Abstract
A cDNA encoding a hepatic isoform of flavin-containing monooxygenase (hFMO) (EF063736) containing an open reading frame of 1792 base pairs (bp) and encoding 554 amino acids was cloned and sequenced from liver mRNA of rainbow trout (Oncorhynchus mykiss). The genomic sequence of hFMO was also characterized and was 4.379 kilobases, possessing 10 exons and 9 introns (EU519462). Structural analysis of the promoter region showed several cis-acting elements including putative glucocorticoid and osmoregulatory response elements, which have been reported to be functionally related to induction of flavin-containing monooxygenase (FMO) proteins in vertebrates. The amino acid sequence showed 74% identity to a putative FMO gene from fugu (Takifugu rubripes; Q6ZZY9), 52 to 55% to zebrafish (Brachydanio rerio; Q5RGM6, Q5RGM3, Q6TLD2, Q7T1D7) FMO5, and 54 and 50% to human FMO1 (Q01740), FMO3 (P49326), and FMO5 (P49326). Southern blot analysis using a 180-bp fragment of the hFMO cDNA indicated at least seven potential genes. Treatment of primary trout hepatocytes with cortisol and sodium chloride for 24 h enhanced hFMO expression. Expression of hFMO was not detected in untreated or solute-treated primary cultures of gill epithelial cells, suggesting tissue-specific expression of hFMO. Induction of hFMO is consistent with the occurrence of cis-osmoregulatory and glucocorticoid response elements identified in the 5′-upstream sequence, indicating regulation of hFMO in response to hypersaline conditions and the osmoregulatory hormone cortisol.
Flavin-containing monooxygenases (FMOs) are a polymorphic family of enzymes found in the smooth endoplasmic reticulum of multiple tissues that catalyze the oxidation of soft nucleophilic heteroatom substances to their respective oxides (Ziegler, 1988; Cashman, 2004; Krueger and Williams, 2005). FMO substrates include alkaloids, pesticides, and pharmaceutical compounds (Cashman, 1995; Rettie and Fisher, 1999). In mammals, five families of distinct FMO genes have been identified and classified based on the amino acid sequence (Lawton et al., 1994). These isoforms differ in tissue distribution, regulation, and substrate specificity (Cashman, 1995; Luo and Hines, 2001; Krueger and Williams, 2005).
FMO activity and protein expression in gill and liver were altered by changes in salinity within euryhaline fish (Schlenk et al., 1996; Larsen and Schlenk, 2001; El-Alfy et al., 2002). The mechanisms surrounding the regulation of FMO by osmotic pressure caused by hypersaline conditions are unclear but may involve endocrine modulation. Previous studies have indicated a role of glucocorticoids and sex steroids in FMO expression within fish and mammals (Lee et al., 1995; El-Alfy and Schlenk 2002; El-Alfy et al., 2002). Both groups of compounds play key roles in the endocrine modulation of osmoregulation. Because of the complexity of the endocrine system, cross-talk between hormonal feedback loops often occurs, preventing accurate determinations of hormonal impact. To evaluate the direct impacts of hormonal regulation and osmotic pressure on FMO, an FMO cDNA was cloned and sequenced in liver from rainbow trout (Oncorhynchus mykiss), which had previously responded with induction in vivo following treatment with hypersaline water. The hypothesis that osmotic pressure plays a direct role on FMO expression was evaluated by analyzing upstream regulatory elements of an FMO gene [hepatic isoform of flavin-containing monooxygenase (hFMO)] isolated by using the hFMO cDNA. Cultured primary cells from liver and gill were treated with NaCl and the osmoregulatory hormone cortisol. Results indicated that hFMO is regulated in a tissue-specific manner in vertebrates, and expression of transcripts can be directly influenced by osmotic pressure and the osmoregulatory hormone cortisol in primary hepatocytes.
Materials and Methods
Organisms
Rainbow trout of approximately 4 months (16 ± 4 cm) were obtained from Jess Ranch Fish Hatchery (Apple Valley, CA). Organisms were maintained in a flow-through living-stream system with dechlorinated carbon-filtered municipal water at 13 to 18°C and acclimatized for 3 weeks before experimental use. Organisms were fed with commercial fish feed (Silver Cup, Murray, UT).
RNA Isolation
Gill and liver mRNA was isolated from tissues obtained from organisms treated for 7 days in hypersaline conditions (14 parts per thousand). Approximately 20-mg aliquots of tissues pooled from three or four individuals (60–80 mg total) were treated with TRIzol (Invitrogen, Carlsbad, CA)/chloroform and ethanol for RNA precipitation. RNA samples were then quantified using a spectrophotometer at 260 nm/280 nm and stored at −80°C until use for reverse transcriptase-polymerase chain reaction.
Cloning and Sequencing of Rainbow Trout FMO cDNAs
Full-length hFMO cDNA was achieved by combining PCR screening of a cDNA pool reverse-transcribed from total RNA isolated from liver of rainbow trout and subsequent rapid amplification of 5′-cDNA ends (RACE). Reverse transcription of total RNA from each tissue (1 μg) using oligo(dT)15 primer (0.5 μg) was performed for 1 h at 42°C. Routinely, reactions in the absence of reverse transcriptase were used as a negative control. PCR amplifications were performed with an aliquot of the cDNA pool (5 μl) in a final volume of 25 μl. Incubations contained 20 mM Tris-HCl, pH 8.4, 50 mM KCl, 3 mM MgCl2, 200 μM deoxyguanosine triphosphate, 200 μM deoxynucleoside-5′-triphosphates, AccuPrime Taq DNA polymerase, thermostable AccuPrime protein and stabilizers, and 0.4 μM each sense and antisense primer using AccuPrime SuperMix I (Invitrogen) and following the manufacturer’s protocol. Primers used for initial screening were designed based on manual alignments of an O. mykiss expressed sequence tag (CA359096) and cDNA sequences from Danio rerio FMO genes (Table 1). Pairs of primers used were (FMO5/CA35-729R) and (CA35-729F/FMO1–5). An initial cycle was initiated at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 68°C, performed in a Bio-Rad iCycler (Bio-Rad, Hercules, CA). PCR products (10 μl) were loaded onto agarose gels (1.2%) containing ethidium bromide (0.5 μg/ml). PCR products were visualized with fluorescent illumination. Amplicons were extracted, purified from gel with the Pure link PCR purification kit (Invitrogen), and subcloned into a pCR2.1-TOPO vector using aliquots (1 μl) of the respective PCR sample. TOP10 chemically competent Escherichia coli was transformed according to the manufacturer’s instructions, and plasmids from clones containing inserts were purified with the QIAprep Miniprep kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Plasmid DNA was sent to a DNA sequencing facility located in the Genomics Core Instrumentation Unit at the University of California, Riverside, for automated sequencing of both strands.
TABLE 1.
Primers and Tm values used in PCR experiments with hFMO
| Name | Nucleotide Sequence (5′–3′) | Tm |
|---|---|---|
| FMO1–5 | ATC ATG GTG CGT ACA GTA GCA GTG | 55° |
| FMO1–3 | TGT TCA AAC TCT CTG AGG | 55° |
| CA35-729F | GGA CAT GAA GTA TAA CAC ACG CTT | 55° |
| CA35-729R | AAG CGT GTG TTA TAC TTC ATG TCC | 55° |
| FMO538-F | ATA AGG GTC CAG AGG ACC TT | 57° |
| FMO538-R | AAG GTC CTC TGG ACC CTT AT | 57° |
| Intr1-F | CCG TGC ACA AAG CGA AGT CCA TAC | 60° |
| FMO5Q | CTG GCT GTT CTT CAC TGA CTA CC | 65° |
| FMO3Q | GCT GGA ACA TGC GTT CAA ACT GG | 65° |
A partial cDNA fragment (hFMO) was identified from liver and used for subsequent RACE using the Smart PCR cDNA Synthesis Kit (Clontech, Palo Alto, CA). Briefly, oligo(dT)-primed cDNA was synthesized from 5 μg of total RNA from trout liver. After synthesis of the second-strand cDNA, the sample was ligated to the Smart cDNA adaptor and the Marathon cDNA adaptor (2 μM) in the presence of T4 ligase (1 U/μl). Adaptor-ligated double-stranded cDNA (1 μl) was then diluted in Tricine/EDTA buffer (250 μl), heated at 94°C for 2 min, and kept at −20°C until use. RACE PCR was performed in a final volume of 25 μl containing Advantage buffer and Advantage 2 polymerase (Clontech) in the presence of an adaptor primer (AP1, 5′-CCA TCC TAA TAC GAC TCA CTA TAG GGC-3′, Clontech) and one specific primer for the amplification of the 5′-end (FMO5/FMO3). PCR conditions were based on the recommendations of the manufacturer. An initial cycle at 95°C for 1 min was followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 120 s at 68°C. When cycling was completed, RACE DNA products were extracted from the gel, purified, subcloned into the pCR2.1-TOPO vector, and sequenced as described above.
Cloning and Sequencing of the hFMO Gene
PCR amplification of the hFMO gene was targeted for sequence analyses using primers designed to amplify the entire hFMO coding region (FMO1–5/CA35729R), (FMO538-F/FMO1–3). Because of its size, intron 1 was analyzed as two overlapping regions using (Intr1-F/FMO538-R) primers. Reactions were performed using 200 ng of genomic DNA and the manufacturer’s protocol of AccuPrime SuperMix II (Invitrogen) as above. The cycle parameters were as follows: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 3.30 min. DNA products were purified, subcloned into the pCR2.1-TOPO vector, and sequenced as described above.
Genomic Southern Blot
Genomic DNA was isolated from the blood of rainbow trout. To avoid polymorphisms, initial studies used DNA obtained from the blood of one fish. Additional analyses with three other individuals were carried out to confirm sequence identity. To isolate the DNA, the DNeasy Mini Kit (Qiagen) was used. A PCR-amplified 180-base pair (bp) fragment of the hFMO cDNA, spanning bp 548 to 728, was used as a probe. The genomic DNA (5 μg) was digested with BamHI, EcoRI, and PstI HindIII, and double digestions were carried out with PstI/BamHI, and HindIII/PstI. The digested genomic DNA was separated on an agarose gel (0.8%) and transferred to Hybond N+ nylon membranes using a 20× standard saline citrate (SSC) solution (3 M NaCl, 0.3 M sodium citrate, 1 mM EDTA). The blot was prehybridized for 2 h with 5× SSC, 0.2% SDS, and 200 mg/ml heparin at 65°C. High stringency hybridization was carried out overnight under the same conditions, with approximately 7 × 106 cpm/ml of the 32P-labeled 180-bp cDNA probe. The blot was washed twice with 0.1 × SSC, 0.2% SDS at 65°C. For low stringency conditions, the blot was prehybridized with 5× SSC, 0.2% SDS, and 200 mg/ml heparin at 45°C and washed with 2× SSC, 0.2% SDS at 42°C. The blots were evaluated using autoradiography and visualized using an Amersham Typhoon 9410 Phosphoimager (Piscataway, NJ).
Structural Analysis of the Promoter Region FMO Gene
The promoter region of hFMO was cloned using the Universal Genome Walker Kit (BD Biosciences, San Jose, CA) and the manufacturer’s protocol. Genomic DNA (100 ng/μl) from trout liver was digested with the blunt-end restriction enzymes DraI, EcoRV, PvuII, and StuI and subsequently ligated to the provided adaptor linkers. Nested PCR with 94°C initial denaturation was then performed using primers derived from the coding region of hFMO (FMO-P1) (Table 1) and adaptor primers (AP1 5′-GTAATACGACTCACTATAGGGC-3′, AP2 5′-ACTATAGGGCACGCGTGGT-3′) with the Advantage 2 PCR kit (BD Biosciences). The cycle parameters were as follows: seven cycles of 94°C for 25 s and 72°C for 3 min, followed by 32 cycles of 94°C for 25 s and 67°C for 3 min and a final extension at 67°C for 7 min. DNA products were purified, subcloned into the pCR2.1-TOPO vector, and sequenced as described above.
Isolation, Culture, and Exposure of Primary Hepatocytes
Fish hepatocytes were isolated using enzymatic digestion with trypsin followed by mechanical disaggregating and gradient centrifugation using Percoll (Amersham Biosciences, Uppsala, Sweden) as described in Sandbacka and Isomaa (2000). Viability was assessed by exclusion of 0.4% trypan blue before starting the treatments. After cell isolation, the cells were seeded in 48-well plates with a density of 1 × 106 cells/well. Cells were allowed to settle for 2 h and then were treated with the water extracts. Three wells were treated for each concentration and control treatment. Cells were exposed to different concentrations of NaCl (8, 16, and 32 mM) and different concentrations of cortisol (0.27, 2.7, and 27 μM) and incubated for 24 h at 18°C. Concentrations of cortisol were determined from previous infusion studies in trout (El-Alfy et al., 2002). The concentrations of NaCl and the duration of exposure were based on range-finding studies. After treatment, cells were resuspended in phosphate-buffered saline (PBS) and centrifuged at 5200g for 5 min, and the pellet was washed twice with PBS. Cells used for quantitative PCR (qPCR) were immediately processed for total mRNA extraction. Total RNA from cells was extracted using QIAshredder (Qiagen) and RNeasy Mini RNA extraction Kit (Qiagen) following the manufacturer’s instructions. RNA samples were then quantified using a spectrophotometer at 260/280 nm and stored at −80°C until use for qPCR.
Cell Viability Assay
The viability of the cells was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) (Sigma-Aldrich, St. Louis, MO) assay, which is based on the reduction of MTT by the mitochondrial dehydrogenase of intact cells to a purple formazan product. Briefly, hepatocytes were collected, and 3 × 105 cells/well were dispensed within 96-well culture plates in 100-μl volumes. Cells were exposed to NaCl (8, 16, and 32 mM) (Fisher Scientific, Fair Lawn, NJ) and cortisol (0.27, 2.7, and 27 μM) (Sigma-Aldrich) for 24 h as described above. An MTT working solution was prepared as 5 mg of MTT/ml in sterile PBS. To each of the above cultured wells, 20 μl of the MTT working solution was added with continuous incubation for 4 h (the last 4 h of the exposure). The water-insoluble formazan that was formed during incubation was solubilized by adding dimethyl sulfoxide/ethanol (1:1 in v/v). The concentration of formazan was determined by measuring the absorbance at 595 nm using an enzyme-linked immunosorbent assay plate reader, and well imperfections were corrected by the measure of the absorbance at 650 nm.
Effect of Cortisol, Sodium Chloride, and Sodium Sulfate on hFMO Expression
After cell isolation, the cells were seeded in 48-well plates with a density of 1.5 × 106 cells/well. Cells were allowed to settle for 3 h and then were treated with increasing concentrations of NaCl and cortisol (Sigma-Aldrich) as described above. Optimal concentrations were determined after testing viability to minimize cytotoxicity. Cells were incubated for 24 h at 18°C and then were resuspended in PBS and centrifuged at 3000 rpm for 5 min, and the pellet was washed twice with PBS and subsequently extracted for total RNA as described above. The qPCR was used to measure expression of hFMO. One microliter of isolated total RNA was reverse-transcribed with 1 U of Super Script Reverse Transcriptase (Invitrogen) in the presence of random hexamers according to the manufacturer’s instructions. One microliter of the reverse transcriptase reaction mixture was used for PCR amplification using primers FMO5Q/FMO3Q (Table 1) as described above. hFMO was quantified with the SYBR Green-based qPCR method using an iCycler iQ apparatus (Bio-Rad). The standard mixture consisted of a 1:10,000 dilution of SYBR Green I (Molecular Probe), 10 mM Tris-Cl, pH 8.5, 40 mM KCl, 2 mM MgCl2, 0.1 mM NTP, 10 pmol of each primer, and 2.5 U of Taq polymerase in a 30-μl reaction volume. The optimal PCR conditions for quantification were determined using melting curve analysis by heating from 55 to 95°C (0.5°C for 10 s per cycle for 80 cycles) with simultaneous detection of the SYBR Green I fluorescence signal. Twenty-eight cycles were used for optimal quantification as this was in the linear range for amplification of the hFMO signal.
Flavin Monooxygenase Nucleotide and Protein Sequence Analysis
The sequences of rainbow trout FMO cDNAs were compared with FMO sequences available in the National Center for Biotechnology Information database for human, fish, and other species using Basic Local Alignment Search Tool. Predicted amino acid sequences for FMOs were aligned using ClustalX software with corresponding human and fish sequences. The degree of amino acid similarity of fish FMO compared with human and other fish species was also analyzed. FMO amino acid sequences were collected from SRS data integration software (LION bioscience AG, Heidelberg, Germany), and transcription factor sequences were analyzed by AliBaba 2 (http://www.gene-regulation.com/pub/programs.html). Conserved motifs were identified using ConSite (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite).
Statistical Analysis
FMO expression in primary cells was measured using three replicates with separate groups of cells from livers and gills of three or four pooled organisms. Between-group differences were determined by oneway analysis of variance (p < 0.05) with Fisher’s least significant difference post hoc test for significance.
Results
Cloning of Rainbow Trout hFMO cDNAs
hFMO (GenBank accession number EF063736) had an open reading frame of 1792 bp and encoded a protein of 554 amino acids. The deduced amino acid sequence of liver hFMO showed 74% identity to the reported putative FMO from Takifugu rubripes (accession number Q6ZZY9), 52 to 55% to zebrafish (Brachydanio rerio; accession number Q5RGM6, Q5RGM3, Q6TLD2, Q7T1D7), and 30 to 45% to Tetraedon nigroviridis (accession number Q4SLVO, Q4T8R2). When compared with mammals, trout hFMO showed between 54% and 50% identities to the sequences of human FMO1, FMO3, and FMO5 (accession number FMO1, Q01740; FMO3, P49326; FMO5, P49326), respectively. Analysis of the hFMO amino acid sequence revealed identical characteristic sequence motifs: 1) FAD-binding domain (GxGxxG), 2) NADPH-binding domain (GxGxxG), and 3) FMO-identifying sequence (FxGxxxHxxxY/F) and the conserved EGLEP and FATGY sequences. Furthermore, the conserved Asn-61 residue involved in the catalytic mechanism of mammalian FMOs was also identified (Eswaramoorthy et al., 2006) (Fig. 1).
Fig. 1.
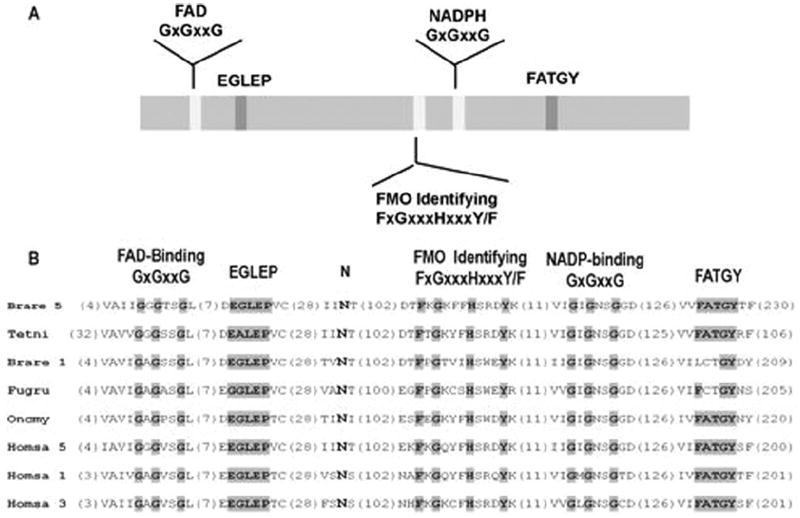
Distribution and alignment of various motifs of FMO proteins. A, schematic drawing of the location of the five motifs along fish FMO proteins. The motifs are shown relative to rainbow trout hFMO. B, alignment of the motifs showing the exact amino acid numbering and the deviation from the consensus shown on the top of the alignment. The shaded residues are identical to the consensus. Brare 5, Danio rerio (Q5RGM4); Tetni, Tetraodon nigroviridis (Q4T8R2); Frugu, Takifugu rubripes (Q802T5); Brare 1, D. rerio (Q5TZD0); Homsa 1, Homo sapiens (Q01740); Homsa 3, H. sapiens (P49326); Homsa 5, H. sapiens (P49326).
Cloning of the Rainbow Trout hFMO Gene (hFMO)
Using the coding sequence of hFMO cDNA, a 4.379-kilobase genomic DNA product was sequenced and assigned the GenBank accession number of EU519462. The genomic FMO sequence (hFMO) possessed a 150-bp 5′-untranslated region. The exon/intron organization of hFMO was determined by overlapping genomic DNA sequence with the cDNA sequence of hFMO. The hFMO gene consisted of 10 exons and 9 introns. The majority of donor and acceptor sites of these introns were GT and AG, respectively, following the GT/AG rule as described by Breathnach and Chambon (1981) (Fig. 2).
Fig. 2.
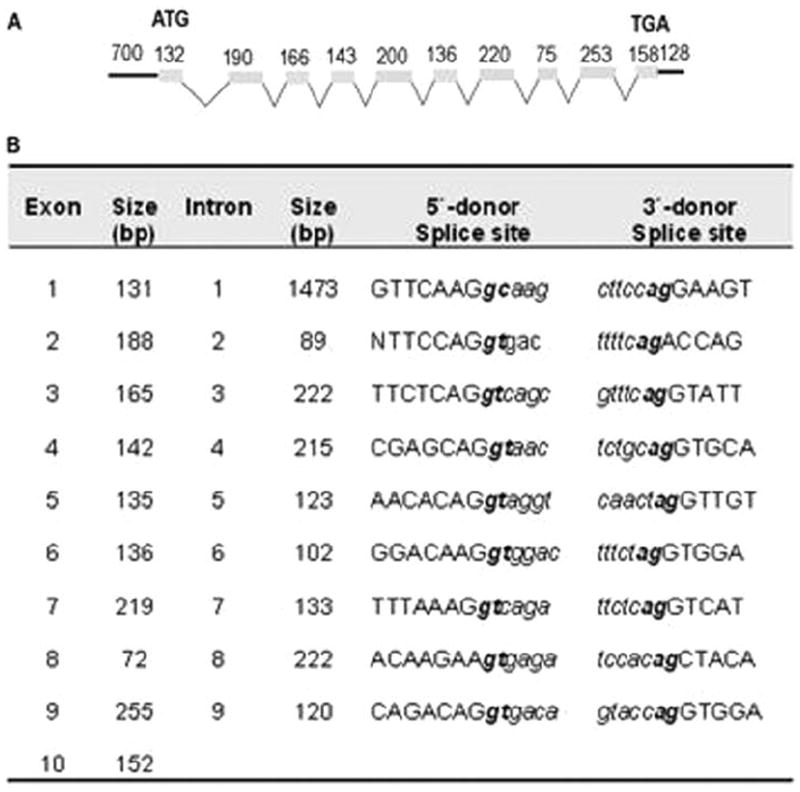
A, genomic structure of the rainbow trout hFMO. Schematic diagram showing the relative positions of introns and exons. Exons are depicted as shaded boxes. Introns are indicated by open triangles. Numbers above the boxes indicate exon size in base pairs; numbers below the triangles indicate intron length. B, exon-intron boundaries of the rainbow trout hFMO gene.
To gain insight into the complexity of FMO gene copy number in the O. mykiss genome, a Southern blot was performed (Fig. 3). At high stringency hybridization conditions, the blot showed either double or multiple bands in each lane. Identical results were obtained using low or moderately low stringency conditions (data not shown). According to the sequence of hFMO after enzymatic digestion with HindIII and PstI, 1200- and 1250-bp fragments would have been predicted, respectively. With the double digestion of BamHI/PstI, a fragment of approximately 800 bp would have been predicted. The occurrence of multiple bands within each digestion suggests that at least seven FMO genes or variants may exist within rainbow trout.
Fig. 3.
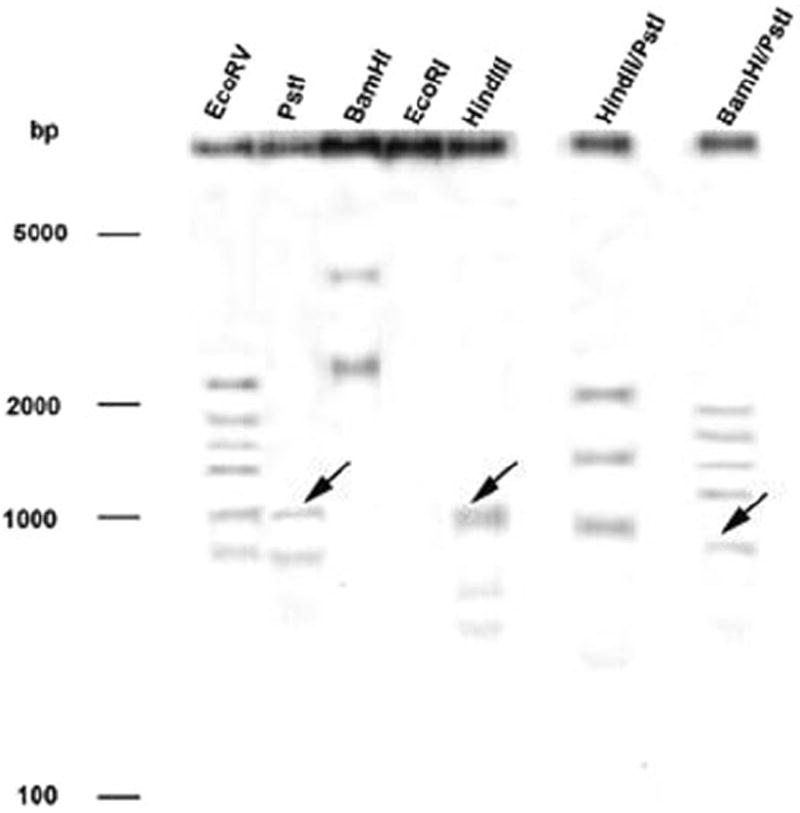
Southern blot of DNA from rainbow trout hybridized with a 180-bp fragment of the rainbow trout hFMO cDNA. Total DNA from a single fish was isolated, and 5 μg was digested with the different restriction enzymes EcoRV, PstI, BamHI, EcoRI, and HindIII and double digestions HindII/PstI and BamHI/PstI; lanes 1 through 7, respectively. A 0.8% agarose gel was run and DNA blotted to a nylon membrane. Hybridization was carried out under high stringency conditions. Arrowheads indicate the predicted fragments generated from digestion of hFMO.
Structural Analysis of the Promoter Region of the FMO Gene
As an initial attempt to investigate the molecular mechanisms regulating the expression of the FMO gene, a structural analysis of the promoter region of hFMO was carried out. An 853-bp segment upstream of the translation start site was evaluated and revealed a putative transcription start site 150 bp upstream from the translation start site. A TATA box was observed at −23 to −29 bp relative to the transcription start site. A CCAAT (enhancer binding protein) binding site (−450) also was observed. Analysis with the phylogenetic foot-printing program ConSite and AliBaba 2.1, with 85% match conservation, identified several additional motifs. Of these, two were predicted as glucocorticoid response elements (GREs), two were consensus sequences for Yin Yang 1 (YY1) core elements, two as sites for Sox-5, one for Sp1, one for Hunchback, and one for forkhead related activator (FREAC) 4 (Fig. 4). An osmotic response element (ORE) consensus site (5′-NGGAAAWDHMC(N)-3′) was also observed at −374 to −363 bp (TGGAAAAACCCA).
Fig. 4.
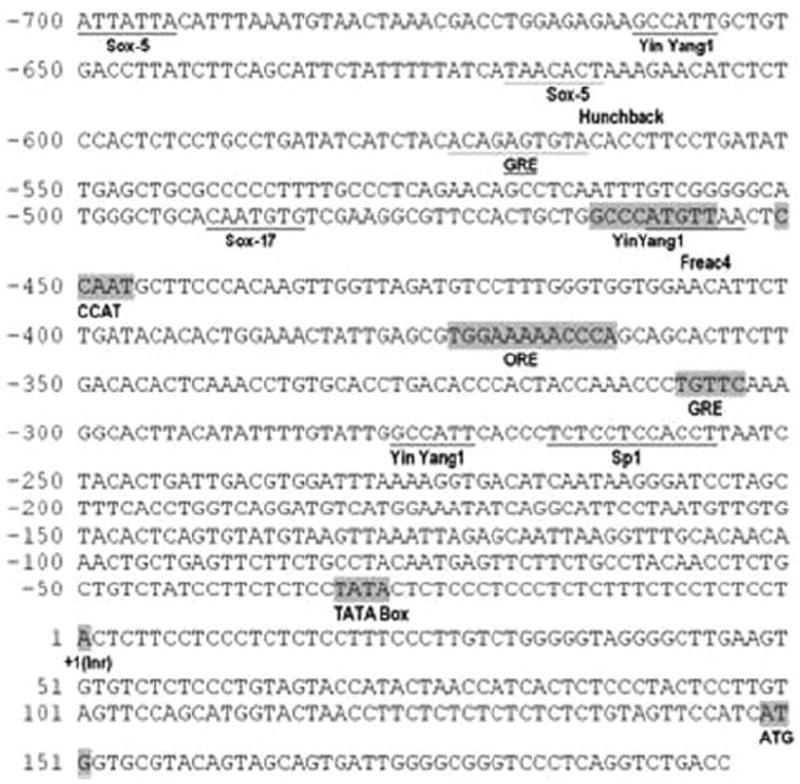
The proximal region of the rainbow trout FMO. Potential start site (+1 initiator sequence, Inr), TATA box, CCAAT, GRE, ORE, and YY1 were detected manually and shown in boxes. Other sites: Sox-5, additional Yin Yang and GREs (underline) were identified by ConSite and AliBaba analysis (85% match conservation).
Effect of Cortisol, Sodium Chloride, and Sodium Sulfate on FMO Expression and Catalytic Activity in Rainbow Trout Hepatocytes and Gill Epithelial Cells
Viability in hepatocytes ranged between 85 and 90% for all the treatments. In gill, viability was 80 to 90%. Cortisol significantly increased hFMO expression in hepatocytes, as did treatment with NaCl (Fig. 5). Reverse transcriptase-PCR from gill cDNA did not produce the amplicon found in hepatocytes.
Fig. 5.
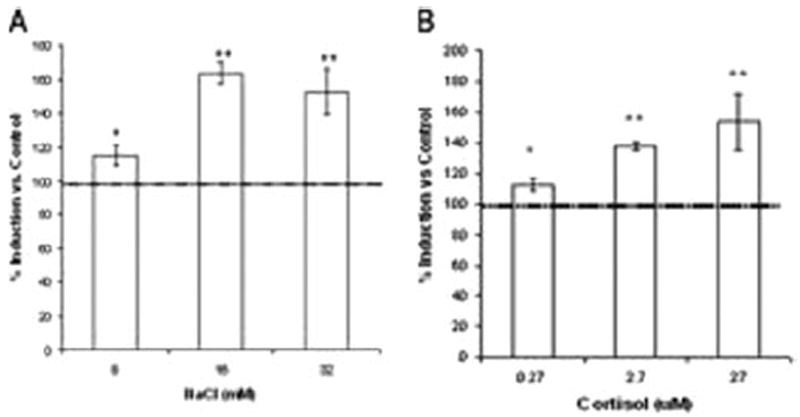
Effect of NaCl (A) and cortisol (B) on hFMO mRNA expression in rainbow trout primary hepatocytes. Each value represents the mean ± S.D. of three replicate experiments with three separate cellular preparations. Values are presented as percentage of control (100 ± 12%). * or **, denotes statistical difference from control (*, p ≤ 0.05; **, p ≤ 0.01).
Discussion
Previous studies in euryhaline fish have indicated up-regulation of FMO expression and catalytic activity by hypersaline conditions (Schlenk et al., 1996; Larsen and Schlenk, 2001; Wang et al., 2001). However, the mechanisms of FMO regulation under hypersaline conditions remain unclear. To better understand mechanisms controlling gene expression, a novel gene encoding a hepatic form of rainbow trout FMO was cloned and sequenced. It is the first report of a salmonid FMO sequence. The derived amino acid sequence of the hFMO cDNA was 55 to 74% similar to two reported fish FMOs. When comparing the sequence with mammalian forms, the sequence was approximately 50% similar to FMO1, FMO3, and FMO5. hFMO possessed the two characteristic FMO pentapeptides (-EGLEP- and -FATGY-), as well as FAD and NADPH biding sites (-GAGPSGL-) and (-GIGNSG-) and the Asp 61, respectively. Southern blot analysis indicated at least seven genes are present in rainbow trout. Attempts to identify additional full-length genes in the gill were unsuccessful, although truncated transcripts were identified (data not shown). Whether these are catalytically active is unclear, but expression studies are underway to see whether any of these additional products are functional or pseudogenes. Splice variants of FMO genes that have recently been identified in humans and shown to have unique catalytic properties (Hines et al., 2002; Lattard et al., 2004). The occurrence of a full-length hFMO transcript in liver but not gill is consistent with tissue-specific expression of FMO in mammals and may indicate unique functional properties.
The hFMO genomic DNA sequence possessed a 150-bp 5′-untranslated region, 10 exons, and 9 introns. In zebrafish, FMO genes are located in chromosome 20 (locus: BX255937; regions: 111695–117217 and 121505–126238) and are distributed over approximately 5000 bp of genomic DNA with a very similar exon-intron organization (accession number CAH68899.1) but with different lengths and boundaries (Fig. 2). In contrast to mammalian FMOs, a distinct TATA box (TATAA) was observed in the hFMO promoter (TATA sequence). The promoter region also contains a preponderance of GC dinucleotides and several other previously observed consensus sequences (Fig. 4). YY1 is recognized by members of the GLI-Krüppel family of zinc-finger proteins and has been found to mediate induction of FMO1*6 during fetus development and the adult human (Luo and Hines, 2001; Hines et al., 2003). Recently, an unidentified GC box binding protein was shown to contribute to human FMO3 regulation (Klick and Hines, 2007). The physiological significance of having both TATA- and GC-rich regions in the trout hFMO promoter is unclear but warrants further study.
Two GREs were also observed in the hFMO promoter and treatment of hepatocytes with cortisol-induced expression of hFMO. Partial GREs have also been observed upstream of mammalian FMOs (Wyatt et al., 1996). Studies in rodents and rabbits have shown that glucocorticoids may contribute to the regulation of FMO2 (Lee et al., 1995). In rainbow trout, infusion of cortisol through a dorsal aortal cannula significantly increased expression of a protein recognized by antibodies to human FMO1 and thiourea oxidase activity in several tissues including liver, gill, and red blood cells (El-Alfy et al., 2002). Hypersaline conditions significantly modulate cortisol expression, and cortisol plays a significant role in saltwater acclimation of salmonids. Up-regulation of hFMO observed in vivo may involve a direct effect of cortisol through the GRE, but a more complex endocrine response, perhaps through the hypothalamus-pituitary axis, should not be overlooked.
In addition to hormonal regulation, several genes involved in osmoregulation may also be regulated directly by changes in the cellular environment, such as redox potential or osmotic pressure. The gene encoding aldose reductase is regulated through interactions at upstream OREs (Ferraris et al., 1996). A cis-ORE was observed upstream of the hFMO and also has been observed in several other FMO genes in rainbow trout that have yet to be fully characterized (data not shown). To determine whether osmotic changes could influence expression, isolated gill epithelium and hepatocytes were treated with an osmotic effector. Treatment of hepatocytes with NaCl clearly showed an increase in expression of hFMO consistent with previous work in vivo, in which an increase of FMO activity, protein expression, and mRNA of FMO1-like proteins and transcripts was observed in gill and liver of rainbow trout liver maintained under hypersaline seawater conditions (Larsen and Schlenk, 2001; Wang et al., 2001). These data are also consistent with the presence of ORE in the 5′-flanking region of the hFMO gene. Further studies are necessary to determine whether ORE-binding proteins or other trans-activating factors that bind OREs previously characterized in mammals and bacteria contribute to FMO regulation.
In conclusion, a hepatic FMO gene (hFMO) and cDNA were cloned and sequenced from rainbow trout. Southern blot analyses indicated at least seven potential genes may be present in trout with some encoding truncated proteins or splice variants that may or may not have catalytic activity. Analysis of 853 bp of the 5′-flanking region of the gene indicated several cis-elements including GREs and an ORE, which may help explain the respective up-regulation of hFMO transcription with cortisol and NaCl treatment in isolated hepatocytes. Additional studies are underway to better understand the mechanisms that control the expression of this toxicologically and physiologically important enzyme in fish.
ABBREVIATIONS
- FMO
flavin-containing monooxygenase
- hFMO
hepatic isoform of flavin-containing monooxygenase
- PCR
polymerase chain reaction
- RACE
rapid amplification of 5′-cDNA end
- bp
base pair
- SSC
standard saline citrate
- PBS
phosphate-buffered saline
- qPCR
quantitative polymerase chain reaction
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- GRE
glucocorticoid response element
- YY1
Yin Yang 1
- ORE
osmotic response element
References
- Breathnach R, Chambon P. Organization and expression of eucaritic split genes coding for proteins. Ann Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cashman JR. Structural and catalytic properties of the mammalian flavin-containing monooxygenase. Chem Res Toxicol. 1995;8:165–181. doi: 10.1021/tx00044a001. [DOI] [PubMed] [Google Scholar]
- Cashman JR. The implication of polymorphisms in mammalian flavin-containing monooxygenases in drug discovery and development. Drug Discov Today. 2004;9:574–581. doi: 10.1016/S1359-6446(04)03136-8. [DOI] [PubMed] [Google Scholar]
- El-Alfy A, Larsen B, Schlenk D. Effect of cortisol and urea on flavin monooxygenase activity and expression in rainbow trout, Oncorhynchus mykiss. Mar Env Res. 2002;54:275–278. doi: 10.1016/s0141-1136(02)00183-6. [DOI] [PubMed] [Google Scholar]
- El-Alfy A, Schlenk D. Effect of 17β-estradiol and testosterone on the expression of flavin containing monooxygenase and the toxicity of aldicarb to Japanese medaka, Oryzias latipes. Toxicol Sci. 2002;68:381–388. doi: 10.1093/toxsci/68.2.381. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. Mechanism of action of a flavin-containing monooxygenase. Proc Natl Acad Sci U S A. 2006;103:9832–9837. doi: 10.1073/pnas.0602398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris JD, Williams CK, Jung KY, Bedford JJ, Burg MB, Garcia-Perez A. ORE, a eukaryotic minimal essential osmotic response element: the aldose reductase gene in hyperosmotic stress. J Biol Chem. 1996;271:18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- Hines RN, Hopp KA, Franco J, Saeian K, Begun FP. Alternative processing of the human hepatic FMO6 gene renders transcripts incapable of encoding a functional flavin containing monooxygenase. Mol Pharmacol. 2002;62:320–325. doi: 10.1124/mol.62.2.320. [DOI] [PubMed] [Google Scholar]
- Hines RN, Luo Z, Hopp KA, Cabacungan ET, Koukouritaki SB, Mccarver DG. Genetic variability at the human FMO1 locus: significance of a basal promoter Yin Yang 1 element polymorphism (FMO1*6) J Pharmacol Exp Ther. 2003;306:1210–1218. doi: 10.1124/jpet.103.053686. [DOI] [PubMed] [Google Scholar]
- Klick DE, Hines RN. Mechanisms regulating human FMO3 transcription. Drug Metab Rev. 2007;39:419–442. doi: 10.1080/03602530701498612. [DOI] [PubMed] [Google Scholar]
- Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106:357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen BK, Schlenk D. Effect of salinity on flavin-containing monooxygenase expression and activity in rainbow trout (Onchorhynchus mykiss) J Comp Physiol B. 2001;171:421–429. doi: 10.1007/s003600100192. [DOI] [PubMed] [Google Scholar]
- Lattard V, Zhang J, Cashman JR. Alternative processing of flavin-containing monooxygenase (FMO) pre-mRNA in human. Mol Pharmacol. 2004;65:1517–1525. doi: 10.1124/mol.65.6.1517. [DOI] [PubMed] [Google Scholar]
- Lawton MP, Cashman JR, Cresteil T, Dolphin CT, Elfarra AA, Hines RN, Hogson E, Kimura T, Ozols J, Phillips IR. A nomenclature for the mammalian flavin-containing monooxygenase gene family based on amino acid sequence identities. Arch Biochem Biophys. 1994;308:254–257. doi: 10.1006/abbi.1994.1035. [DOI] [PubMed] [Google Scholar]
- Lee M-Y, Smiley S, Kadkhodayan S, Hines RN, Williams DE. Developmental regulation of flavin-containing monooxygenase (FMO) isoforms 1 and 2 in pregnant rabbit. Chem Biol Interact. 1995;96:75–85. doi: 10.1016/0009-2797(94)03584-u. [DOI] [PubMed] [Google Scholar]
- Luo Z, Hines RN. Regulation of flavin-containing monooxygenase 1 expression by Ying Yang 1 and hepatic nuclear factors 1 and 4. Mol Pharmacol. 2001;60:1421–1430. doi: 10.1124/mol.60.6.1421. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Fisher MB. In: Handbook of drug metabolism. Woolf TF, editor. Marcel Dekker; New York: 1999. pp. 131–145. [Google Scholar]
- Sandbacka M, Isomaa B. Glutathione S-transferase and UDP-glucuronyltransferase activity in primary cultures of rainbow trout gill epithelial cells. Comp Biochem Physiol C Toxicol Pharmacol. 2000;127:307–315. doi: 10.1016/s0742-8413(00)00156-0. [DOI] [PubMed] [Google Scholar]
- Schlenk D, Peters LD, Livingstone DR. Correlation of salinity with flavin-containing monooxygenase activity but not cytochrome P450 activity in the euryhaline fish (Platichthys flesus) Biochem Pharmacol. 1996;52:815–818. doi: 10.1016/0006-2952(96)00358-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Grisle S, Schlenk D. Effects of salinity on aldicarb toxicity in juvenile rainbow trout (Onchorhynchus mykiss) and stripped bass (Morone saxatilis x chrysops) Toxicol Sci. 2001;64:200–207. doi: 10.1093/toxsci/64.2.200. [DOI] [PubMed] [Google Scholar]
- Wyatt MK, Philpot RM, Carver G, Lawton MP, Nikbakht KN. Structural characteristics of flavin-containing monooxygenase genes one and two (FMO1 and FMO2) Drug Metab Dispos. 1996;24:1320–1327. [PubMed] [Google Scholar]
- Ziegler DM. Flavin-containing monooxygenases: catalytic mechanisms and substrate specificities. Drug Met Rev. 1988;19:1–32. doi: 10.3109/03602538809049617. [DOI] [PubMed] [Google Scholar]


