Abstract
Background
Eosinophilic inflammation is implicated in asthma. Eotaxin 1–3 regulate eosinophil trafficking into the airways along with other chemotactic factors. However, the epithelial and bronchoalveolar lavage (BAL) cell expression of these chemokines in relation to asthma severity and eosinophilic phenotypes has not been addressed.
Objective
To measure the expression of the three eotaxin isoforms in bronchoscopically obtained samples and compare them with clinically relevant parameters between normal subjects and patients with asthma.
Methods
Normal subjects and patients with asthma of varying severity recruited through the Severe Asthma Research Program underwent clinical assessment and bronchoscopy with airway brushing and BAL. Eotaxin 1–3 mRNA/protein were measured in epithelial and BAL cells and compared with asthma severity, control and eosinophilic inflammation.
Results
Eotaxin-2 and eotaxin-3 mRNA and eotaxin-2 protein were increased in airway epithelial brushings from patients with asthma and were highest in cases of severe asthma (p values 0.0155, 0.0033 and 0.0006, respectively), with eotaxin-2 protein increased with age at onset. BAL cells normally expressed high levels of eotaxin-2 mRNA/protein but BAL fluid levels of eotaxin-2 were lowest in severe asthma. Epithelial eotaxin-2 and eotaxin-3 mRNA/protein was associated with sputum eosinophilia, lower forced expiratory volume in 1 s and more asthma exacerbations. Airway epithelial cell eotaxin-2 protein differed by asthma severity only in those with late onset disease, and tended to be highest in those with late onset eosinophilic asthma.
Conclusions
Epithelial eotaxin-2 and 3 are increased in asthma and severe asthma. Their expression may contribute to luminal migration of eosinophils, especially in later onset disease, asthma control and severity.
INTRODUCTION
Severe asthma represents approximately 10% of asthma but disproportionately impacts the mortality, morbidity and costs of asthma.1 Eosinophilic inflammation has long been implicated in severe asthma, with several reports suggesting a more prominent role in a late onset phenotype.2–5
Eosinophil trafficking into the airways is a complex process likely regulated by a host of cells and chemotactic factors, including interleukin 5 (IL-5), eotaxins 1–3 (CCL11, 24 and 26) and regulated on activation normal T-cell expressed and secreted (RANTES) among others.6 IL-5 stimulates proliferation, differentiation and survival of eosinophils in the bone marrow and contributes to airway migration.7 Three recent studies of monoclonal antibodies to IL-5 in eosinophil-predominant asthma demonstrated reductions in blood/sputum eosinophils and improved asthma control.3,7–9 Yet, despite a general reduction in eosinophils, tissue eosinophils remained and eosinophilic exacerbations still occurred.3,10 Thus, it is likely additional factors are involved.
Eotaxin 1–3 are CC chemokines of divergent structure which engage the CCR3 receptor and stimulate eosinophilic chemotaxis. IL-4 and IL-13 induce eotaxins in vitro through activation of the IL-4Rα receptor/STAT-6 pathway, a process generally reported to be attenuated by corticosteroids (CS) in vitro.11–13 All three eotaxins have variably been reported to be increased in epithelial cells from endobronchial biopsies of patients with mild asthma, and in response to allergen challenge.14–17 However, their relative contribution to asthma severity, control or eosinophilic phenotypes remains poorly understood.
We hypothesised that epithelial eotaxin expression would contribute to eosinophil trafficking towards the airway lumen in association with asthma severity/control and in relation to airway eosinophilia. mRNA and protein expression for the three eotaxin isoforms was evaluated in freshly harvested bronchial brushing cells (primarily airway epithelial cells (AECs)) and bronchoalveolar lavage (BAL) cells/fluid from patients with asthma ranging in severity and normal controls (NCs). The relationship of this expression to physiological and clinical markers of severity/control, luminal eosinophilic inflammation and age at asthma onset was addressed.
METHODS
Study participants
Subjects were recruited as part of the National Heart Lung and Blood Institute’s Severe Asthma Research Program (SARP). Asthma was defined using American Thoracic Society criteria and subjects divided into mild, mild/moderate and severe asthma as previously described.18 See online supplement for details.
Bronchoscopy with airway brushing and lavage
Bronchoscopy was performed as previously described.19,20 See online supplement for details.
Sputum induction and processing
Sputum was processed according to the method of Fahy et al.21 Please see online supplement for details.
Real-time quantitative PCR analysis
Eotaxin 1–3 mRNA expression from epithelial and BAL cells was determined by reverse transcription and quantitative real-time PCR, as previously described. Please see online supplement for details.
Enzyme-linked immunosorbent assays
Eotaxin protein levels in AEC and BAL cell lysates and BAL fluid were measured by ELISA with antibodies from R&D Systems (Minneapolis, Minnesota, USA) by ELISATech (Aurora, Colorado, USA), as described in the online supplement.
Eotaxin 1–3 stability to the mast cell protease tryptase
Eotaxin 1–3 (R&D Systems) final concentration 500–700 pg/ml in phosphate-buffered saline were incubated with or without tryptase (recombinant lung, 13 mUnits/ml; Promega, Madison, Wisconsin, USA) and heparin (0.33 units/ml) for 20 h at room temperature. Samples were quantified for eotaxin 1–3 by ELISA as above.
Corticosteroid effects on eotaxin expression in vitro
Primary human bronchial epithelial cells were cultured under air–liquid interface (ALI) as previously described.22 From day 0 of ALI, cells were stimulated with IL-13 (10 ng/ml) every 48 h. On day 6, in addition to IL-13, dexamethasone (100 nM) was added to the cells. Cells were harvested on day 8 and analysed for eotaxin-2 and eotaxin-3 protein using the above methods.
Statistical analyses
Epithelial and BAL cell mRNA and protein data were generally not normally distributed and subsequently analysed non-parametrically. The Kruskal–Wallis variation of the Wilcoxon-rank sum test compared eotaxin 1–3 expression between groups. Intergroup comparisons were made by Wilcoxon rank sum when the overall p value was <0.05. Bonferroni correction was applied with a p value of 0.008 considered significant. Spearman’s rho was used for correlations. In addition, a multivariable non-parametric regression of medians was modelled to test the association between eotaxin-2 protein with age of asthma onset, adjusting for severity, gender, age and age at enrolment.23 A p value <0.05 was regarded as significant. Racial data were analysed by Fisher’s exact t test, taking into consideration cells with ‘zero’ values. Data are presented as medians with 25th–75th percentile ranges. Graphs include maximums and minimums. Data are occasionally presented on a log10 scale for illustrative purposes. Statistics were performed using JMP8 software (SAS).
RESULTS
Subject characteristics
A total of 100 subjects underwent bronchoscopy, including 27 NCs, 9 patients with mild asthma, 20 with mild to moderate asthma on inhaled corticosteroids (mild/mod+ICS) and 44 with severe asthma (table 1). The groups did not differ by sex, race or age at onset but there was a significant difference in age (p=0.004), with patients with severe asthma older than NCs and those with mild/mod+ICS. As expected, patients with severe asthma had the lowest pre-bronchodilator forced expiratory volume in 1 s (FEV1%), but the greatest reversal post bronchodilator. Twenty-one (48%) of the patients with severe asthma used regular systemic corticosteroids (SCS). Atopy was less common in NCs (overall p=0.002), but there were no differences among asthmatic groups.
Table 1.
Subject characteristics
| Normal | Mild | Mild/mod+ICS | Severe | p Value | |
|---|---|---|---|---|---|
| Age (±SD) | 34±12 | 33±15 | 33±11 | 43±12 | 0.004 |
| Race*, n (%) | 22/2/3 (81/7/11) | 8/0/1 (89/0/11) | 12/4/4 (60/20/20) | 34/7/3 (77/16/7) | 0.42 |
| Sex (M/F), n (%) | 14/13 (52/48) | 2/7 (22/78) | 6/14 (30/70) | 11/33 (25/75) | 0.10 |
| FEV1% pred (±SD) | 100±2 | 91±3 | 90±4 | 56±3 | <0.0001 |
| FEV1 reversal % (±SEM) | 6±0.8 | 9±3 | 11±2 | 33±5 | <0.0001 |
| Eosinophils in sputum, % (25th–75th percentiles) | 0.9 (0–3.6) | 5 (0.3–8.3) | 1 (0.4–2.8) | 6.9 (1.8–31.3) | 0.0012 |
| Age at onset, median (25th–75th percentiles) | NA | 12 (5–40) | 5 (2–14) | 8 (2–32) | 0.26 |
| Atopy (Y/N) | 14/13 | 9/0 | 16/2 | 29/8 | 0.002 |
Caucasian/African American/other.
FEV1% pred, forced expiratory volume in 1 s % predicted; ICS, inhaled corticosteroid; NA, not applicable.
Epithelial eotaxin-1 expression is low and does not differ by subject type
Eotaxin-1 mRNA levels were extremely low and did not differ according to asthma or severity (n=74, p=0.76, data not shown). Eotaxin-1 protein was undetectable in epithelial cell lysates from six NCs, seven patients with severe asthma, four with mild/mod+ICS asthma and one with mild asthma. Further studies of eotaxin-1 were not performed.
Epithelial eotaxin-2 expression is increased in severe asthma
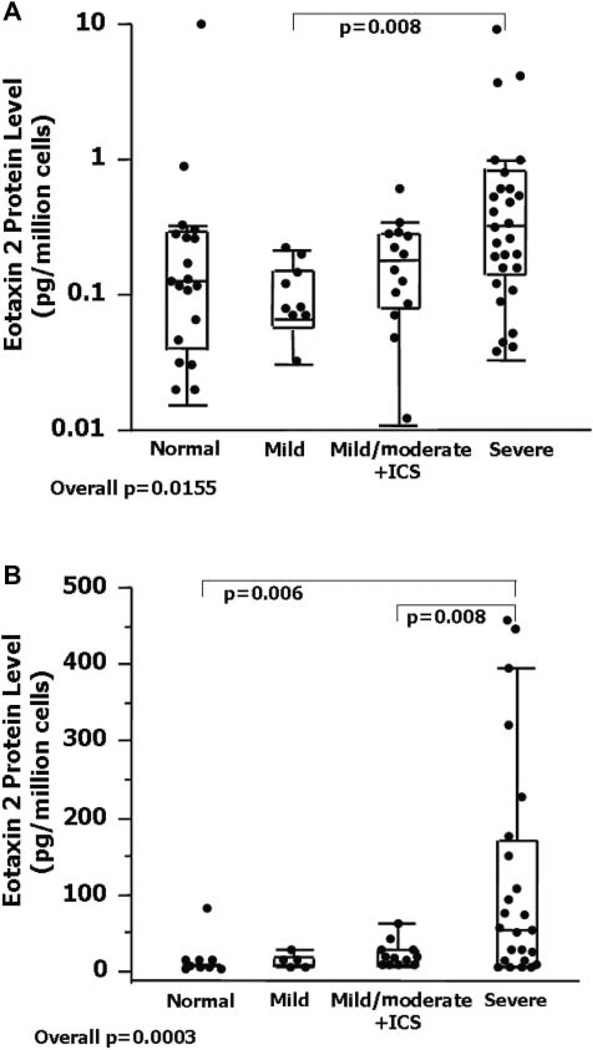
AEC eotaxin-2 mRNA levels differed among groups (n=72, overall p value=0.0155) and were higher in severe than in mild asthma (figure 1A). AEC lysate eotaxin-2 protein was detectable in 19/25 patients with severe asthma, 5/13 with mild/mod +ICS asthma, 1/5 with mild asthma and 4/12 NCs (p=0.0003, figure 1B). Patients with severe asthma expressed higher eotaxin-2 protein levels than NCs (p=0.006) and those with mild/mod+ICS asthma (p=0.008). Patients with asthma on SCS expressed higher eotaxin-2 protein and mRNA compared with those not on SCS (protein: no SCS=5 (5–28) vs SCS=58 (5–151) pg/ml, p=0.006; mRNA/per glyceraldehyde 3-phosphate dehydrogenase (GAPDH): no SCS=0.15 (0.07–0.27) vs SCS=0.34 (0.18–0.82) pg/ml, p=0.004, respectively). Given the high levels of eotaxin-2 protein in those with severe asthma on SCS, the effect of the addition of CS on eotaxin-2 protein was evaluated in ALI-cultured epithelial cells. The addition of dexamethasone decreased eotaxin-2 protein levels by 25+6%, p<0.0001. There were no differences by subject group. AEC eotaxin-2 mRNA and protein modestly correlated (r=0.40, p=0.01).
Figure 1.
(A) Eotaxin-2 mRNA expression in cells obtained by epithelial brushings from patients with asthma and normal controls. (bB) Eotaxin-2 protein levels among the subject groups. Data represent the values of the median, 25th and 75th percentiles with maximum/minimum whiskers. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ICS inhaled corticosteroid.
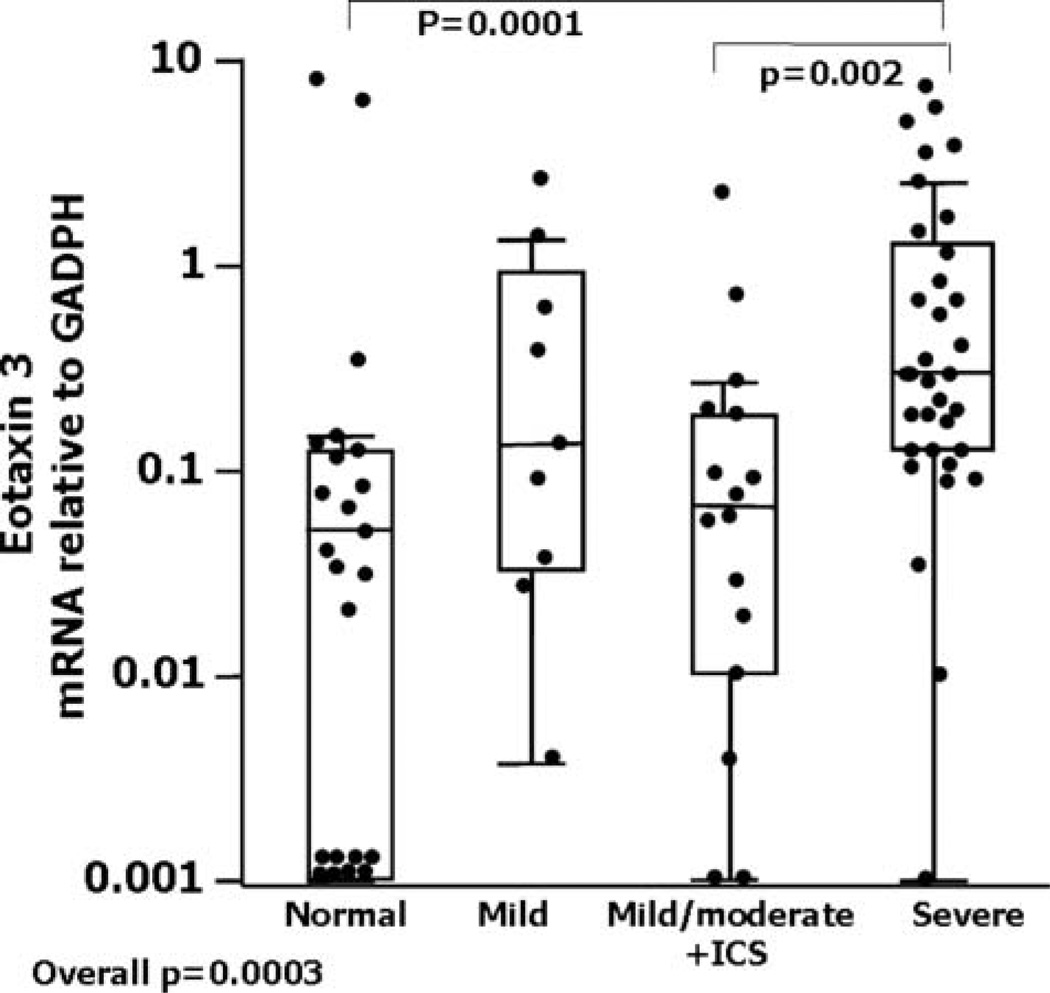
Epithelial eotaxin-3 mRNA is increased in severe asthma
AEC eotaxin-3 mRNA also differed across groups (overall p value=0.0003, figure 2), being higher in severe asthma compared with NCs (p=0.0001) and mild/mod+ICS (p=0.002). Eotaxin-3 protein from 26 AEC lysates was detectable in only five subjects (3/11 with severe asthma). Given the small lysate sample volumes and the low eotaxin-3 protein levels, further eotaxin-3 protein measurements were not performed. Similar to eotaxin-2, eotaxin-3 mRNA levels indexed to GAPDH were higher in those on SCS (0.3 (0.11–0.73)) compared with those not on SCS (0.12 (0.02–0.55)) (p=0.04). Eotaxin-2 and eotaxin-3 mRNA levels only modestly correlated (r=0.24, p=0.04). Similar to eotaxin-2, the impact of CS on eotaxin-3 protein expression was also evaluated in asthmatic epithelial cells in ALI. Dexamethasone decreased eotaxin-3 protein levels by 24+12%, p<0.0001 (n=11), without difference by severity.
Figure 2.
Eotaxin-3 mRNA levels in cells obtained by epithelial brushings from patients with asthma and normal controls (NCs). Data represent the values of the median, 25th and 75th percentiles with maximum/minimum whiskers. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ICS inhaled corticosteroid.
Impact of mast cell tryptase on eotaxin 1–3 protein levels
Eotaxin-2 has previously been reported to be resistant to mast cell proteases, known to be present in the airway epithelium.24 To determine whether, like eotaxin-1, eotaxin-3 might also be susceptible to tryptase and explain the lower protein levels, known levels of eotaxin 1–3 were incubated with tryptase and heparin. Tryptase selectively reduced eotaxin-1 and eotaxin-3 protein levels, while having little effect on eotaxin-2 protein. Eotaxin-1 was reduced by 85%, SEM±0.9% and eotaxin-3 by 70%, SEM±11.5%. Under the same conditions eotaxin-2 remained stable (5% reduction, SEM±1.98%).
BAL cells express high levels of eotaxin-2 mRNA/protein but with variable relationship to severity
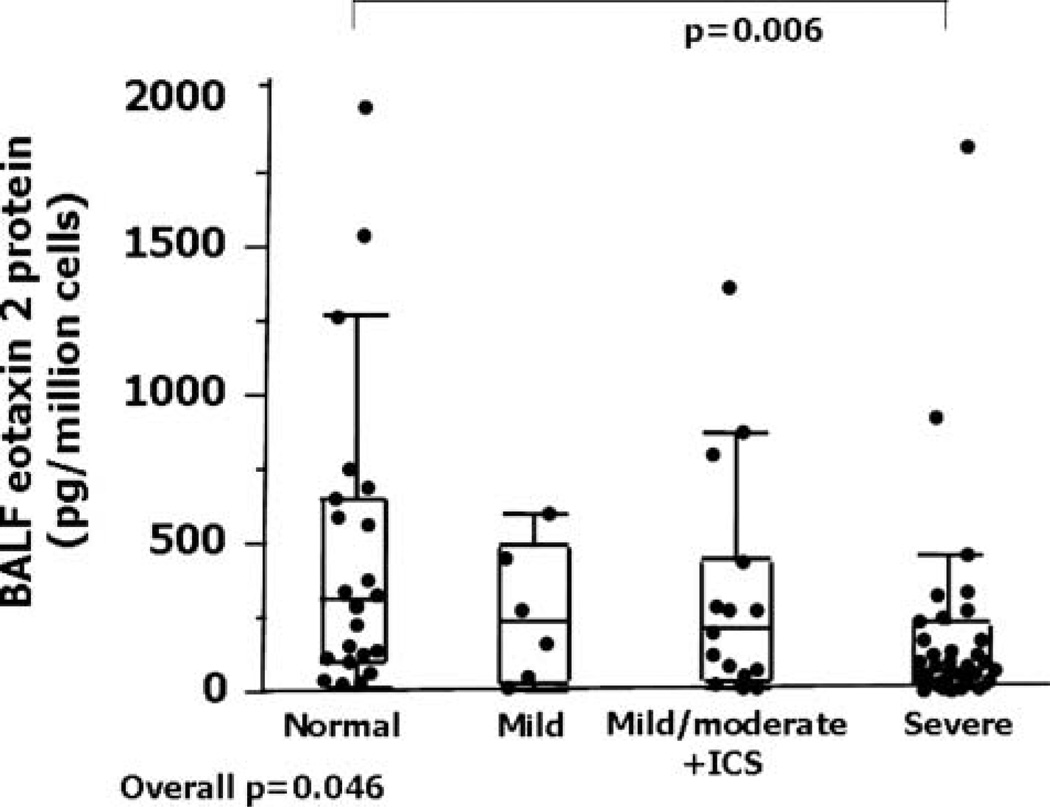
BAL cells expressed high levels of eotaxin-2 mRNA, being 100-fold higher than AEC levels (table 2). Unlike epithelial brushings, the levels did not differ by subject group (overall p=0.83). Eotaxin-2 protein levels in BAL cell lysates and unconcentrated BAL fluid samples were relatively high and strongly correlated (r=0.74, p=0.0002, n=28). Given the limited BAL cell lysate volume and the strong correlations between BAL fluid and BAL lysates, further comparisons among groups were made on BAL fluid samples (n=75). BAL fluid eotaxin-2 levels differed among groups (p=0.046). In contrast to epithelial cells, BAL fluid eotaxin-2 protein levels were lower in severe asthma than in NCs (p=0.006), but not different from the two milder asthma groups (figure 3). BAL cell eotaxin-2 mRNA did not correlate with BAL cell lysate protein (r=0.34, p=0.18, n=17) or BAL fluid eotaxin-2 protein (r=0.08, p=0.66, n=33). BAL fluid eotaxin-2 protein did not correlate with any epithelial cell mRNA or protein levels.
Table 2.
Bronchoalveolar lavage eotaxin levels by severity
| Normal | Mild | Mild/mod+ICS | Severe | Overall p | |
|---|---|---|---|---|---|
| Eotaxin-2 mRNA/GAPDH (n=42) | 19 (1.9–58.7) | 24.7 (4.6–310.9) | 73.5 (3.9–502.2) | 10.3 (3.4–47.5) | 0.83 |
| Eotaxin-2 cell lysate protein (pg/million cells) (n=19) | 1178 (135–3134) | 816 (370–3263) | 795 (456–1195) | 511 (315–2670) | 0.81 |
| Eotaxin-2 BAL fluid (pg/ml) (n=75) | 309 (97–654) | 221 (41–425) | 196 (37–435) | 77 (30–222) | 0.046 |
| Eotaxin-1 mRNA/GAPDH (n=40) | 0.0001 (0–0.04) | 0.0001 (0–0.1) | 0 (0–0.045) | 0 (0–0.0001) | 0.62 |
| Eotaxin-3 mRNA/GAPDH (n=40) | 0.0001 (0–0.06) | 0.0001 (0–0.15) | 0.42 (0–0.11) | 0.05 (0.15–0.25) | 0.14 |
All values are median (25th–75th percentiles).
BAL, bronchoalveolar lavage; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Figure 3.
Bronchoalveolar lavage fluid eotaxin-2 levels from patients with asthma and normal controls. Data represent medians with 25th–75th percentiles and maximum/minimum whiskers. BALF, bronchoalveolar lavage fluid; ICS inhaled corticosteroid.
BAL cell eotaxin-1 and eotaxin-3 expression
Similar to epithelial cells, BAL cells expressed low levels of eotaxin-1 mRNA which did not differ across subject groups (table 2, p=0.62, n=40). Eotaxin-1 protein was not measurable in any BAL cell lysate. BAL cell eotaxin-3 mRNA levels were low and did not differ across groups (p=0.14, n=40). However, the pattern was similar to epithelial cells, with levels numerically higher in asthma and severe asthma (table 2). Eotaxin-3 protein was not detected in BAL cell lysates or in BAL fluid. Unlike eotaxin-2, BAL cell eotaxin-3 mRNA marginally correlated with epithelial cell eotaxin-3 mRNA (r=0.31, p=0.07).
Eotaxin-2 and eotaxin-3 expression: relation to control characteristics
In patients with asthma only, AEC eotaxin-2 and eotaxin-3 mRNA levels inversely correlated with FEV1% predicted (r=−0.39, p=0.0005 and −0.31, p=0.005 respectively). AEC eotaxin-2 protein also inversely correlated with FEV1% predicted (r=−0.35, p=0.013). There were no correlations of eotaxin levels in any compartment with reversibility. However, higher eotaxin-2 mRNA and protein associated with three or more oral CS bursts in the previous year (p=0.006 and p=0.004, respectively) (table 3). Higher eotaxin-3 mRNA was marginally associated with a history of three or more CS bursts (p=0.051). Higher eotaxin-2 (mRNA and protein) and eotaxin-3 mRNA levels all associated with an emergency room visit in the previous year (table 3). BAL fluid eotaxin-2 mRNA or protein levels did not correlate with any control characteristics.
Table 3.
Epithelial eotaxins in relation to exacerbations
| No ER visit | ER visit | p Value | <3 oral CS bursts | ≥3 oral CS bursts | p Value | |
|---|---|---|---|---|---|---|
| Eotaxin-2 protein (pg/million cells) | 5 (5–28) | 30 (5–94) | 0.04 | 5 (5–27) | 42 (5–137) | 0.004 |
| Eotaxin-2 mRNA/GAPDH | 0.16 (0.07–0.27) | 0.26 (0.12–0.26) | 0.06 | 0.15 (0.08–0.29) | 0.32 (0.18–0.85) | 0.006 |
| Eotaxin-3 mRNA/GAPDH | 0.10 (0.03–0.38) | 0.34 (0.13–0.88) | 0.01 | 0.09 (0.15–0.66) | 0.3 (0.12–0.71) | 0.051 |
All values median (25th–75th percentiles).
CS, corticosteroid; ER, emergency room; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Relation to eosinophilia and age at onset
Evaluating patients with asthma only, AEC eotaxin-2 and eotaxin-3 mRNA modestly correlated with sputum eosinophils (r=0.3, p=0.048 and p=0.038, respectively) while eotaxin-2 mRNA also correlated with BAL eosinophils (r=0.32, p=0.04). Epithelial eotaxin-2 protein positively correlated with sputum eosinophils (r=0.48, p=0.01, n=34), while BAL fluid eotaxin-2 protein was marginally inversely correlated (r=−0.34, p=0.046). There were no correlations with BAL eosinophils.
AEC eotaxin-2 protein correlated with age at onset (in epithelial and BAL cell lysates (r=0.58, p=0.0001 and ρ=0.53, p=0.01 respectively)) and age at enrolment (r=0.50, p=0.0001 epithelial lysates only). Using a median regression (nonparametric) model incorporating severity, gender, age and age at enrolment, increasing age at onset was the only significant contributor to epithelial eotaxin-2 protein levels. Additionally, AEC eotaxin-2 protein levels in patients with asthma with age at onset under 12 years did not differ by asthma severity (p=0.28, n=23, severe vs milder asthma). In contrast, AEC eotaxin-2 protein levels in later onset asthma (onset ≥12 years of age) were highest in those with severe (94 (52–275) pg/million cells, n=13) compared with milder asthma (5 (5–37) pg/million cells, n=5) (p=0.006).
As previously reported,4,25 sputum eosinophilia (>2%) was also more common in later onset asthma (≥12 years of age n=18) (83%) than in early onset asthma (onset <12 years of age n=32) (44%) (p=0.005). AEC eotaxin-2 protein levels tended to be higher in late onset eosinophilic (>2% eosinophils) asthma (53 (17–298) pg/million cells, n=9) compared with early onset eosinophilic asthma (23 (5–36) pg/million cells, n=10) (p=0.08).
DISCUSSION
This study identified differential expression of the three eotaxin isoforms from freshly brushed epithelial cells of patients with asthma and normal controls. While eotaxin-1 expression was negligible, eotaxin-2 and eotaxin-3 mRNA were higher in patients with asthma and highest in those with severe asthma. At the protein level, eotaxin-2 was measurable and significantly higher in patients with severe asthma compared with those with milder asthma. Eotaxin-2 and eotaxin-3 expression related to lower lung function, asthma exacerbations and importantly eosinophilia. Eotaxin-2 protein was strongly predicted by increasing age at asthma onset, suggesting a distinct role for eotaxin-2 in the eosinophilia associated with later onset asthma.4,25 Unlike epithelial cells, BAL cells constitutively expressed eotaxin-2 mRNA and protein, with generally lower BAL fluid eotaxin-2 protein levels in severe asthma.
In contrast to eotaxin-2 and 3, eotaxin-1 mRNA expression in epithelial and BAL cells was nearly undetectable. This result differs from previous studies.12,15,26,27 The reasons for this difference are not clear, although highly quantifiable quantitative PCR and ELISA were used here. Interestingly, a previous study did not find eotaxin-1 expression in primary human epithelial cells in either submerged or ALI cultures.28
The current study suggests eotaxin-2 and 3 are relevant to luminal migration of eosinophils. Epithelial eotaxin-2, at the biologically relevant protein level, was quantifiable, differentiated severe from milder asthma and was associated with eosinophilic inflammation. This is consistent with a previous report of increased eotaxin-2 and 3 (as measured by immuno-histochemistry) in the epithelium of patients with asthma following allergen challenge.14 However, this study expands on those results by evaluating a broad range of carefully phenotyped patients with chronic asthma recruited through SARP.
Despite similar absolute epithelial mRNA levels for eotaxin-2 and 3 measured in this study, the levels poorly correlated. Eotaxin-2 and 3 are induced in human AECs by T helper 2 cytokines, but their expression differs by epithelial differentiation. Eotaxin-2 is generated by less differentiated cells in submerged culture, while eotaxin-3 is generated by fully differentiated cells in ALI.28 In addition to different patterns of mRNA expression, only eotaxin-2 was consistently detectable at the protein level in epithelial and BAL cell lysates and BAL fluid. Our inability to consistently detect eotaxin-3 in the epithelial or BAL cell lysates/fluid despite significant differences in epithelial mRNA is likely due to enzymatic degradation by mast cell tryptase, as our results show that tryptase selectively degrades eotaxin-124,29 and 3, while eotaxin-2 remains stable.
Surprisingly, eotaxin-2 protein in AEC and BAL cell lysates strikingly correlated with age at asthma onset, and the levels were higher in patients with ‘late onset’ (≥12 years) disease. In fact, the differences in eotaxin-2 protein by severity were dependent on age at onset, with the differences by severity only observed in those with later onset asthma. Later/adult onset asthma has been associated with persistent eosinophilic inflammation despite high-dose ICS and even SCS.4,25 Although eotaxin-2 and 3 (mRNA and/or protein) associated and correlated with eosinophilic inflammation, eotaxin-2 protein levels were more strongly associated with the eosinophilia observed in late as opposed to early onset asthma (p=0.08). Thus, these findings are the first to suggest a unique mechanism for the eosinphilia associated with later onset asthma but the mechanisms for the increase are not clear.
The high levels of eotaxin-2 and 3 in severe asthma suggest a relative CS resistance. AEC eotaxin-2 and 3 were higher in patients with asthma on SCS compared with those not on SCS. This sustained expression is surprising given the reported suppression of IL-4/IL-13-stimulated eotaxins by CS in vitro and in those with mild asthma treated with oral CS.11,30,31 To address this, IL-13-treated epithelial cells were treated with dexamethasone for 48 h. Although IL-13-stimulated eotaxin-2 and eotaxin-3 protein were consistently suppressed by CSs, the overall decrease was small (25%). Thus, although in simple cell culture systems there is consistent suppression, the translation of this to the more complicated in vivo setting is unclear. For instance, continued eotaxin expression could involve the presence of other cell types or cytokines beyond IL-4/IL-13, including tumour necrosis factor α, IL-1ß and others, which may not be suppressed by CS.16
In addition to being associated with increased asthma severity, eotaxin-2 and 3 were associated with poor asthma control, as measured by lower FEV1% predicted and frequent exacerbations. However, the relative importance of these CC chemokines, compared with IL-5, eosinophilic inflammation and asthma remains poorly understood. The recent positive clinical trials of specific monoclonal antibodies to anti-IL-5 suggest that IL-5, in isolation, has substantial impact on lung eosinophils, asthma control, and in one case, FEV1.3,8,9 However, anti-IL-5 also incompletely reduced airway eosinophilia, suggesting other pro-eosinophilic factors remain.3,10 The expression of eotaxins in these studies was not reported, but they were not likely to have been inhibited. Interestingly, murine models of allergic inflammation suggest that reductions in both IL-5 and eotaxins are required to eliminate the eosinophilia post-allergen challenge.32
While epithelial cell expression of eotaxin-2 and 3 increased with increasing asthma severity and eosinophilia, BAL cells (primarily monocytes/macrophages) constitutively expressed high levels of eotaxin-2 mRNA and protein (but not eotaxin-1), a finding previously reported by normal human monocytes in vitro.33 The expression in BAL cell lysates strongly correlated with BAL fluid levels, supporting these cells as the primary source. Unlike epithelial cells, BAL fluid levels were lower in severe asthma and did not correlate with eosinophilia. Thus, although BAL cell/fluid eotaxin-2 protein may play a role in lung eosinophilia in some conditions, it does not appear to play a prominent role in asthma-related eosinophilia.
Limitations of the current study include the lack of data for each eotaxin isoform from each subject, across all cell samples. This impacted the measurement of eotaxin-2 protein in the BAL cell lysates. However, the strong correlations with BAL fluid eotaxin-2 imply that the findings in BAL fluid generally reflect the BAL cell lysates. The extremely low levels of eotaxin-1 mRNA/protein and eotaxin-3 mRNA in the BAL cells suggest that analysing greater numbers of these small volume samples would not add to the findings. Finally, sputum samples were not available on every subject due to FEV1 restrictions and inability to produce sputum. Greater numbers of sputum samples may have improved our ability to address the specificity of the eotaxin-2 increases to late onset eosinophilic asthma.
In summary, this study shows significant increases in eotaxin-2 and 3 in the epithelium in human severe asthma in association with poor asthma control and sputum eosinophilia. Higher eotaxin-2 protein levels (in the epithelial and BAL cell compartment) were strongly associated with older age at asthma onset, and marginally with the eosinophilia associated with late onset asthma. Whether eotaxin-2 plays a specific contributory role in this persistent and often CS-refractory eosinophilic phenotype awaits further study.
Supplementary Material
Key messages.
What is the key question?
-
►
Do epithelial and/or bronchoalveolar lavage cell expression of eotaxin isoforms contribute to asthma severity and eosinophilic phenotypes?
What is the bottom line?
-
►
Epithelial eotaxin-2 and 3 are increased in patients with asthma, highest in those with severe asthma and are associated with eosinophilia. Eotaxin-2, in particular, strongly correlates with late onset asthma and its associated greater degree of eosinopihilia.
Why read on?
-
►
Epithelial eotaxins may contribute to the eosinophilia associated with asthma, with eotaxin-2 a potentially important contributor to the greater degree of eosinophilia associated with later onset asthma.
Acknowledgments
Funding National Institute of Health, National Heart Lung and Blood Institute. HL-69174, AI40600, AI67780 and CTSI UL1 RR024153.
Footnotes
Additional supplementary files are published online only. To view these files please visit the journal online (http://dx.doi.org/10.1136/thoraxjnl-2012-201634).
Competing interests None.
Ethics approval University of Pittsburgh and National Jewish Health Institutional Review Boards.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–2351. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel SE, Schwartz LB, Langmack EL, et al. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med. 1999;160:1001–1008. doi: 10.1164/ajrccm.160.3.9812110. [DOI] [PubMed] [Google Scholar]
- 3.Haldar PBC, Hargadon B, Gupat S, et al. Mepolizumab (anti-IL 5) and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miranda C, Busacker A, Balzar S, et al. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol. 2004;113:101–108. doi: 10.1016/j.jaci.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 5.ten Brinke A, Zwinderman AH, Sterk PJ, et al. Factors associated with persistent airflow limitation in severe asthma. Am J Respir Crit Care Med. 2001;164:744–748. doi: 10.1164/ajrccm.164.5.2011026. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg HF, Phipps S, Foster PS. Eosinophil trafficking in allergy and asthma. J Allergy Clin Immunol. 2007;119:1303–1310. doi: 10.1016/j.jaci.2007.03.048. quiz 1311–1302. [DOI] [PubMed] [Google Scholar]
- 7.Humbert M, Corrigan CJ, Kimmitt P, et al. Relationship between IL-4 and IL-5 mRNA expression and disease severity in atopic asthma. Am J Respir Crit Care Med. 1997;156:704–708. doi: 10.1164/ajrccm.156.3.9610033. [DOI] [PubMed] [Google Scholar]
- 8.Nair P, Pizzichini M, Kjarsgaard MIM, et al. Mepolizumab in prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360:985–993. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 9.Castro M, Mathur S, Hargreave F, et al. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo-controlled study. Am J Respir Crit Care Med. 2011;184:1125–1132. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 10.Flood-Page P, Menzies-Gow A, Phipps S, et al. Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J Clin Invest. 2003;112:1029–1036. doi: 10.1172/JCI17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stellato C, Matsukura S, Fal A, et al. Differential regulation of epithelial-derived C-C chemokine expression by IL-4 and the glucocorticoid budesonide. J Immunol. 1999;163:5624–5632. [PubMed] [Google Scholar]
- 12.Matsukura S, Stellato C, Georas SN, et al. Interleukin-13 upregulates eotaxin expression in airway epithelial cells by a STAT6-dependent mechanism. Am J Respir Cell Mol Biol. 2001;24:755–761. doi: 10.1165/ajrcmb.24.6.4351. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann N, Hershey GK, Foster PS, et al. Chemokines in asthma: cooperative interaction between chemokines and IL-13. J Allergy Clin Immunol. 2003;111:227–42. doi: 10.1067/mai.2003.139. quiz 243. [DOI] [PubMed] [Google Scholar]
- 14.Ravensberg AJ, Ricciardolo FL, van Schadewijk A, et al. Eotaxin-2 and eotaxin-3 expression is associated with persistent eosinophilic bronchial inflammation in patients with asthma after allergen challenge. J Allergy Clin Immunol. 2005;115:779–785. doi: 10.1016/j.jaci.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 15.Komiya A, Nagase H, Yamada H, et al. Concerted expression of eotaxin-1, eotaxin-2, and eotaxin-3 in human bronchial epithelial cells. Cell Immunol. 2003;225:91–100. doi: 10.1016/j.cellimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Lilly CM, Nakamura H, Belostotsky OI, et al. Eotaxin expression after segmental allergen challenge in subjects with atopic asthma. Am J Respir Crit Care Med. 2001;163:1669–1675. doi: 10.1164/ajrccm.163.7.9812044. [DOI] [PubMed] [Google Scholar]
- 17.Berkman N, Ohnona S, Chung FK, et al. Eotaxin-3 but not eotaxin gene expression is upregulated in asthmatics 24 hours after allergen challenge. Am J Respir Cell Mol Biol. 2001;24:682–687. doi: 10.1165/ajrcmb.24.6.4301. [DOI] [PubMed] [Google Scholar]
- 18.Moore WC, Bleecker ER, Curran-Everett D, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trudeau J, Hu H, Chibana K, et al. Selective downregulation of prostaglandin E2-related pathways by the Th2 cytokine IL-13. J Allergy Clin Immunol. 2006;117:1446–1454. doi: 10.1016/j.jaci.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 20.Moore WC, Evans MD, Bleecker ER, et al. Safety of investigative bronchoscopy in the Severe Asthma Research Program. J Allergy Clin Immunol. 2011;128:328–336.e3. doi: 10.1016/j.jaci.2011.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahy JV, Liu J, Wong H, et al. Cellular and biochemical analysis of induced sputum from asthmatic and from healthy subjects. Am Rev Respir Dis. 1993;147:1126–1131. doi: 10.1164/ajrccm/147.5.1126. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, O’Donnell VB, Balzar S, et al. 15-Lipoxygenase 1 interacts with phosphatidylethanolamine-binding protein to regulate MAPK signaling in human airway epithelial cells. Proc Natl Acad Sci USA. 2011;108:14246–14251. doi: 10.1073/pnas.1018075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gould W. Quantile regression with bootstrapped standard errors. Stata Tech Bull. 1993;2:19–21. [Google Scholar]
- 24.Balzar S, Fajt ML, Comhair SA, et al. Mast cell phenotype, location, and activation in severe asthma. Data from the Severe Asthma Research Program. Am J Respir Crit Care Med. 2011;183:299–309. doi: 10.1164/rccm.201002-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haldar P, Pavord ID, Shaw DE, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178:218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lilly CM, Nakamura H, Kesselman H, et al. Expression of eotaxin by human lung epithelial cells: induction by cytokines and inhibition by glucocorticoids. J Clin Invest. 1997;99:1767–1773. doi: 10.1172/JCI119341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying S, Robinson DS, Meng Q, et al. Enhanced expression of eotaxin and CCR3 mRNA and protein in atopic asthma. Association with airway hyperresponsiveness and predominant co-localization of eotaxin mRNA to bronchial epithelial and endothelial cells. Eur J Immunol. 1997;27:3507–3516. doi: 10.1002/eji.1830271252. [DOI] [PubMed] [Google Scholar]
- 28.van Wetering S, Zuyderduyn S, Ninaber DK, et al. Epithelial differentiation is a determinant in the production of eotaxin-2 and -3 by bronchial epithelial cells in response to IL-4 and IL-13. Mol Immunol. 2007;44:803–811. doi: 10.1016/j.molimm.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Pang L, Nie M, Corbett L, et al. Mast cell beta-tryptase selectively cleaves eotaxin and RANTES and abrogates their eosinophil chemotactic activities. J Immunol. 2006;176:3788–3795. doi: 10.4049/jimmunol.176.6.3788. [DOI] [PubMed] [Google Scholar]
- 30.Banwell ME, Tolley NS, Williams TJ, et al. Regulation of human eotaxin-3/CCL26 expression: modulation by cytokines and glucocorticoids. Cytokine. 2002;17:317–323. doi: 10.1006/cyto.2002.1021. [DOI] [PubMed] [Google Scholar]
- 31.Scheicher ME, Teixeira MM, Cunha FQ, et al. Eotaxin-2 in sputum cell culture to evaluate asthma inflammation. Eur Respir J. 2007;29:489–495. doi: 10.1183/09031936.00060205. [DOI] [PubMed] [Google Scholar]
- 32.Pope SM, Fulkerson PC, Blanchard C, et al. Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J Biol Chem. 2005;280:13952–13961. doi: 10.1074/jbc.M406037200. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe K, Jose PJ, Rankin SM. Eotaxin-2 generation is differentially regulated by lipopolysaccharide and IL-4 in monocytes and macrophages. J Immunol. 2002;168:1911–1918. doi: 10.4049/jimmunol.168.4.1911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.