Abstract
LIM domain Only 2 (Lmo2) is frequently deregulated in sporadic and gene therapy-induced acute T-cell lymphoblastic leukemia (T-ALL) where its overexpression is an important initiating mutational event. In transgenic and retroviral mouse models, Lmo2 expression can be enforced in multiple hematopoietic lineages but leukemia only arises from T cells. These data suggest that Lmo2 confers clonal growth advantage in T-cell progenitors. We analyzed proliferation, differentiation, and cell death in CD2-Lmo2 transgenic thymic progenitor cells to understand the cellular effects of enforced Lmo2 expression. Most impressively, Lmo2 transgenic T-cell progenitor cells were blocked in differentiation, quiescent, and immortalized in vitro on OP9-DL1 stromal cells. These cellular effects were concordant with a transcriptional signature in Lmo2 transgenic T-cell progenitor cells that is also present in hematopoietic stem cells and Early T-cell Precursor ALL. These results are significant in light of the crucial role of Lmo2 in the maintenance of the hematopoietic stem cell. The cellular effects and transcriptional effects have implications for LMO2-dependent leukemogenesis and the treatment of LMO2-induced T-ALL.
Keywords: T cells, hematopoiesis, stem cells, leukemia
Introduction
LIM domain Only-2 (LMO2) is one of the most frequently deregulated oncogenes in human acute T-cell lymphoblastic leukemia (T-ALL) where it is mono-allelically or bi-allelically overexpressed by diverse chromosomal rearrangements and other transcriptional mechanisms(1),(2-6). Despite elegant and extensive studies on Lmo2’s normal role in hematopoiesis, the precise mechanism by which it transforms T cells remains unknown. LMO2 overexpression is an early and important mutational event in T-ALL since chromosomal rearrangements are clonal in newly diagnosed patients and are maintained in relapsed disease(2, 7). Our laboratory found that Lmo2 is a frequent clonal retroviral integration in murine T-ALLs(8). One intriguing finding from our studies and other transgenic mouse models is that Lmo2 expression can be enforced ubiquitously yet leukemia only develops in the T-cell lineage(9, 10). This was impressively demonstrated in the T-ALLs that developed as a result of gammaretroviral gene therapy for severe combined immunodeficiency (SCID-X1)(11). Four out of 20 patients treated developed T-ALL due to integration of the retroviral vector 5′ of the LMO2 gene thereby activating it(12). Most remarkably, the LMO2 integrations could be detected in the original hematopoietic stem and progenitor cells (CD34+ HSPCs) that were transduced ex vivo prior to their infusion back to the patients. The marked clones steadily expanded only in the T-cell lineage until leukemia developed 2-3 years after retroviral transduction with the accumulation of other oncogenic mutations. The gene therapy-induced leukemias and the mouse models suggest that enforced expression of LMO2 confers a clonal growth advantage over normal thymic progenitor cells but the cellular effects behind this are not clear.
Recently, McCormack et al reported that Lmo2-overexpressing thymocytes were radio-resistant and had increased self-renewal. These findings are consistent with a stem cell function for Lmo2. Indeed, Lmo2 is required for the maintenance of the embryonic and adult hematopoietic stem cell (HSC). Lmo2−/− mice die at E10.5 due to the lack of erythropoiesis (13). Blastocyst complementation experiments showed that Lmo2−/− ES cells contributed to all tissues except blood(14). The molecular role of Lmo2 in the HSC is unknown, but it is probably part of a large macromolecular protein complex comprised of Tal1 or Lyl1, E47, Ldb1, Gata2, Ssbp2 and other proteins best characterized in erythroid progenitor cells (15) (16). Lmo2 has two Zn-coordinated LIM domains that can bind Gata proteins and class II basic helix-loop-helix (bHLH) factors such as Tal1 and Lyl1, thereby bridging these transcription factors at regulatory sequences to activate or repress target genes (17-20). Most importantly, many of these same proteins are co-expressed or co-mutated in T-ALL suggesting that the functional role of Lmo2 in HSCs may be recapitulated in T-ALL(21, 22).
In this study, we present data on the preleukemic effects of Lmo2 overexpression in T-lineage progenitor cells. We analyzed the developmental phenotype and cell cycling and show that Lmo2 overexpression induced a differentiation arrest and triggered a proliferative defect consistent with quiescence in vivo. Furthermore, Lmo2 overexpressing cells were immortalized in in vitro assays and expressed a transcriptional signature found in HSCs and a highly treatment-resistant form of T-ALL. Our results imply that Lmo2 induces stem cell-like features in T-cell progenitors.
Materials and Methods
Mouse studies
B6.CD2-Lmo2 mice are transgenics created at NCI-Frederick and maintained at Vanderbilt University under IACUC approval and monitoring. The Lmo2 transgenic construction with the CD2 minigene will be described elsewhere but in brief the mice were created by pronuclear injection into B6C3HF2 hybrid zygotes; one founder was backcrossed to pure C57BL/6 for 11 generations and maintained in that manner (23, 24). For studies on T-cell progenitors, TG and WT littermates were age-matched and analyzed at 8-10 weeks. To analyze for the engraftment of T-cell progenitors immortalized by Lmo2, termed LTO cells, 15 NSG mice (an immunocompromised strain that supports engraftment of diverse leukemias available from Jackson Labs, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) were tail vein injected with 1×106 to 2×106 LTO cells. Mice were bled every 4 weeks and followed by FACS for CD25, CD44, CD4, and CD8 expression. Cell profiles were analyzed by Hemavet (Drew Scientific Inc.). B6.CD45.1 congenic mice were injected with 4×106 LTO cells retro-orbitally or intravenously after sublethal irradiation (550Gy). Spleens, bone marrow and thymi were harvested for secondary transplant.
Cell culture and OP9 assay
OP9-DL1 cells were maintained in culture as described(25). Briefly, cells were cultured in alpha-MEM media with 20% FCS and 1% penicillin/streptomycin in 10cm plates. Stem cells from E15.5 fetal livers were harvested and Lin− cells were selected using Stem Cell Technologies mouse hematopoietic progenitor enrichment kit per manufacturer’s instructions (cat# 19756). Kit+Lin−Sca-1+ (KLS) cells were enriched using Stem Cell Technologies’ mouse CD117 selection cocktail (cat# 18757). KLS enriched cells were cultured in 24-well plates containing 75% confluent, irradiated OP9-DL1 or OP9–GFP with 6 ng/ml IL-7 and Flt-3. Cells were collected, washed and plated on fresh OP9 cultures every 7 days.
Gene expression analysis
Total RNA was purified by TRIzol (Invitrogen Life sciences) per manufacturer’s instructions. First strand cDNA was synthesized using oligo-dT primer and Superscript II transcriptase enzyme (Invitrogen). Sybr green (Bio-Rad) and TaqMan real time analysis were performed as previously described. Primers are available upon request. Transcriptome analysis by RNA-seq is presented in supplemental methods.
Cell Cycle Analysis and Flow cytometry
Bromodeoxyuridine (BrdU) incorporation was analyzed per manufacturer’s instructions (BD Biosciences). Briefly, B6 and B6.Cd2-Lmo2 mice were injected IP with 100ug BrdU. Two hours post-injection, thymocytes were collected and treated with erythrocyte lysis buffer (Qiagen). For analysis of double negative populations, cells expressing CD4 and/or CD8 were sorted using Dynal magnetic separation beads per manufacturer’s instructions (Invitrogen). Antibodies for flow analysis were purchased from BD Pharmingen. FACS assays were performed on a BD FACSAria and analyzed with CellQuest (BD Biosciences). Pyronin Y (Sigma) and Hoechst 33342 (Invitrogen) staining was performed per BD Bioscience protocols. Briefly, 2×106 cells/ml were stained with 10mg/ml Hoechst 33342 at 37°C for 45 minutes. After washing with PBS, cells were fixed with 5% PFA overnight at 4°C. Pyronin Y was then added at 1μg/ml for 30min on ice in the dark. Cells were washed with PBS and analyzed by FACS. Annexin V/PI staining was performed via manufacturer’s instructions (BD Pharmingen, Cat# 556547).
PCR and Bisulfite Pyrosequencing
Bisulfite conversion was performed using the Zymo Research EZ DNA Methylation kit per instructions (cat# D5005). Bisulfite-converted gDNA was amplified with primers listed in the supplemental data file. Amplicons were subjected to pyrosequence to analyze p16 promoter DNA methylation level as previously described (26).
Results
Lmo2 transgenic thymocytes have increased DN3 progenitor cells
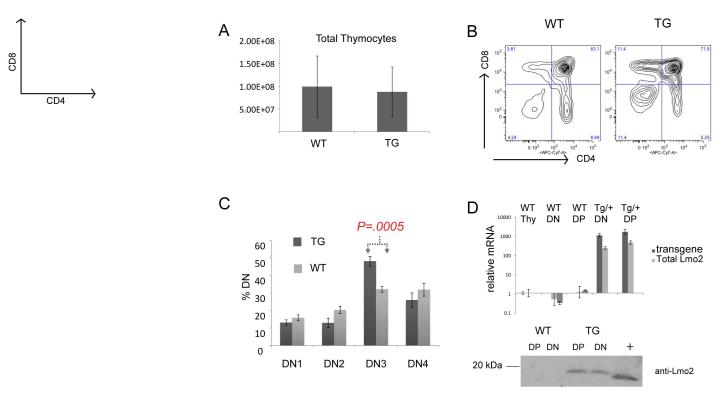
We hypothesized that Lmo2 confers a growth advantage on T-lineage progenitor cells before the onset of leukemia. To test this, we analyzed thymocytes from wild type (WT) C57BL6 and B6.CD2-Lmo2 transgenic mice (TG). First, we analyzed the thymic cellularity and T-cell progenitor subsets using FACS and antibodies against CD4, CD8, CD44, and CD25. The total thymic cellularity was not statistically different in TG and WT mice at 8-10 weeks of age (Figure 1A). Hence, for subsequent experiments, we harvested and assayed the same number of cells from TG and WT thymi and statistically compared the proportions of subsets. The FACS profiles of CD4 and CD8 expressing TG and WT thymocytes were similar (Figure 1B) but TG thymocytes consistently showed increased proportions of double negative (DN, CD4−CD8−) cells, varying from 5-15%. We analyzed the DN1-4 subsets which are well characterized differentiation stages preceding the co-expression of CD4+CD8+ in double positive (DP) thymocytes and found a statistically significant increase in the DN3 subset in TG compared to WT mice (Figure 1C)(27). The DN3 subset comprised an average of 48% of total TG DN cells and an average of 32% of total WT DN cells, a statistically significant difference (Student t-test, P=0.0005). A comparison of the absolute number of DN3 cells also showed a statistically significant difference (P=.04) between WT and TG. We analyzed the mRNA and protein of WT and TG DN and DP thymocytes and confirmed that Lmo2 was overexpressed (Figure 1D). Lmo2 mRNA and protein was not detectable in WT thymocytes and, in TG thymi, showed no change from DN to DP differentiation (Figure 1D).
Figure 1. Lmo2 transgenic mice show increased T-cell progenitors at DN3 stage.
(A) bar graph shows counts from WT (n=8) and CD2-Lmo2 transgenic (TG, n=6) thymocytes with S.D. shown with error bars. (B) shows representative contour plots of WT and TG thymi stained for CD4 and CD8. (C) WT (n=8) and TG (n=9) thymocyte double negative thymocytes were subtyped for DN1-4 by CD44 and CD25 staining; percentage of each is shown. P value is from two-tailed Student t-test. (D) graph shows qRT-PCR for Lmo2, total and transgenic. Bottom panel shows a western blot of whole protein lysate from TG or WT DN or DP thymocytes probed with anti-Lmo2 antibody; “+” denotes 293T lysate transfected with an Lmo2 expression plasmid. Error bars show the SD.
Thymic progenitor cells expressing Lmo2 have fewer cycling cells in vivo
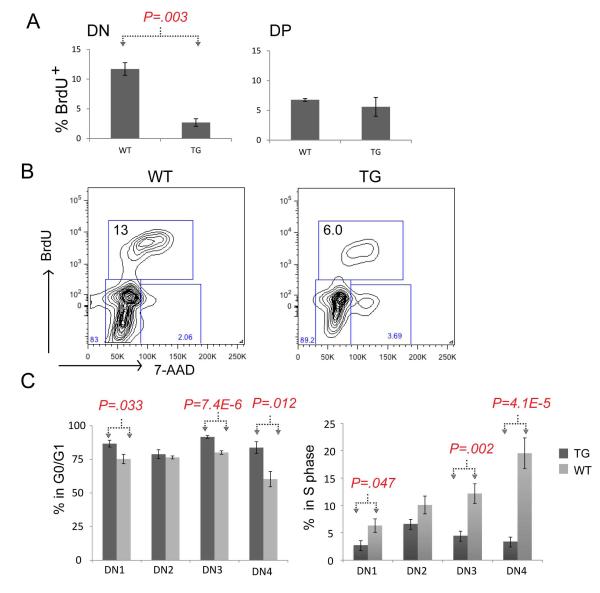
We thought that the increase in the DN3 subset population could be due to increased proliferation induced by Lmo2 overexpression. To analyze this, we performed in vivo BrdU labeling in age-matched TG and WT littermates. We analyzed T-cell progenitors by anti-BrdU/7-AAD co-staining to discern cell cycle stages. On average, 11.7% of WT DN thymocytes were in S-phase compared to 2.7% of TG DN cells, a statistically significant difference (Figure 2A; P=0.003). There was no difference between TG and WT DP thymocytes in S-phase. A representative FACS profile of BrdU/7-AAD staining is shown in Figure 2B. Next, we gated on the various DN subsets along with BrdU/7-AAD. The TG thymocytes showed increased proportions of cells in G0/G1 at DN1 (86% v. 75% in WT), DN3 (91% v. 80% in WT), and DN4 (83% v. 60% WT) stages (Figure 2C). These same subsets showed comparatively lower proportions of TG cells in S-phase: DN1 (2.7% v. 6.3% in WT), DN3 (4.5% v. 12.1% in WT), DN4 (3.4% v. 22.8% in WT). The pairwise comparisons of these cell cycle stages were statistically significant (Figure 2C). Despite a striking difference in proportions as shown in Figure 2C, WT and TG DN3 cells showed the same absolute number of cells in S-phase (data not shown). This is interesting since it may explain the lack of difference in overall thymic cellularity and peripheral mature T cells between WT and TG mice.
Figure 2. Lmo2 transgenic T-cell progenitors have decreased BrdU uptake.
(A) Bar graphs show the percentage of cells staining with anti-BrdU antibody after in vivo labeling of WT (n=4) and TG (n=5) mice. P value is a result of Student t-test; DN, double negative; DP, double positive thymocytes were electronically gated. (B) shows representative FACS contour plot of in vivo BrdU labeling of DN (electronically gated) thymocytes from WT and TG mice; y-axis is anti-BrdU and x-axis shows 7-AAD staining. Box shows the proportion of cells in S phase in these WT and TG mice. (C) Bar graph shows the mean percentage of DN1-4 subsets in G0/G1 (left panel) or in S-phase. Dark gray denotes TG (n=9) T-cell progenitors and light gray denotes WT (n=8); error bars show the SD. Brackets and P values show two-tailed Student t-test analysis.
Thymic progenitor cells expressing Lmo2 are predominantly in G0
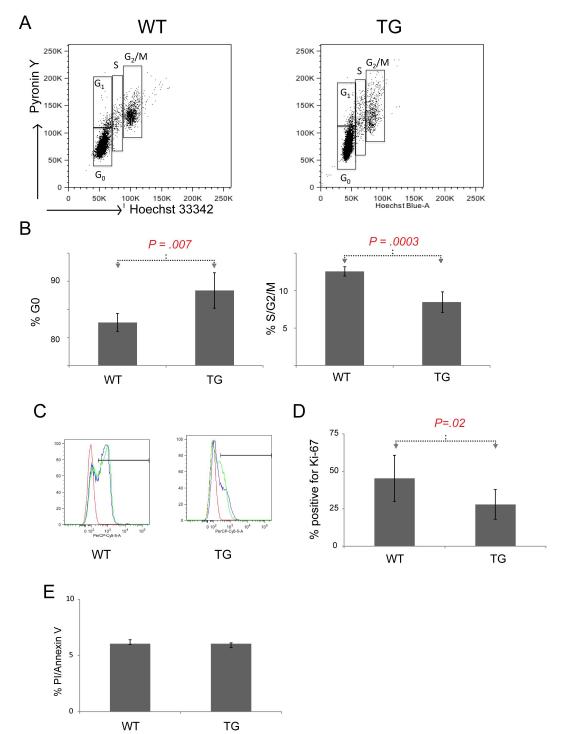
Our in vivo BrdU labeling showed that Lmo2-overexpressing TG thymocytes had a marked defect in cell cycling in various DN subsets compared to WT cells. Since BrdU and 7-AAD could not discriminate between G0 and G1, we next stained the DN populations of TG and WT thymocytes with Hoechst 33342 and pyronin Y. As noted, the total DN population showed a statistically significant difference in cells in the S-phase as measured by BrdU incorporation (Figure 2A). DNA content analysis using Hoechst 33342 demonstrated a similar result with 8.5% of TG DN thymocytes in S/G2/M phases compared with 12.6% of WT cells, a statistically significant result (Figure 3A and 3B; P=0.0003). Combined staining with pyronin Y and by applying the same gates on TG and WT DN thymocytes, we observed that 88.4% of TG cells were in the G0 phase compared to 82.7% of WT cells (P=0.007). We did not find G1 arrest in TG thymocytes.
Figure 3. Lmo2 transgenic T-cell progenitors are quiescent.
(A) representative FACS plots for DN thymocytes isolated from TG or WT mice stained for pyronin Y (y-axis) or Hoechst are shown. Boxes denote cell cycle phases. (B) Bar graph shows the mean of cell cycle phases for TG (n=5) and WT (n=5) DN thymocytes in G0 (left panel) or in combined S/G2/M phases (right panel). Error bars show the SD. Brackets and P values are from two tailed Student t-test analysis. (C) A representative FACS plot shows the staining of TG and WT DN thymocytes with anti-Ki67 antibody. The bar shows positive staining. Red curve is isotype control; blue and green are specific antibody. (D) Bar graph shows the mean percentage of DN thymocytes staining positive for Ki-67 antigen in TG (n=10) and WT (n=6) mice. (E) Bar graph shows the mean proportion of positive cells in TG and WT. Error bars show the SD. Bracket and P value are from two-tailed Student t-test analysis.
To further confirm that TG cells were mostly in G0, we stained TG and WT thymocytes for Ki-67. This nuclear antigen is present in actively cycling cells so TG cells in G0 would be expected to have less staining than WT cells (28). As shown in Figure 3C, 28% of TG DN cells stained for Ki-67 compared with 45% of WT DN cells, a significant difference (P=0.02). Although the DN3 population was more quiescent, it was proportionately increased in number which could be due to increased survival or decreased apoptosis. Therefore, we stained the DN populations of TG and WT thymocytes for Annexin V and propidium iodide (PI) to analyze for apoptosis and death but found no significant difference, even when we gated on the various DN1-4 subsets (Figure 3E).
In vitro T-cell differentiation recapitulates the differentiation block but not the in vivo proliferative defect
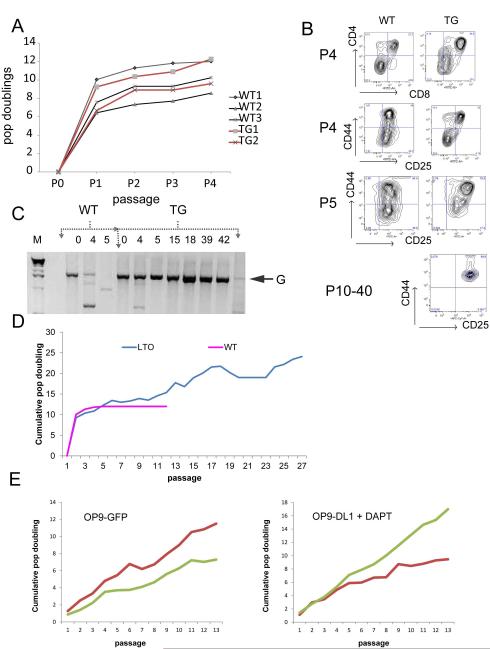
To further explore the mechanism of the differentiation block in TG thymocytes, we co-cultured Lineage− Sca-1+ c-Kit+ (LSK)-enriched fetal liver hematopoietic stem and progenitor cells (HSPCs) with OP9-DL1 stromal cells in the presence of Flt3L and IL-7(25). We passaged cells on to fresh, irradiated OP9-DL1 cells weekly and observed differentiation to CD4+CD8+ (DP) by passage 4 for both TG and WT LSK cells (top panel, Figure 4B). LSK cells from TG expanded in vitro to the same extent as WT LSK cells as shown in the growth curve of cumulative population doublings for a representative group of in vitro cultures (Figure 4A). We found no difference in BrdU uptake or Annexin V/PI staining between TG and WT cultures at early passages (P3-5, data not shown). This was in marked contrast to the in vivo cell cycle analysis. We consistently observed an accumulation of cells at the DN2 stage only in cultures of TG LSK cells. Some LSK TG cultures were blocked at DN1, but never at DN3, which was the major subset of differentiation arrest observed in vivo.
Figure 4. In vitro T-cell differentiation recapitulates differentiation block and immortalizes Lmo2 expressing T-cell progenitors.
(A) Graph shows population doublings versus passage number for WT and TG LSK cells plated on irradiated OP9-DL1 stromal cell line. (B) These representative FACS contour plots show the appearance of WT and TG T cells arising in OP9-DL1 culture at various passages (P). Top panel shows CD4/CD8 staining; DN cells were electronically gated as CD4−CD8− and assayed for CD44 and CD25. WT T-cell progenitors could not be recovered for staining beyond P5. TG lines that were immortalized and termed LTOs resembled DN2 cells as shown for P10-40. (C) Agarose gel shows PCR of Jβ2 region from gDNA derived from WT or TG T cells on OP9-DL1 cultures. Column numbers show the passage number. G denotes 2 kb germline band. (D) Line graph shows the cumulative population doublings versus passage number for WT and TG (LTO) T cells growing on OP9-DL1. The graph is representative of 4 independent experiments where TG cells immortalized. (E) Late passage LTOs were plated on to OP9-GFP stromal line or OP9-DL1 cells in the presence of 10 μM DAPT. Y-axis shows cumulative population doublings and x-axis shows passage number.
The DN2 block could be replicated in OP9-DL1 co-cultures of WT LSKs transduced with MSCV-Lmo2-ires-GFP retrovirus (data not shown) and was not unique to the transgenic model. We performed Jβ2 PCR on genomic DNA from WT and TG cultures to further analyze the differentiation state of the progenitor cells. The WT cultures completely differentiated to DP cells by passage 5 with loss of the germline Jβ2 (Figure 4C) band but the TG cells showed some rearrangement at passage 4, the point at which there were the most DP cells (Figure 4B), but then showed a stronger signal for the germline band. This germline band corresponded with the accumulation of DN2 cells in culture. The DN2 stage is the first stage at which T-cell commitment occurs prior to beta selection in DN3. These cells expressed Sca-1 and Thy1.2 verifying that we were observing T cells in culture; but, interestingly, the cells were also positive for B220, an antigen of the B-cell lineage, and 24% also expressed Gr-1, a marker of the myeloid lineage (data not shown). These data show that TG LSK cells in OP9-DL1 co-culture become blocked at the DN2 stage at early T-cell commitment preceding beta selection with expression of some markers of other lineages. Notably, B-cell and myeloid antigens are seen in Lmo2-induced T-ALL in humans and mouse models (8, 29).
The discrepancy in the stages of block between in vivo and OP9-DL1 culture was also observed in conditional knockouts of Hes1 and Bcl11b (30, 31). These blocks were attributed to the lower strength of the Notch1 signal delivered by OP9-DL1 compared to the in vivo microenvironment. We decided to test this concept in our TG DN2 blocked progenitors by transducing them with the intracellular Notch1 fragment (MIG-ICN1). These cells proliferated after transduction enriching the culture for GFP+ cells in comparison to those transduced with MIG alone. Most remarkably, the TG DN2 cells transduced with MIG-ICN1 progressed to DP cells leaving very few DN cells in culture (data not shown). Thus, the Lmo2-induced differentiation block could be overcome by ICN1 expression.
T-cell precursors overexpressing Lmo2 become immortalized in vitro
In serial replating of T-cell progenitor /OP9-DL1 co-cultures, we observed a growth plateau at P5 that continued through P12 for WT cultures (Figure 4D). In contrast, the TG thymocytes showed some doublings during this same timeframe. Most remarkably, we observed immortalization of thymocytes derived from TG LSK cells in four independent experiments (representative growth plot shown in Figure 4D). We estimate the transformation efficiency at 10%. We never observed continuous growth of WT thymocytes beyond P12. The four independent lines of TG thymocytes were serially passaged for over one year. These TG cells, termed LTOs (i.e. Lmo2 transgenic OP9 cultures), were DN2 blocked (see lower panel Figure 4B), B220+, and had germline Tcr Jβ2 configuration(Figure 4C). The LTO cells continued to proliferate even when transferred at P20 to OP9-GFP cells which do not express the Notch1 ligand, DL1. Similarly, LTO cells on OP9-DL1 showed no growth inhibition when Notch1 intramembranous cleavage was inhibited by the gamma secretase inhibitor DAPT (32).
Given the Notch-independent growth observed, we amplified and sequenced Notch1 exons from LTO genomic DNA but found no mutations. We isolated RNA from the LTOs and analyzed the transcripts of well-defined Notch1 targets by qRT-PCR and RNA-seq. Notch1, Hes1, Dtx1, and c-Myc transcripts were no different between WT and TG cultures (data not shown). Additionally, the LTOs continued to require IL-7 and Flt3L in culture and could not be passaged without OP9 cells. We injected the LTOs into immunodeficient and sublethally irradiated congenic CD45.1 mice but observed no engraftment and no leukemia development.
Long term cultures of Lmo2 transgenic thymocytes show deregulation of Cdkn2a
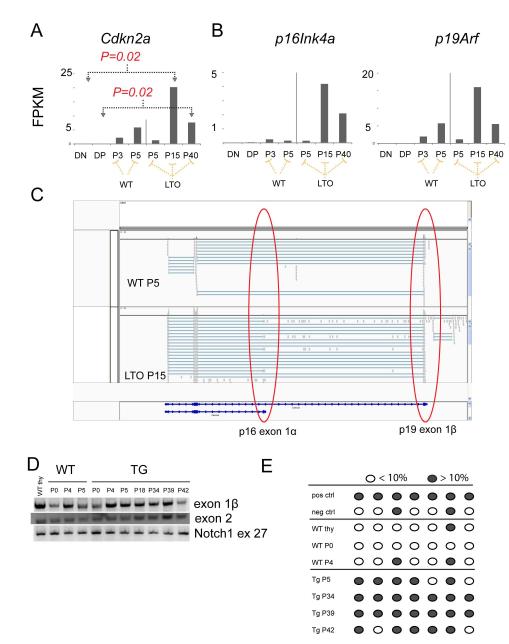
Since we observed no difference in apoptosis or proliferation between WT and TG OP9-DL1 co-cultures, we considered whether the Lmo2-overexpressing thymocytes became immortalized by suppressing or bypassing senescence. This was particularly relevant for the in vitro culture system since primary cultures of murine cells are limited in their doubling capacity but can bypass senescence by loss of p53 or p19Arf (33). Most WT T-cell progenitors did not proliferate beyond P5 and eventually died in culture. The TG progenitors behaved like WT through P5 (Figure 4A), showed limited growth until P15 (Figure 4D), and were immortalized and stable by P40 (Figure 4D). We analyzed the early and late passages of WT (P3, P4) and TG (P5, P15, P40) (Figure 4B shows FACS profiles for these cells) thymocytes differentiated on OP9-DL1 stroma by RNA-seq(34, 35). In human T-ALL, the CDKN2A locus is deleted in over 70% of cases suggesting that expression of the two tumor suppressor genes (p14 or p19Arf in mouse and p16Ink4a) was induced during the development of leukemia(36). We analyzed Cdkn2a expression in RNA-seq shown as fragments per kilobase pair of gene per million reads analyzed or FPKM (Figure 5A). Normal DN and DP thymocytes showed few Cdkn2a transcripts (0.077 and 0.12 FPKMs respectively), but mRNA was detectable in WT progenitor cells that differentiated in vitro on OP9-DL1 stroma at P3 (2.22 FPKM) and P5 (5.9 FPKM). In the LTO cultures, there was a statistically significant increase in Cdkn2a expression at P15 (20.2). By P40 when the LTO cultures were immortalized the Cdkn2a transcripts (7.6) were higher than normal DN levels but were not statistically different.
Figure 5. Lmo2 expressing T-cell progenitors deregulate Cdkn2a expression concordant with immortalization.
(A) Bar graphs show the normalized quantification of total Cdkn2a transcripts in FPKM (fragments per kilobase per million reads) for DN and DP thymocytes from WT mice; passage 3 and 5 of WT T cells in OP9-DL1 culture; and, passages 5, 15, and 40 of TG T-cell progenitors. P values are from comparisons of bracketed values and are corrected for multiple hypothesis testing. FPKMs for p16Ink4a and p19Arf are shown for the same samples. P values were not statistically significant and are not shown. (C) This schematic is a snapshot of the Integrated Genome Viewer (IGV) that shows WT P5 and TG (LTO) P15 RNA-seq reads. The red circles show the exon 1β of p19 and exon 1α of p16. The latter was only found in the LTO samples. (D) Agarose gel shows PCR of gDNA for exon 1β, exon 2 of Cdkn2a and of exon 27 of Notch1. Numbers denote passage number for WT and TG cells. (E) Schematic shows analysis of CpGs in the promoter of p16 analyzed by PCR of bisulfite-converted gDNA from WT or TG T cells from various passages. Dark gray circles show greater than 10% methylation whereas open circles show less than 10% methylation as analyzed by pyrosequencing.
RNA-seq allowed us to visualize those mRNAs arising from p16Ink4a and those arising from p19Arf. In Figure 5C, we show a snapshot of the integrated genome browser (IGV) that shows high quality reads (encircled gray boxes) mapping to exon 1α of p16Ink4a and exon 1β of p19Arf. The p16Ink4a expression was only detectable at P15 in the LTO cultures and not in WT cells (Figure 5C). As noted, p16Ink4a was decreased by P40. We isolated genomic DNA from the various passages and could amplify Cdkn2a exons ruling out widespread deletion of the locus (Figure 5D); and, we did not find mutations in Cdkn2a. We next analyzed the p16Ink4a promoter by pyrosequencing of bisulfite-converted genomic DNA from a single culture of LTO and WT progenitor cells. As shown in Figure 5E, late passage LTO cells showed hypermethylation of CpGs at the p16Ink4a promoter whereas WT T-cell progenitor cells under the same conditions did not. Thus, Cdkn2a transcription was dynamic in LTO cells compared with the absolute repression observed in DN and DP thymocytes and the relative repression in WT T-cell progenitors grown in vitro.
Lmo2 expressing progenitor cells express a transcriptional program similar to that seen in hematopoietic stem and progenitor cells
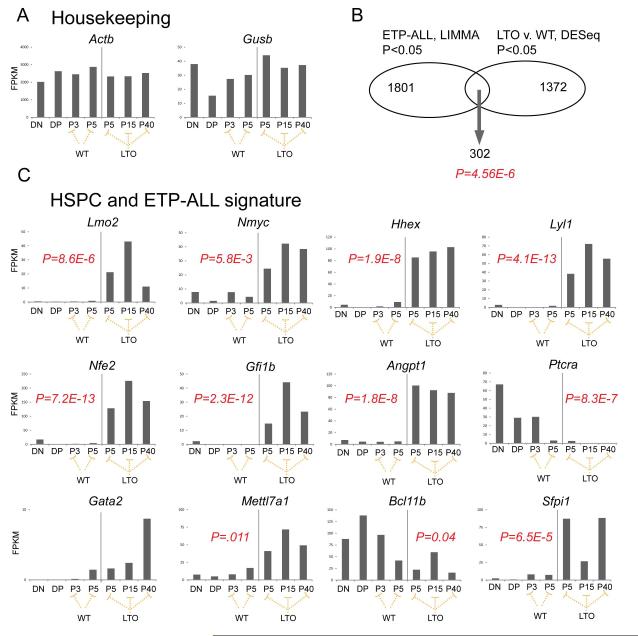
We analyzed the RNA-seq data for differential gene expression between WT and LTO cells grown on OP9-DL1. We performed pairwise comparisons and also class comparisons by grouping the LTO samples and WT samples to lend more power to the statistical analysis. We suspected that the LTOs may model the cell of origin of Early T-cell Precursor (ETP)-ALL because both show DN block and express markers of myeloid or B-cell lineages. To test this, we compared those genes that were differentially expressed in the mouse studies shown here (RNA-seq, LTO v. WT, P<0.05) with the transcriptome of ETP-ALL (Affymetrix gene expression array, ETP v. non-ETP-ALL, P<0.05) and found 302 genes that were present in both datasets (Figure 6B), a remarkably significant result (Fisher exact test, P=4.56 × 10−6). Most strikingly, the ETP-ALL cases overexpressed a group of transcription factor/co-factor genes, Lmo2, Lyl1, Hhex, Gfi1b, Nfe2, Gata2, and Ldb1 that have specific regulatory regions occupied by Ldb1 in ChIP-seq analysis in HSPCs (Figure S1). Importantly, these genes were significantly upregulated only in the LTOs and not in WT cells cultured under the same conditions. We analyzed the 302-gene dataset by gene set enrichment analysis. As expected, the significant pathways were related to T cells and T-cell signaling (Table S1). The fourth most significant pathway was KEGG_Hematopoietic_Cell_Lineage (FDR=8.95×10−6). The upregulation of this HSC/ETP-ALL transcriptional signature was maintained in T-ALLs that developed in Lmo2 transgenic mice (manuscript in preparation, U.P. Davé).
Figure 6. Lmo2 expressing T-cell progenitors show a transcriptional profile of genes important in HSPC and ETP-ALL.
(A) Bar graph show RNA-seq analysis of DN and DP cells from WT mice; WT T cells from passages 3 and 5; and, TG (LTO) T cells from passages 5, 15, and 40. FPKMs are normalized read counts from sequencing. Housekeeping genes are shown. (B) Venn diagram denotes the overlap between two datasets: LIMMA analysis of ETP-ALL versus non-ETP-ALL patients; number of genes with raw P <0.05 are shown; the right circle shows the dataset generated from DESeq differential gene expression between LTO versus WT T cells growing on OP9-DL1. As shown 302 genes were present in the overlap, a highly significant result; P value was generated from Fisher exact test and confirmed independently by permutation analysis. (C) Bar graphs show FPKMs for several representative genes differentially expressed between LTO and WT T cells by RNA-seq analysis. P values are generated from DESeq application and are corrected for multiple hypothesis testing.
We examined cell cycle genes that may be differentially expressed or mutated between LTO and WT cultures. We did not find mutation or differential expression for Trp53, Rb1, or Rbl1 (see Figure S2). We found upregulation of Ccnd1, Ccnd2, and Cdkn1a in both WT and LTO compared to DN and DP thymocytes. Interestingly, E2f2 was significantly (P=8×10−5) downregulated in the LTO cultures but not in WT; Cdk6 was significantly upregulated in the LTO but not in WT (P=0.03). Cytokines and their cognate receptors were deregulated in several instances; Il7r (P=4.9×10−12) was significantly downregulated in the LTOs but Kitl (P=0.02), Igf2 (P=3.3×10−6), Fgf3 (4.1×10−13), and Igf1r (P=5.1×10−3) were upregulated; other cytokines were upregulated in both the WT and LTO (Figure S3). Also, LTOs showed significant downregulation of several death receptor genes (Tnfrsf8, Tnfrsf4, and Tnfrsf18) and the Fasl gene compared to WT (Figure S4). These were upregulated in WT cultures with Fasl strikingly increased in late passage WT cells. Apoptotic regulators such as Bcl2 and Bcl2l1 (Bcl-xL) were not differentially expressed in LTO and WT cells (Figure S4).
Lmo2 expressing progenitors require Ldb1 for differentiation arrest
The transcription factor genes with specific Ldb1 occupancy suggested to us that the differentially expressed genes and the resultant phenotype were induced by Lmo2/Ldb1 complexes. Indeed, we and others have data that HHEX is a direct target of an LMO2/LDB1 protein complex in human T-ALL (manuscript in preparation, U.P. Davé) (37).
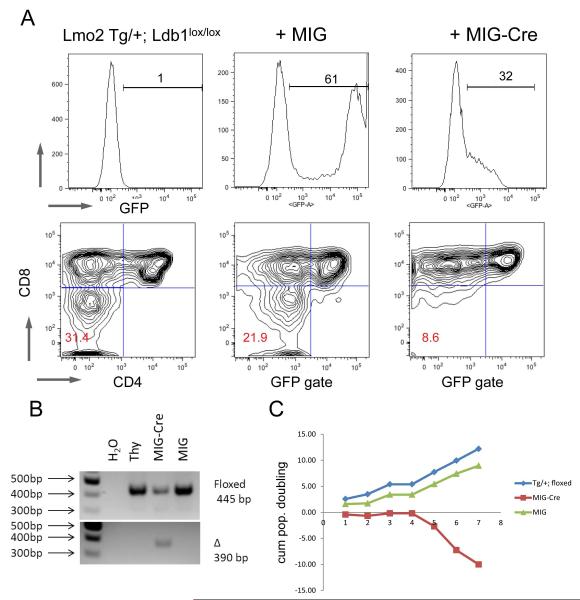
To directly test whether Lmo2 requires Ldb1 to induce T-cell progenitor phenotypes, we bred the B6.CD2-Lmo2 transgenic mice on to floxed Ldb1 strain to generate TG/+; Ldb1lox/lox mice so that we could control the induction of knockout by transduction with retrovirus expressing Cre recombinase. We isolated DN thymocytes from these mice and transduced them with MIG or MIG-Cre retroviruses. We plated the cells on OP9-DL1 after transduction and observed proliferation and differentiation into DP cells. One week after transduction, gating on GFP+ cells, we observed both DN and DP cells in culture. The TG cells were blocked at DN stages and this population was comparable between untransduced and those transduced with MIG. Most remarkably, on P2, the cells transduced with MIG-Cre showed loss of the DN cells. Thus, DN cells from TG/+; Ldb1lox/lox differentiated in culture to DP cells but the DN cells were not maintained and were lost by the second passage. We plotted the absolute number of GFP+ cells on serial passages of DN thymocytes. Untransduced T-cell progenitors and those transduced with empty MIG differentiated and proliferated on OP9-DL1 cells (Figure 7C). In striking contrast, T-cell progenitor cells transduced with MIG-Cre differentiated into DP cells but did not expand. These MIG-Cre transduced cells underwent growth arrest and died in culture as shown by the plot of absolute number of GFP positive cells.
Figure 7. Lmo2 requires Ldb1 to induce and maintain DN progenitors.
(A) DN thymocytes were sorted from TG/+; Ldb1lox/lox mice and transduced with MIG or MIG-Cre retroviruses and plated on OP9-DL1 and passaged weekly; FACS plots in the top panel show the proportion of cells positive for GFP emission in the three groups. Bottom panel shows FACS contour plots of CD4 and CD8 staining. Red numbers show the proportion of cells in the DN quadrant. (B) Agarose gel of gDNA PCR from the same experiment shown in (A) shows floxed and deleted amplicons. (C) shows a line graph of cumulative population doublings (y-axis) of GFP+ cells in culture versus passage number (x-axis). Blue shows untransduced TG thymocytes; green line shows MIG transduced; and, red line shows MIG-Cre transduced cells. The FACS plots are representative of three independent transductions.
Ldb1lox/lox thymocytes showed minimal DN differentiation arrest that did not change with MIG-Cre transduction; however, MIG-Cre transduced cells were not maintained in culture (Figure S5).
Discussion
In this study, we show that CD2-Lmo2 transgenic T-cell progenitor cells are arrested in differentiation and quiescent, have increased self-renewal in vitro and express a transcriptional signature found in HSPCs and ETP-ALL. First, we consider mechanistic interpretations for these cellular features and how our in vivo and in vitro data suggest an order by which cellular events are acquired in T-cell leukemogenesis. Then, we discuss how the cellular features are HSC-like and offer insight into the clinical presentation and management of T-ALL.
CD2-Lmo2 transgenic progenitor cells are blocked at the DN3 stage in vivo although the total thymic cellularity and DP differentiation were similar to WT. DN block is a defining trait of enforced Lmo2 expression that has been attributed to functional deficiency of E2a (2, 38-40). Most notably, E2a−/− thymocytes have DN arrest at the same stages as Lmo2 overexpression and spontaneously develop T-ALL (41). Lmo2 protein does not bind E2A proteins directly but does bind class II bHLH proteins such as Tal1 and Lyl1 that heterodimerize with E2A (42). Lmo2/bHLH complexes may redirect E2A proteins away from their normal targets or recruit co-repressors in place of co-activators (38). In TAL1/LMO1 and Tal1/Lmo2 double transgenic mice, DN3 arrest is more pronounced and the thymic cellularity markedly reduced compared to CD2-Lmo2 transgenic thymi (43, 44). The total thymic cellularity in CD2-Lmo2 transgenics may be preserved because the absolute number of DN3 S-phase cells was the same in TG and WT. The phenotype of Lmo2 overexpression we describe cannot be completely attributed to E2A deficiency. Most importantly, E2a knockout T-cell progenitors show increased proportions of cells in S-phase in contrast to relative quiescence observed in the Lmo2 transgenic progenitors(45) .
Lmo2 transgenic and WT progenitors cycled alike in OP9-DL1 co-culture but only Lmo2 transgenic thymocytes showed DN2 arrest similar to thymocytes transduced with Lmo2 retrovirus(46). Lmo2-overexpressing progenitors are blocked at the DN2 stage which is prior to beta selection (DN3) where pre-T-cell receptor assembly with Ptcra and signaling occur(43) . The pattern of DN2 block on OP9-DL1 cultures and DN3 in vivo occurs in thymocytes from conditional knockout of Hes1 and Bcl11b genes but not in Ptcra−/− which are blocked at DN3 in vitro and in vivo(30, 31, 47). In these reports, this discrepancy (DN2 v. DN3) was attributed to attenuated Notch1 signaling in in vitro culture. This idea is consistent with our finding that the Lmo2-induced DN block was alleviated by the enforced expression of ICN1 (Figure S2). Both Hes1 and Bcl11b were repressed in Lmo2 transgenic T-cell progenitor cells (Figure 6 and S2B) and may be direct or indirect targets of Lmo2. Attenuated Notch1 signaling may be important for Lmo2-induced DN block. ICN1 expression accelerates differentiation towards DP cells which are resistant to Lmo2 transformation (48). Of note, Lmo2 transgenic progenitors resemble ETP-ALL, a leukemia where NOTCH1 mutation is uncommon compared to other T-ALL subtypes (49, 50).
We noted marked proliferative defects at multiple DN stages in the TG T-cell progenitor cells in vivo. Apoptosis was not a prominent process in these thymocytes. Pyronin Y staining with Hoechst 33342 staining and Ki-67 antigen analysis showed that the Lmo2-overexpressing T-cell progenitor cells were more quiescent than their wild type counterparts. This quiescent phenotype explains the radio-resistance observed in Lmo2-overexpressing DN cells (51). Interestingly, quiescence was not preserved in the OP9-DL1 co-culture which could be due to cell cycle entry promoted by serum or cytokines in vitro. Alternatively, the thymic microenvironment may play a role in maintaining progenitors in G0. E2f2 was markedly repressed in LTOs compared to WT progenitors maintained in the same culture conditions. This may explain the increased proliferation in vitro since E2f2−/− thymocytes show increased S-phase entry (52, 53). E2F2 is significantly downregulated in ETP-ALL cases in comparison to non-ETP-ALL cases (FDR=6.65 × 10−6).
Most remarkably, the DN2-blocked progenitors co-cultured with OP9-DL1 became immortalized in vitro. They remained cytokine-dependent and no longer required Notch1 signaling but they could not engraft into sublethally irradiated B6 mice or immunosuppressed strains. Transcriptional profiling of Lmo2-overexpressing T-cell progenitors revealed early establishment of a signature seen in ETP-ALL and HSPCs and deregulation of Cdkn2a in late passage LTOs. These data suggest a specific order in which cellular and molecular effects accumulate in Lmo2-induced T-ALL. In this model, quiescence without differentiation and the HSC transcriptional signature are established early by LMO2/LDB1 complexes and proliferative mutations are secondary events. Proliferation in the setting of Lmo2 expression induces a senescence checkpoint with upregulation of both p16 Ink4a and p19Arf, which must be bypassed by deletion or transcriptional repression. Cdkn2a promoter hypermethylation was found in the LTOs and may be present in human ETP-ALLs cases where CDKN2A deletion is not as common as other forms of T-ALL(50). Recently, whole genome sequencing of ETP-ALL cases revealed common recurrent mutations in genes that drive proliferation (i.e. IL7R, JAK1, JAK3, NRAS, IGF1R) (50).
Finally, Lmo2 overexpression induces cellular effects that are hallmarks of HSCs: quiescence without differentiation, self-renewal, and expression of Lmo2, Nmyc, Hhex, Lyl1, and Ldb1. The resemblance is even more significant in that definitive hematopoiesis and the maintenance of HSPCs require most of these same genes: Lmo2, Ldb1 and either Tal1 or Lyl1 (14, 54, 55). We show that Lmo2 required Ldb1 to induce and maintain differentiation blocked T-cell progenitors (Figure 7). Most importantly, many of the genes in the ETP-ALL signature are regulated by Ldb1 in HSPCs and we are testing whether this same cohort of genes is regulated in T-cell progenitors. We hypothesize that Ldb1 is required for Lmo2 to induce T-ALL and the genetic experiments to test this are ongoing. Important questions remain as to how stem cell like features confer competitive growth advantage or predispose to leukemia. Addressing these questions will provide insights into the function of Lmo2 and the physiology of leukemia-initiating cells.
Supplementary Material
Figure S1. Ldb1 ChIP-seq of Lineage negative HSPCs. Sequence data from Ldb1 ChIP-seq study (55) was loaded onto the UC Santa Cruz genome browser as a custom track. Genes are shown that are also upregulated in LTO cells from the current study. The black boxes show the raw tags of sequences from the Ldb1 chromatin immunoprecipitation. The red arrows show genomic regions with high sequence tags denoting occupancy by Ldb1 protein. The RefSeq mRNA for each gene in shown in blue at the bottom of each browser snapshot.
Figure S2. RNA-seq data on cell cycle regulators. WT and TG cells were co-cultured with OP9-DL1 stroma. WT progenitor cells did not survive past passage 5 but TG cells became immortalized. We performed RNA-seq on the WT and LTO RNA. Bar graphs show the FPKM values for each mRNA. Statistical comparisons were made using the DESeq program. The absence of an asterisk denotes no statistical significance between values. An asterisk shows statistical significance (with correction) and downward pointing arrows show which sample was compared. For example, for Ccnd1, DN cells were compared pairwise to all other samples and those with asterisks were significantly different (P<.05). The P values showed in red were generated by comparing grouped LTO samples versus grouped WT samples.
Figure S3. RNA-seq data on cytokines and their receptors. Same description as above. P values shown in red are derived from group comparisons, LTO v. WT. Arrows denote specific pairwise comparisons but not grouped comparisons (e.g. Csf1r).
Figure S4. RNA-seq data on apoptosis regulators. Same description as above. P values shown in red are derived from group comparisons, LTO v. WT.
Figure S5. MIG and MIG-Cre transductions into Ldb1lox/lox T-cell progenitor cells as a control. Same experimental setup as in Figure 7. Ldb1lox/lox DN cells were sorted and transduced with MIG or MIG-Cre and plated on OP9-DL1 stroma. After 1-2 weeks of co-culture, the progenitor cells were analyzed for GFP (top panel); the GFP+ cells were gated and CD4/CD8 expression analyzed (bottom panel). The GFP expression was steadily lost during culture. Few progenitors were blocked at DN stage and MIG-Cre did not change this effect.
Acknowledgements
We thank Dr. Juan Carlos Zuniga-Pflucker for OP9-DL1 and OP9-GFP cells and Dr. Monica Justice for MIG-Hhex plasmid. We thank Drs. M. Koury, S. Hiebert, and S. Zinkel for critically reviewing the manuscript and for helpful discussions. We thank Dr. Kevin Weller and the Flow Cytometry Core of Vanderbilt University. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). This work was supported by NIH K08HL089403, the Leukemia & Lymphoma Society, the Vanderbilt Ingram Cancer Center (P30 CA68485), Monforton family grant, and the T.J. Martell Foundation (U.P. D.). This work was also supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH.
Footnotes
Conflict of Interest The authors have no conflicts of interest to declare.
Authorship contributions Susan Cleveland: collection and/or assembly of data, data analysis and interpretation, manuscript writing
Stephen Smith: collection and/or assembly of data
Rati Tripathi: collection and/or assembly of data
Elizabeth Mathias: collection and/or assembly of data
Charnise Goodings: collection and/or assembly of data
Dunfa Peng; collection and/or assembly of data
Wael El-Rifai: data analysis and interpretation
Dajun Yi: data analysis and interpretation
Xi Chen: data analysis and interpretation
Liqi Li: collection and/or assembly of data
Charles Mullighan: provision of study material
James Downing: provision of study material
Paul Love: data analysis and interpretation
Utpal Davé: conception and design, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript
References
- 1.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 2.Nam CH, Rabbitts TH. The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion. Mol Ther. 2006;13:15–25. doi: 10.1016/j.ymthe.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 3.P., Beverloo H, Buijs-Gladdines J, et al. Monoallelic or biallelic LMO2 expression in relation to the LMO2 rearrangement status in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2007;22:1434–1437. doi: 10.1038/sj.leu.2405063. [DOI] [PubMed] [Google Scholar]
- 4.Van Vlierberghe P, Beverloo H, Buijs-Gladdines J, et al. Monoallelic or biallelic LMO2 expression in relation to the LMO2 rearrangement status in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2007;22:1434–1437. doi: 10.1038/sj.leu.2405063. [DOI] [PubMed] [Google Scholar]
- 5.Van Vlierberghe P, van Grotel M, Beverloo HB, et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood. 2006;108:3520–3529. doi: 10.1182/blood-2006-04-019927. [DOI] [PubMed] [Google Scholar]
- 6.Ferrando AA, Herblot S, Palomero T, et al. Biallelic transcriptional activation of oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Blood. 2004;103:1909–1911. doi: 10.1182/blood-2003-07-2577. [DOI] [PubMed] [Google Scholar]
- 7.Mullighan CG, Phillips LA, Su X, et al. Genomic Analysis of the Clonal Origins of Relapsed Acute Lymphoblastic Leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dave UP, Akagi K, Tripathi R, et al. Murine leukemias with retroviral insertions at Lmo2 are predictive of the leukemias induced in SCID-X1 patients following retroviral gene therapy. PLoS Genet. 2009;5:e1000491. doi: 10.1371/journal.pgen.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam C-H, Rabbitts TH. The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;13:15–25. doi: 10.1016/j.ymthe.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Neale GA, Rehg JE, Goorha RM. Disruption of T-cell differentiation precedes T-cell tumor formation in LMO-2 (rhombotin-2) transgenic mice. Leukemia. 1997;11(Suppl 3):289–290. [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S,C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science (New York, N.Y.) 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. The Journal of clinical investigation. 2008 doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren AJ, Colledge WH, Carlton MB, et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 14.Yamada Y, Warren AJ, Dobson C, et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meier N, Krpic S, Rodriguez P, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Meng X, Cai Y, et al. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadman IA, Osada H, Grutz GG, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. Embo J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Y, Xu Z, Xie J, et al. Eto2/MTG16 and MTGR1 are heteromeric corepressors of the TAL1/SCL transcription factor in murine erythroid progenitors. Biochem Biophys Res Commun. 2009;390:295–301. doi: 10.1016/j.bbrc.2009.09.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S, Qiu Y, Stein RW, et al. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- 20.Osada H, Grutz GG, Axelson H, et al. LIM-only protein Lmo2 forms a protein complex with erythroid transcription factor GATA-1. Leukemia. 1997;11(Suppl 3):307–312. [PubMed] [Google Scholar]
- 21.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 22.Grutz GG, Bucher K, Lavenir I, et al. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. Embo J. 1998;17:4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love PE, Shores EW, Lee EJ, et al. Differential effects of zeta and eta transgenes on early alpha/beta T cell development. The Journal of experimental medicine. 1994;179:1485–1494. doi: 10.1084/jem.179.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy A. Manipulating the mouse embryo : a laboratory manual. x. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2003. p. 764. [Google Scholar]
- 25.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 26.Peng DF, Razvi M, Chen H, et al. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut. 2009;58:5–15. doi: 10.1136/gut.2007.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul WE. Fundamental immunology. xxi. Lippincott Williams & Wilkins; Philadelphia: 2003. p. 1701. [Google Scholar]
- 28.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Fisch P, Boehm T, Lavenir I, et al. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- 30.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wendorff AA, Koch U, Wunderlich FT, et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010;33:671–684. doi: 10.1016/j.immuni.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science (New York, N.Y.) 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 33.Sherr CJ. The INK4a/ARF network in tumour suppression. Nature reviews. Molecular cell biology. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 34.Marioni JC, Mason CE, Mane SM, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 36.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol. 2008;73:461–467. doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 37.Oram SH, Thoms JA, Pridans C, et al. A previously unrecognized promoter of LMO2 forms part of a transcriptional regulatory circuit mediating LMO2 expression in a subset of T-acute lymphoblastic leukaemia patients. Oncogene. 2010;29:5796–5808. doi: 10.1038/onc.2010.320. [DOI] [PubMed] [Google Scholar]
- 38.O’Neil J, Shank J, Cusson N, et al. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 39.Herblot S, Steff A-M, Hugo P, et al. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T[alpha] chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 40.Chervinsky DS, Zhao XF, Lam DH, et al. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: parallels with E2A-deficient mice. Mol Cell Biol. 1999;19:5025–5035. doi: 10.1128/mcb.19.7.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bain G, Engel I, Robanus Maandag EC, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadman I, Li J, Bash RO, et al. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. Embo J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herblot S, Steff AM, Hugo P, et al. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 44.Tatarek J, Cullion K, Ashworth T, et al. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood. 2011;118:1579–1590. doi: 10.1182/blood-2010-08-300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. Embo J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treanor LM, Volanakis EJ, Zhou S, et al. Functional interactions between Lmo2, the Arf tumor suppressor, and Notch1 in murine T-cell malignancies. Blood. 2011;117:5453–5462. doi: 10.1182/blood-2010-09-309831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garbe AI, Krueger A, Gounari F, et al. Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newrzela S, Cornils K, Li Z, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 49.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCormack MP, Young LF, Vasudevan S, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 52.Infante A, Laresgoiti U, Fernandez-Rueda J, et al. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle. 2008;7:3915–3927. doi: 10.4161/cc.7.24.7379. [DOI] [PubMed] [Google Scholar]
- 53.Murga M, Fernandez-Capetillo O, Field SJ, et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- 54.Souroullas GP, Salmon JM, Sablitzky F, et al. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell stem cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Jothi R, Cui K, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Ldb1 ChIP-seq of Lineage negative HSPCs. Sequence data from Ldb1 ChIP-seq study (55) was loaded onto the UC Santa Cruz genome browser as a custom track. Genes are shown that are also upregulated in LTO cells from the current study. The black boxes show the raw tags of sequences from the Ldb1 chromatin immunoprecipitation. The red arrows show genomic regions with high sequence tags denoting occupancy by Ldb1 protein. The RefSeq mRNA for each gene in shown in blue at the bottom of each browser snapshot.
Figure S2. RNA-seq data on cell cycle regulators. WT and TG cells were co-cultured with OP9-DL1 stroma. WT progenitor cells did not survive past passage 5 but TG cells became immortalized. We performed RNA-seq on the WT and LTO RNA. Bar graphs show the FPKM values for each mRNA. Statistical comparisons were made using the DESeq program. The absence of an asterisk denotes no statistical significance between values. An asterisk shows statistical significance (with correction) and downward pointing arrows show which sample was compared. For example, for Ccnd1, DN cells were compared pairwise to all other samples and those with asterisks were significantly different (P<.05). The P values showed in red were generated by comparing grouped LTO samples versus grouped WT samples.
Figure S3. RNA-seq data on cytokines and their receptors. Same description as above. P values shown in red are derived from group comparisons, LTO v. WT. Arrows denote specific pairwise comparisons but not grouped comparisons (e.g. Csf1r).
Figure S4. RNA-seq data on apoptosis regulators. Same description as above. P values shown in red are derived from group comparisons, LTO v. WT.
Figure S5. MIG and MIG-Cre transductions into Ldb1lox/lox T-cell progenitor cells as a control. Same experimental setup as in Figure 7. Ldb1lox/lox DN cells were sorted and transduced with MIG or MIG-Cre and plated on OP9-DL1 stroma. After 1-2 weeks of co-culture, the progenitor cells were analyzed for GFP (top panel); the GFP+ cells were gated and CD4/CD8 expression analyzed (bottom panel). The GFP expression was steadily lost during culture. Few progenitors were blocked at DN stage and MIG-Cre did not change this effect.