 2.26 | (1.33,3.83) | 66 | 63 | 1.97 | (1.10,3.53) | |
| (20 – 28) | 17 | 32 | 4.05 | (1.91,8.59) | 39 | 55 | 2.87 | (1.54,5.35) | 23 | 35 | 3.09 | (1.52,6.25) | |
Adjusted for age, center, race, and sex.
Discussion
The importance of the TGF-β-signaling pathway in the etiology of colon and rectal cancer is well recognized. In this study, we have shown that numerous components of the pathway contribute independently to colon and rectal cancer risk, and that risk increases with increasing genetic susceptibility. Likewise, lifestyle factors play an important role in the composite risk. Our data suggest that while lifestyle factors can modify risk of colon and rectal cancer at low levels of genetic risk, they lose their protective benefits when high levels of genetic risk exist.
We have used a candidate pathway approach, identifying genetic variants in components of the TGF-β-signaling pathway that involves BMP, RUNX, MAPK1, eIF4E, and Smad as well as the non-Smad related signaling regulators that include AKT, PTEN, mTOR, and NFκB1. Using this candidate pathway approach we identified genetic variants that independently influenced colon and rectal cancer risk after consideration of other genes in the pathway. Given our goal of identifying composite risk associated with the pathway, we developed a polygenic risk score that took into account most appropriate model of inheritance and number of “at-risk” alleles for each individual. Refinement of the score taking into account the ratio of the strength of the association using the beta coefficient for each SNP and the precision of the estimate by factoring in the variance associated with each individual SNP, did not appreciably alter the risk estimates presented. Thus, we present data from the simpler method of calculation that assigns risk based on number of at-risk alleles for each genotype that independently were associated with colon and rectal cancer. A limitation is that individuals without complete data were dropped from the analysis. However, the numbers excluded because of incomplete genotype data are few and were dropped non-differentially between cases and controls.
In contrast to our approach, others have used gene-set analysis as a method to identify important disease pathways.13 While we hypothesized that the TGF-β-signaling pathway was important, others have identified it as important using the gene-set analysis approach.11 This supports our hypothesis of the importance of the pathway. While the gene-set analysis returns a p value that shows the degree of statistical significance, our approach has been to identify the magnitude of the association with the pathway, reporting risk estimates and 95% confidence intervals associated with those estimates. In doing this, it should be recognized that cut-points are arbitrary resulting in estimates of association varying depending on where cut-points are set. Our goal was to establish a referent group with stability for estimation of risk, thus we targeted approximately the bottom 10% of the distribution as those with lowest genetic risk. However, despite the variation in exact risk estimate based on cut-points, a significant linear trend was observed across all cut-points evaluated suggests that mutational load is important in a linear fashion. It also is of interest to note, that despite variation in ORs based on cut-points, the lower bound of the CI remained consistent, providing additional support for the estimated association.
Our approach has many advantages as a way to estimate the magnitude of risk and consolidate data across a candidate pathway. This approach has reduced the number of individual comparisons with diet and lifestyle factors. We have estimated the magnitude of the association, not just a p value for significance. We have focused on the pathway as important, rather than tagSNPs for which we have limited information on their functionality. Our composite score provides a tool to evaluate how the pathway interacts with lifestyle factors, under the hypothesis that the candidate pathway is the primary object of importance.
Our identified composite lifestyle risk was of similar magnitude to that observed with our composite genotype risk. By looking at these factors together, we increase our understanding of the importance of lifestyle factors in modifying the risk associated with genetic factors. For both colon and rectal cancer associations were stronger for genotype score than lifestyle, although having the high risk score of both resulted in risk that was greater than additive for the independent risk.
We further examined three lifestyle factors that have previously been identified as independently interacting with specific genotypes within the pathway, NSAIDS, estrogen status and cigarette smoking. Both NSAID and estrogen reduced the risk associated with genetic risk. This was most evident at low levels of genetic risk. Likewise, cigarette smoking increased risk at every level of genotype risk, except at the highest level of risk. This implies that lifestyle factors are important in reducing inherited genetic susceptibility. Unfortunately when the genetic risk is more extreme, the lifestyle factors have less effect. This is in line with inherited susceptibility associated with APC or BRCA1, where the lifestyle and environmental cannot offset the inherited risk.
In summary, our data support the importance of the TGF-β-signaling pathway in the etiology of colon and rectal cancer. Our data suggest that risk increases with increasing number of deleterious variations resulting in an overall unstable or dysfunctional pathway. Our results further suggest that the integrity of the pathway, while diminished with additional at-risk genotypes, can in part be offset by lifestyle factors. These findings need replication in other studies.
Acknowledgements
This study was funded by NCI grants CA48998. This research also was supported by the Utah Cancer Registry, which is funded by Contract #N01-PC-67000 from the National Cancer Institute, with additional support from the State of Utah Department of Health, the Northern California Cancer Registry, and the Sacramento Tumor Registry. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We would like to acknowledge the contributions of Sandra Edwards, Roger Edwards, Leslie Palmer, Donna Schaffer, Dr. Kristin Anderson, Dr. John Potter, and Judy Morse for data management and collection.
Footnotes
Author Contributions:
Dr. Slattery oversaw all data collection, oversaw data analysis, and prepared manuscript.
Abbie Lundgreen provided statistical analysis and approved final paper
Jennifer Herrick provided statistical analysis and managed study data and approved final paper
Roger Wolff conducted genetic analysis and approved final paper
Bette Caan was involved in data collection and approved final paper
References
- 1.Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Zhu BaK, Natasha . Transforming Growth Factor-B and Cancer. San Francisco: John Wiley & Sons, Ltd; 2007. [Google Scholar]
- 3.Slattery ML, Lundgreen A, Herrick JS, et al. Genetic variation in bone morphogenetic protein and colon and rectal cancer. Int J Cancer. 2012;130:653–664. doi: 10.1002/ijc.26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KS, Hong SH, Bae SC. Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-beta and bone morphogenetic protein. Oncogene. 2002;21:7156–7163. doi: 10.1038/sj.onc.1205937. [DOI] [PubMed] [Google Scholar]
- 5.Cameron ER, Blyth K, Hanlon L, et al. The Runx genes as dominant oncogenes. Blood Cells Mol Dis. 2003;30:194–200. doi: 10.1016/s1079-9796(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 6.Ito Y, Osato M, Ito K. RUNX and cancer. Ann Acad Med Singapore. 2003;32:S6–S57. [PubMed] [Google Scholar]
- 7.Rajasekhar VK, Holland EC. Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene. 2004;23:3248–3264. doi: 10.1038/sj.onc.1207546. [DOI] [PubMed] [Google Scholar]
- 8.Javelaud DaMA. Smad Signal Transduction. Interplays between the Smad and Map Kinase Signaling Pathways. Protein and Cell Regulation. 2006;5:317–334. [Google Scholar]
- 9.Aggarwal BB, Shishodia S, Takada Y, et al. TNF blockade: an inflammatory issue. Ernst Schering Res Found Workshop. 2006:161–186. doi: 10.1007/3-540-37673-9_10. [DOI] [PubMed] [Google Scholar]
- 10.Ahn KS, Sethi G, Aggarwal BB. Simvastatin potentiates TNF-alpha-induced apoptosis through the down-regulation of NF-kappaB-dependent antiapoptotic gene products: role of IkappaBalpha kinase and TGF-beta-activated kinase-1. J Immunol. 2007;178:2507–2516. doi: 10.4049/jimmunol.178.4.2507. [DOI] [PubMed] [Google Scholar]
- 11.Chen LS, Hutter CM, Potter JD. Insights into Colon Cancer Etiology via a Regularized Approach to Gene Set Analysis of GWAS Data. Am J Hum Genet. 2010;86:860–871. doi: 10.1016/j.ajhg.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slattery ML, Herrick J, Curtin K, et al. Increased risk of colon cancer associated with a genetic polymorphism of SMAD7. Cancer Res. 2010;70:1479–1485. doi: 10.1158/0008-5472.CAN-08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slattery ML, Herrick JS, Lundgreen A, Wolff RK. Genetic variation in the TGF-beta signaling pathway and colon and rectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2011;20(1):57–69. doi: 10.1158/1055-9965.EPI-10-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slattery ML, Lundgreen A, Herrick JS, Wolff RK, Caan BJ. Genetic variation in the transforming growth factor-beta signaling pathway and survival after diagnosis with colon and rectal cancer. Cance. r. 2011;117:4175–4183. doi: 10.1002/cncr.26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slattery ML, Lundgreen A, Herrick JS, Caan BJ, Potter JD, Wolff RK. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and riskof colon and rectal cancer: additional support for a TGF-beta-signaling pathway. Carcinogenesis. 2011;32:318–326. doi: 10.1093/carcin/bgq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slattery ML, Potter J, Caan B, et al. Energy balance and colon cancer--beyond physical activity. Cancer Res. 1997;57:75–80. [PubMed] [Google Scholar]
- 17.Slattery ML, Wolff RK, Herrick JS, Caan BJ, Samowitz W. Calcium, vitamin D, VDR genotypes, and epigenetic and genetic changes in rectal tumors. Nutr Cancer. 2010;62:436–442. doi: 10.1080/01635580903441204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slattery ML, Benson J, Edwards S, Schaffer D. Plant foods, fiber, and rectal cancer. Am J Clin Nutr. 2004;79:276–283. doi: 10.1093/ajcn/79.2.274. [DOI] [PubMed] [Google Scholar]
- 19.Slattery ML, Samowitz W, Hoffman M, Ma KN, Levin TR, Neuhausen S. Aspirin, NSAIDs, and colorectal cancer: possible involvement in an insulin-related pathway. Cancer Epidemiol Biomarkers Prev. 2004;13:538–545. [PubMed] [Google Scholar]
- 20.Slattery ML, Neuhausen SL, Hoffman M, et al. Dietary calcium, vitamin D, VDR genotypes and colorectal cancer. Int J Cancer. 2004;111:750–756. doi: 10.1002/ijc.20330. [DOI] [PubMed] [Google Scholar]
- 21.Slattery ML, Edwards S, Curtin K. Physical activity and colorectal cancer. Am J Epidemiol. 2003;158:2142–2224. doi: 10.1093/aje/kwg134. [DOI] [PubMed] [Google Scholar]
- 22.Slattery ML, Boucher KM, Caan BJ, Potter JD, Ma KN. Eating patterns and risk of colon cancer. Am J Epidemiol. 1998;148:4–16. doi: 10.1093/aje/148.1.4-a. [DOI] [PubMed] [Google Scholar]
- 23.Slattery ML, Ballard-Barbash R, Edwards S, Caan BJ, Potter JD. Body mass index and colon cancer: an evaluation of the modifying effects of estrogen (United States) Cancer Causes Control. 2003;14:75–84. doi: 10.1023/a:1022545017867. [DOI] [PubMed] [Google Scholar]
- 24.Zha Y, Leung KH, Lo KK. TGFB1 as a susceptibility gene for high myopia: a replication study with new findings. Arch Ophthalmol. 2009;127:541–548. doi: 10.1001/archophthalmol.2008.623. [DOI] [PubMed] [Google Scholar]
- 25.Edwards S, Slattery ML, Mori M, et al. Objective system for interviewer performance evaluation for use in epidemiologic studies. Am J Epidemiol. 1994;140:1020–1028. doi: 10.1093/oxfordjournals.aje.a117192. [DOI] [PubMed] [Google Scholar]
- 26.Slattery ML, Caan BJ, Duncan D, Berry TD, Coates A, Kerber R. A computerized diet history questionnaire for epidemiologic studies. J Am Diet Assoc. 1994;94:761–766. doi: 10.1016/0002-8223(94)91944-5. [DOI] [PubMed] [Google Scholar]
- 27.Slattery ML, David R, Jacobs J. Assessment of ability to recall physical activity of several years ago. Ann Epidemiol. 1995;5:292–296. doi: 10.1016/1047-2797(94)00095-b. [DOI] [PubMed] [Google Scholar]
- 28.Liu K, Slattery M, Jacobs D, Jr, et al. A study of the reliability and comparative validity of the cardia dietary history. Ethn Dis. 1994;4:15–27. [PubMed] [Google Scholar]
- 29.Reeves GK, Travis RC, Green J. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304:426–434. doi: 10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]