Abstract
Two single nucleotide polymorphisms (SNPs) at 6q25.1, near the ESR1 gene, have been implicated in the susceptibility to breast cancer for Asian (rs2046210) and European women (rs9397435). A genome-wide association study in Europeans identified two further breast cancer susceptibility variants: rs11249433 at 1p11.2 and rs999737 in RAD51L1 at 14q24.1. Although previously identified breast cancer susceptibility variants have been shown to be associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers, the involvement of these SNPs to breast cancer susceptibility in mutation carriers is currently unknown. To address this, we genotyped these SNPs in BRCA1 and BRCA2 mutation carriers from 42 studies from the Consortium of Investigators of Modifiers of BRCA1/2. In the analysis of 14 123 BRCA1 and 8053 BRCA2 mutation carriers of European ancestry, the 6q25.1 SNPs (r2 = 0.14) were independently associated with the risk of breast cancer for BRCA1 mutation carriers [hazard ratio (HR) = 1.17, 95% confidence interval (CI): 1.11–1.23, P-trend = 4.5 × 10−9 for rs2046210; HR = 1.28, 95% CI: 1.18–1.40, P-trend = 1.3 × 10−8 for rs9397435], but only rs9397435 was associated with the risk for BRCA2 carriers (HR = 1.14, 95% CI: 1.01–1.28, P-trend = 0.031). SNP rs11249433 (1p11.2) was associated with the risk of breast cancer for BRCA2 mutation carriers (HR = 1.09, 95% CI: 1.02–1.17, P-trend = 0.015), but was not associated with breast cancer risk for BRCA1 mutation carriers (HR = 0.97, 95% CI: 0.92–1.02, P-trend = 0.20). SNP rs999737 (RAD51L1) was not associated with breast cancer risk for either BRCA1 or BRCA2 mutation carriers (P-trend = 0.27 and 0.30, respectively). The identification of SNPs at 6q25.1 associated with breast cancer risk for BRCA1 mutation carriers will lead to a better understanding of the biology of tumour development in these women.
INTRODUCTION
Genome-wide association studies (GWASs) have identified multiple common alleles that are associated with breast cancer risk in the general population (1–7). Such alleles provide plausible candidates as modifiers of cancer risk for BRCA1 and BRCA2 mutation carriers. Nine of these polymorphisms have been investigated as risk modifiers to date (8–10); single nucleotide polymorphisms (SNPs) in FGFR2, TOX3, MAP3K1, LSP1, 2q35, SLC4A7 and 5p12 have been shown to be associated with breast cancer risk for BRCA2 mutation carriers, but only SNPs in TOX3 and 2q35 were associated with the risk for BRCA1 mutation carriers. The differential patterns of associations between BRCA1 and BRCA2 mutation carriers appear to be in line with the differential effects of these polymorphisms for oestrogen receptor-positive and oestrogen receptor-negative breast cancer in the general population (10,11). More recently, a GWAS restricted to BRCA1 mutation carriers identified a locus at 19p13 which modified breast cancer risk for BRCA1 mutation carriers and the risk of oestrogen receptor (ER) negative and triple negative (oestrogen, progesterone receptor (PR) and Human Epidermal growth factor receptor 2 (HER2) negative) breast cancer in the general population (12). A separate GWAS in BRCA2 mutation carriers suggested that another locus at ZNF365 may modify the risk of breast cancer for BRCA2 mutation carriers (13). Candidate gene studies have also suggested that a SNP in CASP8 is also associated with the risk of breast cancer for BRCA1 mutation carriers (14). Each of these polymorphisms confers modest relative risks for breast cancer, but evidence so far suggests that they interact multiplicatively on the breast cancer risk for mutation carriers and the range of the combined risks of these SNPs is ∼6-fold (10). Since BRCA1 and BRCA2 mutations confer high risks of breast cancer, these relative risks result in substantial differences in the absolute risk of developing breast cancer between SNP genotype categories, and such differences could potentially influence the clinical management of mutation carriers (15). However, several other variants identified through population-based GWAS have not yet been evaluated as modifiers of cancer risk for BRCA1 and BRCA2 mutation carriers. Identifying further modifiers of risk could enhance risk prediction and will lead to a better understanding of the biology of tumour development in BRCA1 and BRCA2 mutation carriers.
Using data from the Shanghai Breast Cancer Study, Zheng et al. (7) identified a polymorphism at 6q25.1 through a GWAS on the risk of breast cancer among Chinese women. SNP rs2046210 was located upstream of the gene encoding for ER α-ESR1: 29 kb upstream of the first untranslated exon and 180 kb upstream of the first coding exon. Each copy of the minor allele of the SNP was estimated to confer an Odds Ratio (OR) of 1.29 among Chinese women and the authors reported a stronger association with ER-negative than ER-positive breast cancer. The same study also found an association between rs2046210 and the risk of breast cancer for European women, but a subsequent larger study among Europeans suggested that the association in Europeans is primarily due to another weakly correlated SNP in the region (rs9397435) (16). In a separate GWAS, Thomas et al. identified two further SNPs associated with the risk of the breast cancer in the Cancer and Genetic Markers of Susceptibility (CGEMS) study: rs11249433 at 1p11.2 in a linkage disequilibrium block neighbouring NOTCH2 and FCGR1B, and rs999737 at 14q24.1 in RAD51L1 (6). SNP rs11249433 was mainly associated with ER-positive disease.
To evaluate the associations between these four SNPs and breast cancer risk for BRCA1 and BRCA2 mutation carriers, we genotyped these SNPs in BRCA1 and BRCA2 mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA).
RESULTS
Characteristics of the eligible mutation carriers, after quality control exclusions, are summarised in Table 1. The primary analysis included only mutation carriers of self-reported white European ancestry, and included data from 11 604 women considered affected (first breast cancer diagnosis) and 10 572 considered as unaffected (censored at bilateral prophylactic mastectomy, ovarian cancer or age at last observation).
Table 1.
Summary characteristics for the 22 176 eligible BRCA1 and BRCA2 carriersa used in the analysis
| Characteristic |
BRCA1 |
BRCA2 |
||
|---|---|---|---|---|
| Unaffected | Breast cancer | Unaffected | Breast cancer | |
| Number | 6930 | 7193 | 3642 | 4411 |
| Person-years follow-up | 294 555 | 296 222 | 160 459 | 194 052 |
| Median age at censure (IQR) | 41 (34–50) | 40 (34–47) | 43 (34–52) | 43 (37–50) |
| Age at censure, N (%) | ||||
| <30 | 1016 (14.7) | 603 (8.4) | 510 (14.0) | 209 (4.7) |
| 30–39 | 1990 (28.7) | 2816 (39.1) | 978 (26.9) | 1366 (31.0) |
| 40–49 | 2075 (29.9) | 2528 (34.1) | 1005 (27.6) | 1687 (38.3) |
| 50–59 | 1218 (17.6) | 940 (13.1) | 647 (17.8) | 812 (18.4) |
| 60–69 | 448 (6.5) | 244 (3.4) | 344 (9.5) | 274 (6.2) |
| 70+ | 183 (2.6) | 62 (0.9) | 158 (4.3) | 63 (1.4) |
| Year of birth, N (%) | ||||
| <1920 | 31 (0.5) | 38 (0.5) | 24 (0.7) | 46 (1.0) |
| 1920–1929 | 143 (2.1) | 220 (3.1) | 111 (3.1) | 191 (4.3) |
| 1930–1939 | 419 (6.1) | 577 (8.0) | 255 (7.0) | 484 (11.0) |
| 1940–1949 | 935 (13.5) | 1522 (21.2) | 502 (13.8) | 1018 (23.1) |
| 1950–1959 | 1590 (22.9) | 2256 (31.4) | 760 (20.9) | 1361 (30.9) |
| 1960+ | 3812 (55.0) | 2580 (35.9) | 1990 (546.8) | 1311 (29.7) |
| Mutation class, N (%) | ||||
| Class 1b | 4581 (66.1) | 4363 (60.7) | 3426 (94.1) | 4092 (92.8) |
| Class 2b | 1964 (28.3) | 2252 (31.3) | 75 (2.0) | 115 (2.6) |
| Other | 385 (5.6) | 578 (8.0) | 141 (3.9) | 204 (4.6) |
IQR, interquartile range.
aCarriers of self-reported European ancestry only.
bSee methods for definitions.
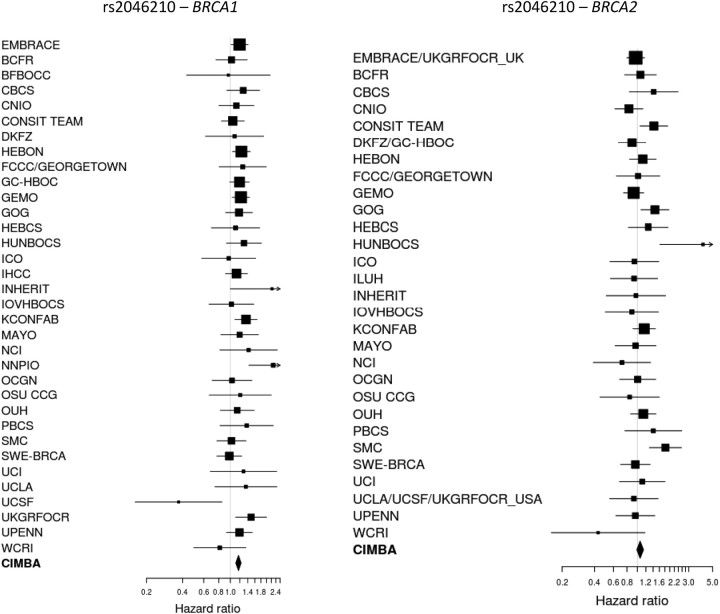
The association results with breast cancer risk are summarized in Table 2. The minor allele of SNP rs2046210, at 6q25.1, was associated with an increased risk of breast cancer for BRCA1 mutation carriers (per-allele HR = 1.17, 95% CI: 1.11–1.23, P-trend = 7.5 × 10−9). In contrast, there was little evidence of association with the risk of breast cancer for BRCA2 mutation carriers (per-allele HR = 1.06, 95% CI: 0.99–1.14, P-trend = 0.09). There was no evidence for heterogeneity in the HRs across studies for BRCA1 mutation carriers (P-heterogeneity = 0.47), but there was marginal evidence for heterogeneity for BRCA2 mutation carriers (P-heterogeneity = 0.03; Fig. 1). This was mainly due to data from the HUNBOCS study. After excluding this study from the analysis, there was no longer evidence for heterogeneity (P = 0.14) and the estimated HR for BRCA2 mutation carriers was virtually unchanged (per-allele HR = 1.05, 95%CI: 0.98–1.13, P-trend = 0.13). There was no evidence that the HR for BRCA2 mutation carriers varied by age (P = 0.87), but there was evidence that the per-allele HR for BRCA1 mutation carriers decreased with age (P-trend = 0.0036). To investigate this further, we fitted models allowing for a separate HR for each decade of age (Supplementary Material, Table S2). This analysis revealed significant associations between SNP rs2046210 and the risk of breast cancer for BRCA1 mutation carriers at ages <50 years (HR for the age group 20–29 = 1.24, 95% CI: 1.08–1.42; HR for the age group 30–39 = 1.24, 95% CI: 1.16–1.33; HR for the age group 40–49 = 1.11, 95% CI: 1.04–1.21) but not for ages 50 or over (HR for the age group 50–59 = 1.02, 95% CI: 0.90–1.15; HR for the age group 60–69 = 1.19; 95% CI: 0.96–1.47; HR for the age group 70–79 = 0.82, 95% CI: 0.50–1.36).
Table 2.
SNP genotype distributions and associations with breast cancer risk
| Mutation | Genotype | Unaffected, N (%) | Affecteda, N (%) | HR | 95% CI | P-value |
|---|---|---|---|---|---|---|
| 6q25.1 (rs2046210) | ||||||
| BRCA1 | CC | 2282 (43.0) | 2067 (37.5) | 1 | ||
| TC | 2361 (44.5) | 2669 (48.4) | 1.23 | 1.14–1.33 | ||
| TT | 659 (12.4) | 779 (14.1) | 1.32 | 1.18–1.47 | ||
| 2-df test | 7.5 × 10−9 | |||||
| Per-allele | 1.17 | 1.11–1.23 | 4.5 × 10−9 | |||
| BRCA2 | CC | 1144 (40.8) | 1321 (39.1) | 1 | ||
| TC | 1312 (46.7) | 1574 (46.5) | 1.02 | 0.92–1.13 | ||
| TT | 351 (12.5) | 486 (14.4) | 1.16 | 1.00–1.34 | ||
| 2-df test | 0.13 | |||||
| Per-allele | 1.06 | 0.99–1.14 | 0.09 | |||
| 6q25.1 (rs9397435) | ||||||
| BRCA1 | AA | 5361 (86.5) | 5282 (82.9) | 1.00 | ||
| AG | 802 (12.9) | 1043 (16.4) | 1.31 | 1.19–1.43 | ||
| GG | 38 (0.6) | 49 (0.8) | 1.37 | 0.92–2.06 | ||
| 2-df test | 5.3 × 10−8 | |||||
| Per-allele | 1.28 | 1.18–1.40 | 1.3 × 10−8 | |||
| BRCA2 | AA | 2786 (84.1) | 3141 (82.6) | 1.00 | ||
| AG | 510 (15.4) | 631 (16.6) | 1.11 | 0.98–1.26 | ||
| GG | 17 (0.5) | 32 (0.8) | 1.56 | 0.91–2.67 | 0.077 | |
| 2-df test | 0.077 | |||||
| Per-allele | 1.14 | 1.01–1.28 | 0.031 | |||
| 1p11.2 (rs11249433) | ||||||
| BRCA1 | TT | 1833 (34.4) | 1961 (35.1) | 1 | ||
| CT | 2584 (48.5) | 2732 (48.9) | 1.00 | 0.90–1.10 | ||
| CC | 911 (17.1) | 890 (15.9) | 0.92 | 0.83–1.03 | ||
| 2-df test | 0.21 | |||||
| Per-allele | 0.97 | 0.92–1.02 | 0.20 | |||
| BRCA2 | TT | 1016 (35.9) | 1135 (33.2) | 1 | ||
| CT | 1377 (48.7) | 1698 (49.6) | 1.07 | 0.96–1.19 | ||
| CC | 434 (15.4) | 590 (17.2) | 1.20 | 1.04–1.38 | ||
| 2-df test | 0.05 | |||||
| Per-allele | 1.09 | 1.02–1.17 | 0.015 | |||
| RAD51L1 (rs999737b/rs10483813c) | ||||||
| BRCA1 | CC/TT | 2725 (62.3) | 2849 (63.6) | 1 | ||
| TC/AT | 1461 (33.4) | 1439 (32.1) | 0.93 | 0.86–1.01 | ||
| TT/AA | 186 (4.3) | 195 (4.3) | 1.01 | 0.84–1.22 | ||
| 2-df test | 0.25 | |||||
| Per-allele | 0.96 | 0.90–1.03 | 0.27 | |||
| BRCA2 | CC/TT | 1609 (61.1) | 1950 (62.2) | 1 | ||
| TC/AT | 869 (33.0) | 1039 (33.1) | 0.98 | 0.88–1.09 | ||
| TT/AA | 154 (5.9) | 147 (4.7) | 0.86 | 0.69–1.08 | ||
| 2-df test | 0.43 | |||||
| Per-allele | 0.96 | 0.88–1.04 | 0.30 | |||
Analysis restricted to mutation carriers of European ancestry.
aBreast cancer.
bGenotyped using iPLEX.
cGenotyped using Taqman, pair-wise r2 = 1 between rs999737 and rs10483813 based on HapMap data.
Figure 1.
Study-specific per-allele HR estimates for BRCA1 and BRCA2 mutation carriers for SNP rs2046210 at 6q25.1 near ESR1. The area of the square is proportional to the inverse of the variance of the estimate. Horizontal lines indicate 95% CIs.
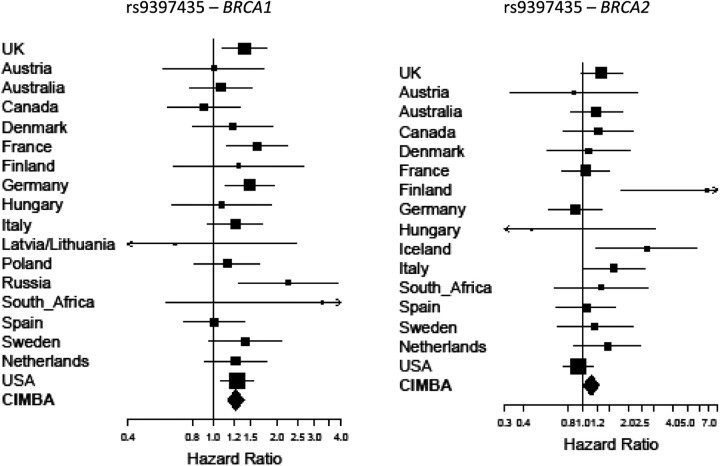
SNP rs9397435 was associated with the risk of breast cancer for both BRCA1 and BRCA2 mutation carriers, but the evidence of association was stronger for BRCA1 (per-allele HR = 1.28, 95% CI: 1.18–1.40, P-trend = 1.3 × 10−8) than for BRCA2 (HR = 1.14, 95% CI: 1.14, 95% CI: 1.01–1.28, P-trend = 0.031). There was some evidence of heterogeneity in the HRs across studies when considered individually (P-heterogeneity = 0.023). However, this was mainly due to studies with small numbers of mutation carriers and the low frequency of the minor allele of this SNP (minor allele frequency among unaffected = 6.7%). Repeating the analysis by grouping all studies within each country, there was no evidence of heterogeneity in the country-specific HRs (P-heterogeneity = 0.26; Fig. 2). Similarly, there was no evidence of heterogeneity in the HRs across countries for BRCA2 mutation carriers (P-heterogeneity = 0.12; Fig. 2). There was no evidence that the HR for BRCA1 mutation carriers varied by age (P-trend = 0.34), but there was evidence that the HR for BRCA2 mutation carriers decreased with age (P-trend = 0.0025). The estimated age-specific HRs for rs9397435 among BRCA2 mutation carriers were all >1 for ages <50, but there was no evidence of an increased risk for ages >50 years (Supplementary Material, Table S2).
Figure 2.
Country-specific per-allele HR estimates for BRCA1 and BRCA2 mutation carriers for SNP rs9397435 at 6q25.1. The area of the square is proportional to the inverse of the variance of the estimate. Horizontal lines indicate 95% CIs. Due to the low minor allele frequency at this SNP and small study sample we were unable to obtain study-specific estimates for all studies. Studies were therefore grouped by country of origin.
SNPs rs9397435 and rs2046210 are located in the same region at 6q25.1 and were only weakly correlated (pair-wise r2 = 0.14 based on the current data set). In an analysis for the joint effects of these SNPs on breast cancer risk for BRCA1 mutation carriers (based on 9347 carriers with genotypes at both SNPs), the most parsimonious model included the effects of both SNPs (P for inclusion = 1.4 × 10−5 and 0.0037 for rs2046210 and rs9397435, respectively; 2-degree of freedom (df) P = 5.8×10−10 for the inclusion of both SNPs compared with the null model).
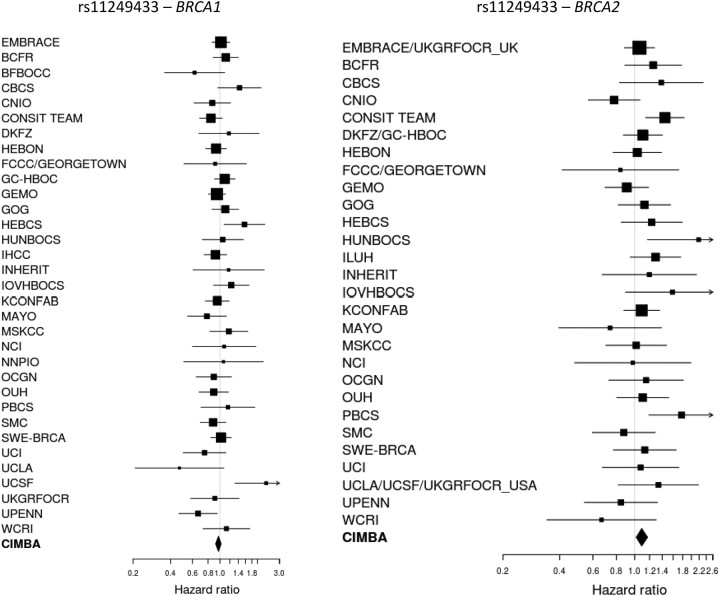
The minor allele of SNP rs11249433 at 1p11.2 was associated with the risk of breast cancer for BRCA2 mutation carriers (HR = 1.09, 95% CI: 1.02–1.17, P-trend = 0.015), but was not associated with the risk of breast cancer for BRCA1 mutation carriers (HR = 0.97, 95% CI: 0.92–1.02, P-trend = 0.20). There was no evidence that these HRs varied across the study groups for either BRCA1 or BRCA2 mutation carriers (p-heterogeneity = 0.10 and 0.14, respectively, Fig. 3) or that the HRs varied by age (P = 0.41 for BRCA1; P = 0.93 for BRCA2).
Figure 3.
Study-specific per-allele HR estimates for BRCA1 and BRCA2 mutation carriers for SNP rs11249433 at 1p11.2. The area of the square is proportional to the inverse of the variance of the estimate. Horizontal lines indicate 95% CIs.
Carriers in each study were genotyped for either rs999737 or rs10483813 in the RAD51L1 region, but none were genotyped for both SNPs. Since rs999737 and rs10483813 are perfectly correlated (pair-wise r2 = 1), the genotypes across studies were combined and analysed as a single locus. There were no significant associations between this SNP and the risk of breast cancer for either BRCA1 or BRCA2 mutation carriers (BRCA1: per-allele HR = 0.96, 95% CI: 0.90–1.03, P-trend = 0.27; BRCA2: per-allele HR = 0.96, 95% CI: 0.88–1.04, P-trend = 0.30). The HR estimates were consistent across the studies for both BRCA1 (P-heterogeneity = 0.11) and BRCA2 mutation carriers (P-heterogeneity = 0.42). There was no evidence that the HRs varied by age for BRCA1 (P = 0.50) or BRCA2 (P = 0.60) mutation carriers.
The associations were not altered after excluding long-term survivors (Supplementary Material, Table S3) and there was no evidence of differences in the associations between class 1 and class 2 BRCA1 mutation carriers (P for difference > 0.15 for all SNPs, Supplementary Material, Table S3).
BRCA1 and BRCA2 mutations also confer high risks of ovarian cancer. To determine whether the three polymorphisms modify ovarian cancer risk in mutation carriers, we analysed the associations within a competing risk analysis framework by estimating simultaneously the HRs for breast and ovarian cancer. There was no evidence of association with the risk of ovarian cancer for any of the SNPs (Table 3). The estimated HRs for breast cancer were similar to those from the primary analysis. SNPs rs2046210 and rs9397435 remained significantly associated with the risk of breast cancer for BRCA1 mutation carriers (P-trend = 6.7 × 10−8 and 7.8 × 10−7) and there was a slightly stronger evidence of association between SNP rs11249433 and the risk of breast cancer for BRCA2 mutation carriers (P-trend = 0.0052).
Table 3.
Competing risk analysis
| Genotype | Unaffected, N (%) | Breast cancer, N (%) | Ovarian cancer, N (%) | Breast cancer |
Ovarian cance r |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |||||
| 6q25.1 (rs2046210) | ||||||||||
| BRCA1 | CC | 1658 (42.5) | 2046 (37.4) | 645 (44.7) | 1 | 1 | ||||
| TC | 1740 (44.6) | 2648 (48.4) | 642 (44.5) | 1.23 | 1.13–1.33 | 0.98 | 0.86–1.12 | |||
| TT | 505 (12.9) | 777 (14.2) | 156 (10.8) | 1.30 | 1.16–1.47 | 0.89 | 0.72–1.10 | |||
| 2-df test | 9.8 × 10−8 | 0.54 | ||||||||
| Per-allele | 1.16 | 1.10–1.23 | 6.7 × 10−8 | 0.95 | 0.87–1.05 | 0.31 | ||||
| BRCA2 | CC | 988 (40.8) | 1317 (39.1) | 160 (40.4) | 1 | 1 | ||||
| TC | 1130 (46.6) | 1567 (46.5) | 189 (47.7) | 1.01 | 0.91–1.13 | 0.99 | 0.78–1.27 | |||
| TT | 305 (12.6) | 485 (14.4) | 47 (11.9) | 1.14 | 0.98–1.33 | 0.88 | 0.61–1.26 | |||
| 2-df test | 0.22 | 0.76 | ||||||||
| Per-allele | 1.05 | 0.98–1.13 | 0.15 | 0.95 | 0.81–1.12 | 0.57 | ||||
| 6q25.1 (rs9397435) | ||||||||||
| BRCA1 | AA | 4116 (86.1) | 5245 (82.9) | 1282 (87.5) | 1.00 | 1.00 | ||||
| AG | 633 (13.2) | 1034 (16.3) | 178 (12.1) | 1.28 | 1.16–1.41 | 0.90 | 0.75–1.08 | |||
| GG | 33 (0.7) | 49 (0.8) | 5 (0.3) | 1.25 | 0.81–1.94 | 0.47 | 0.18–1.18 | |||
| 2-df test | 2.8 × 10−6 | 0.15 | ||||||||
| Per-allele | 1.25 | 1.14–1.37 | 7.8 × 10−7 | 0.87 | 0.73–1.03 | 0.10 | ||||
| BRCA2 | AA | 2410 (83.9) | 3131 (82.5) | 386 (85.4) | 1.00 | 1.00 | ||||
| AG | 466 (15.5) | 631 (16.6) | 64 (14.2) | 1.11 | 0.97–1.26 | 0.93 | 0.69–1.26 | |||
| GG | 15 (0.5) | 32 (0.8) | 2 (0.4) | 1.50 | 0.85–2.62 | 0.65 | 0.16–2.68 | |||
| 2-df test | 0.12 | 0.75 | ||||||||
| Per-allele | 1.13 | 1.00–1.27 | 0.047 | 0.92 | 0.70–1.20 | 0.52 | ||||
| 1p11.2 (rs11249433) | ||||||||||
| BRCA1 | TT | 1363 (34.8) | 1945 (35.1) | 486 (33.5) | 1 | 1 | ||||
| CT | 1863 (47.5) | 2714 (49.0) | 739 (50.9) | 1.02 | 0.94–1.11 | 1.12 | 0.97–1.29 | |||
| CC | 695 (17.7) | 880 (15.9) | 226 (15.6) | 0.91 | 0.81–1.01 | 0.91 | 0.75–1.11 | |||
| 2-df test | 0.08 | 0.05 | ||||||||
| Per-allele | 0.96 | 0.91–1.02 | 0.18 | 0.98 | 0.90–1.08 | 0.74 | ||||
| BRCA2 | TT | 889 (36.4) | 1129 (33.1) | 133 (33.7) | 1 | 1 | ||||
| CT | 1182 (48.4) | 1695 (49.7) | 198 (50.1) | 1.10 | 0.98–1.22 | 1.12 | 0.88–1.44 | |||
| CC | 372 (15.2) | 588 (17.2) | 64 (16.2) | 1.23 | 1.06–1.42 | 1.23 | 0.88–1.72 | |||
| 2-df test | 0.02 | 0.44 | ||||||||
| Per-allele | 1.11 | 1.03–1.19 | 0.0052 | 1.11 | 0.94–1.31 | 0.20 | ||||
| RAD51L1 (rs999737/rs10483813) | ||||||||||
| BRCA1 | CC/TT | 2014 (61.9) | 2828 (63.6) | 732 (63.4) | 1 | 1 | ||||
| TC/AT | 1100 (33.8) | 1426 (32.1) | 374 (32.4) | 0.92 | 0.84–1.01 | 0.95 | 0.81–1.10 | |||
| TT/AA | 138 (4.2) | 194 (4.4) | 49 (4.2) | 1.02 | 0.83–1.24 | 1.00 | 0.69–1.45 | |||
| 2-df test | 0.17 | 0.76 | ||||||||
| Per-allele | 0.96 | 0.89–1.03 | 0.21 | 0.97 | 0.85–1.10 | 0.60 | ||||
| BRCA2 | CC/TT | 1384 (61.1) | 1942 (62.1) | 233 (62.0) | 1 | 1 | ||||
| TC/AT | 753 (33.2) | 1036 (33.2) | 119 (31.6) | 0.98 | 0.87–1.09 | 0.95 | 0.74–1.21 | |||
| TT/AA | 130 (5.7) | 147 (4.7) | 24 (6.4) | 0.88 | 0.70–1.11 | 1.16 | 0.71–1.90 | |||
| 2-df test | 0.56 | 0.72 | ||||||||
| Per-allele | 0.96 | 0.88–1.05 | 0.35 | 1.01 | 0.83–1.23 | 0.92 | ||||
Associations with breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers. Analysis restricted to mutation carriers of European ancestry.
DISCUSSION
Several common variants identified through GWASs in the general population and BRCA1 and BRCA2 mutation carriers have been demonstrated to be associated with the risk of breast cancer for BRCA1 and/or BRCA2 mutation carriers (8–10,12,13). In this study, we evaluated the associations of four additional variants, identified through population-based GWAS or subsequent follow-up mapping studies, with the risk of breast and ovarian cancer for BRCA1 and BRCA2 mutation carriers.
We found strong evidence that SNP rs2046210 at 6q25.1 was associated with the risk of breast cancer for BRCA1 mutation carriers, but there was no clear evidence of association with the risk of breast cancer for BRCA2 mutation carriers (P for difference: 0.027). The observed association with BRCA1 breast cancer risk was unaltered after the exclusion of prevalent breast cancer cases, and did not vary by the predicted functional effect of the mutations. This polymorphism was identified through a GWAS in Chinese women (7), in whom the authors estimated a per-allele OR of 1.29 (95% CI: 1.21–1.37) for breast cancer in this population. This OR estimate was greater than our estimated HR for BRCA1 mutation carriers. However, the Chinese study reported a further replication of their findings among women of European ancestry for whom the per-allele OR was estimated to be 1.15, similar to the HR based on our analysis of BRCA1 mutation carriers who were also of self-reported European ancestry. A more recent study by the same group also found evidence for association with breast cancer in an independent sample of European-ancestry American cases and controls (17). Both studies (7,17) also reported a stronger association with ER-negative disease than ER-positive in particular among Asian women, although the SNP was associated with both disease subtypes. This is consistent with our finding that this SNP is predominantly associated with the risk of breast cancer for BRCA1 mutation carriers, the majority of whom present with ER-negative tumours (18), and hence conforms to the general pattern we have observed previously that the breast cancer susceptibility SNPs confer a similar relative risk in carriers to that in the general population, once receptor status is taken into account (10,11). A more recent study by Stacey et al. (16) evaluated the associations of SNP rs2046210 in a larger set of women of European ancestry, but failed to replicate the association. The authors concluded that this SNP does not confer a substantial risk of breast cancer in Europeans and they postulated that this is due to the different linkage disequilibrium structures between the causal variant and SNP rs2046210 in Europeans. They found a different SNP, rs9397435 (pair-wise r2 = 0.08 based on CEU HapMap data, r2 = 0.71 based on CHB + JPT HapMap data), accounting for the association in both the Europeans and Chinese. SNP rs9397435 was also strongly associated with the risk of breast cancer for BRCA1 mutation carriers in our data set and exhibited weak association with the risk for BRCA2 mutation carriers (P for difference between BRCA1 and BRCA2 = 0.10). Our joint analysis of SNPs rs9397435 and rs2046210 among BRCA1 mutation carriers demonstrated that a model that includes both SNPs fits significantly better than a model that includes either SNP on its own, and are not therefore consistent with the conclusions of Stacey et al. (16) who suggest that the association is primarily due to SNP rs9397435. Our results suggest that either the observed associations are driven by another causative variant that is partially associated with both SNPs, or that more than one causative variant is located in this region. Further comprehensive genotyping of variants across the region will be required to determine which of these hypotheses is correct. A potential explanation for the observed differences between our study and that of Stacey et al. (16) could be the fact the BRCA1 tumours are predominantly ER-negative, whereas the majority of cases in Stacey et al. were ER-positive. This result could have been observed if rs2046210 was mainly associated with ER-negative breast cancer and rs9397435 was associated with both ER-negative and ER-positive breast cancer. Stacey et al. (16) did not present the associations of rs2046210 by tumour ER status, but reported that rs9397435 was associated with both ER-positive and ER-negative disease, with a higher per-allele OR for ER-negative breast cancer consistent with our observation of a larger HR estimate for BRCA1 mutation carriers than BRCA2 carriers for rs9397435. Although we found no significant evidence of association between rs2046210 and the risk of breast cancer for BRCA2 mutation carriers, the estimated association was in the same direction.
Our results also suggest that rs2046210 is associated with higher relative risks of breast cancer at younger ages among BRCA1 mutation carriers. A similar pattern was observed for rs9397435 among BRCA2 mutation carriers. Stacey et al. (16) also found that rs9397435 was associated with an earlier age at diagnosis in Europeans from the general population.
SNPs rs2046210 and rs9397435 are located close to ESR1, which encodes ER α mediator of oestrogen action (19). Elevated oestrogen levels have been associated with increased breast cancer risk (20), and although it is assumed that the action of oestrogen is via ER in ER-positive tumours, two studies have recently provided evidence that the size and repopulating ability of the mammary stem cell compartment in mice are controlled by17β-estradiol and progesterone via a paracrine-signalling mechanism from steroid receptor-positive luminal cells to steroid receptor-negative stem cells (21,22). This may explain the apparently paradoxical observation that a SNP in ESR1 could modify the risk of breast cancer in BRCA1 carriers, in which the tumour phenotype is usually ER-negative. The cell of origin of basal ER-negative tumours in BRCA1 mutation carriers is likely to be a luminal progenitor cell that is dependent on steroid hormone signalling (23). There is also indirect evidence that steroid hormones regulate breast cancer stem cells in humans where the same paracrine regulation probably occurs, perhaps mediated via the Receptor Activator of NF-κB) (RANK) ligand (24). Other studies have also provided evidence that oophorectomy decreases the risk of breast cancer in BRCA1 mutation carriers (26,27) and tamoxifen treatment may decrease the risk of contralateral breast cancer for BRCA1 mutation carriers. Both of these findings suggest the potential ER involvement in BRCA1 associated disease (28).
There are currently limited data in the literature on the impact of these variants on expression levels of ESR1 in breast tumour samples. Stacey et al. found some evidence that breast tumours from 11 GG homozygote carriers of rs9397435 expressed higher mean levels of ESR1 compared with tumours from over one thousand carriers of the ‘A' allele (16). However, Dunbier et al. (29) recently reported that ESR1 is co-expressed in tumour biopsies along with three uncharacterized open reading frames located upstream of ESR1. It is therefore currently uncertain whether rs9397435 or correlated causal variant(s) affect breast cancer risk through modulating ESR1 expression levels or those of additional genes in the region. If the ESR1 gene is found to be the target, this would provide direct evidence that ER signalling is important in the development of ER-negative breast cancer (and breast cancer in BRCA1 carriers in particular).
We also found evidence that SNP rs11249433 at 1p11.2 was associated with the risk of breast cancer for BRCA2, but not BRCA1 mutation carriers (P for difference: 0.007). The estimated HR was slightly greater (1.15 versus 1.09) when BRCA2 mutation carriers with prevalent breast cancer were excluded from the analysis, although the difference in the estimates was small. This could potentially arise if the SNP is also associated with survival after breast cancer diagnosis. In this case, the inclusion of prevalent cases could lead to an attenuation of the HR. Future studies will aim to evaluate the associations of this SNP with breast cancer prognosis. The observed association with the risk for BRCA2 mutation carriers is consistent with the observation of Thomas et al. (6) that this SNP is mainly associated with the risk for ER-positive breast cancer.
We found no significant evidence of association between the RAD51L1 locus and the risk of breast cancer for BRCA1 or BRCA2 mutation carriers. However, the OR estimate from the original breast cancer GWAS (0.94) is only slightly different from the HR estimates for both BRCA1 and BRCA2 carriers (0.96) and is included in the CIs for both estimates (5). If the relative risk associated with each copy of the minor allele of this SNP is between 0.90 and 1.00, we have limited power to detect these associations given our sample size (30). RAD51L1 is known to be essential to DNA repair via homologous recombination; therefore, if the breast cancer association seen in the general population was mediated through RAD51L1, an absence of an association in BRCA1 and BRCA2 mutation carriers (i.e. a ‘negative interaction' with BRCA status) could also be plausible. It is interesting to note, however, that a rare allele in the RAD51 gene, in the same pathway, was previously associated with an increased risk of breast cancer for BRCA2 mutation carriers (25). Future studies with a larger number of mutation carriers, and analysis of the causal variant once it has been identified, may help to clarify the involvement of this locus in breast cancer for mutation carriers.
Including the SNPs from the present study, five loci are now known to modify the risk of breast cancer for BRCA1 mutation carriers (CASP8, TOX3, 2q35, 19p13 and 6q25.1) (8–10,12,14) and nine loci are known to modify the risk of breast cancer for BRCA2 mutation carriers (FGFR2, TOX3, MAP3K1, LSP1, 2q35, SLC4A7, 5p12, ZNF365 and 1p11.2) (8–10,13). These loci are estimated to account for ∼3.0% of the genetic variance in the risk of breast cancer in BRCA1 mutation carriers and 5.6% of the variance in BRCA2 mutation carriers. Although these variants account for a small proportion of the variability in risk, it has been demonstrated that these SNPs have implications for the absolute risk prediction in mutation carriers (10), and could therefore be relevant in the genetic counselling of women carrying mutations (15). There are also suggestions from candidate gene studies that other variants may modify cancer risks for mutation carriers, which are currently being investigated in larger sample sizes (31,32). The three associated polymorphisms presented here, in conjunction with previously identified risk-modifying polymorphisms and other risk- modifying factors, can be used to improve risk prediction in BRCA1 and BRCA2 mutation carriers.
Data from the general population indicate that chemopreventive agents have different effects on the risk of ER-positive and ER-negative breast cancer (33). Ongoing and future CIMBA studies will aim to clarify the involvement of these polymorphisms in ER-positive and ER-negative breast cancer risk, as well as other tumour subtypes, in BRCA1 and BRCA2 mutation carriers, which should lead to further improvements in risk prediction. Since BRCA1 and BRCA2 mutations confer high risks of breast cancer, these SNPs, taken together with other risk factors such as mammographic breast density (34), will result in substantial differences in the absolute risk of developing breast cancer between combined SNP and risk factor categories (10,35). These will enable preventive therapies, including chemoprevention and prophylactic surgery, to be targeted at mutation carriers most likely to benefit.
MATERIALS AND METHODS
Subjects
All carriers participated in clinical or research studies at the host institutions under ethically approved protocols and data were analysed anonymously. Subjects were BRCA1 and BRCA2 mutation carriers recruited by 42 study centres in 22 countries through the CIMBA initiative (Supplementary Material, Table S1). The large majority of carriers were recruited through cancer genetics clinics offering genetic testing, and enrolled into national or regional studies. Some carriers were identified by population-based sampling of cases, and some by community recruitment (e.g. in Ashkenazi Jewish populations). Eligibility to participate in CIMBA is restricted to female carriers of pathogenic BRCA1 or BRCA2 mutations who were 18 years old or older at recruitment. Information collected included the year of birth; mutation description, including nucleotide position and base change; age at the last follow-up; ages at breast and ovarian cancer diagnoses; and age or date at bilateral prophylactic mastectomy. Information was also available on the country of residence, which was defined to be the country where the carrier family was recruited to the study. Related individuals were identified through a unique family identifier. Women were included in the analysis if they carried mutations that were pathogenic according to generally recognized criteria (25) (Breast Cancer Information Core). Further details of the CIMBA initiative can be found elsewhere (36).
Women who carried pathogenic mutations in both BRCA1 and BRCA2 were excluded from the current analysis. The primary analysis was restricted to women self-reported as ‘white of European ancestry', but additional analyses were performed which were restricted to mutation carriers of non-European ancestry. We investigated possible overlap of carriers between studies by comparing the year of birth, exact mutation description, and the reported ages, to identify potential duplicate individuals. Where possible we also used SNP genotype data available within the CIMBA database to find hidden duplicates. When a potential duplicate was identified, we contacted the relevant centres for further information about these individuals, in a manner that protected the identity of the individuals in question, in order to determine precisely the extent of true overlap in subjects and families appearing more than once in the data set. Duplicated mutation carriers were included only once in the analysis. To avoid inclusion of families extending over several studies, we included only the individual with the most complete version of the family history in the study.
Genotyping
The genotyping platforms used by each study are shown in Supplementary Material, Table S1. Genotyping for the four SNPs was performed in two stages. Stage 1 involved SNPs rs2046210, rs11249433 and the RAD51L1 SNPs rs999737 and rs10483813. DNA samples from 11 studies were genotyped using the iPLEX Mass Array platform at a single genotyping centre. All remaining studies used the 5′ endonuclease assay (Taqman), with reagents supplied by Applied Biosystems and tested centrally. A Taqman assay could not be adequately designed for SNP rs999737 and studies using this platform genotyped the surrogate SNP rs10483813 (pair-wise r2 = 1 with rs999737 based on HapMap data). Stage 2 involved SNP rs9397435 and all samples were genotyped using the iPLEX Mass Array platform at four genotyping centres. All centres included at least 2% of the samples in duplicate, no template controls in every plate and a random mixture of affected and unaffected carriers. Samples that failed for more than two of the SNPs genotyped (or ≥20% of the SNPs typed if more than three SNPs were analysed using multiplex genotyping) were excluded from the analysis. A study was included in the analysis only if the call rate was over 95% after samples that failed at multiple SNPs had been excluded. The concordance between duplicates had to be at least 98%. To assess the accuracy of genotyping across genotyping centres, all centres genotyped 95 DNA samples from a standard test plate (Coriell Institute) for all three SNPs. If the genotyping was inconsistent for more than one sample in the test plate, the study was excluded from the analysis of that SNP. On the basis of these criteria, two studies were excluded from the analysis of rs2046210, eight studies were excluded from the analysis of rs999737/rs10483813 and three studies were excluded from the analysis of rs11249433. As an additional genotyping quality-control check, we also evaluated the deviation from Hardy–Weinberg equilibrium (HWE) for unrelated subjects separately for each SNP and study. Seven studies had HWE P-values in the range 0.003–0.05 (one study for the rs2046210 SNP, two for rs9397435 and four studies for rs11249433). Upon examination of the cluster plots for these studies and SNPs, none revealed any unusual patterns and these studies were included in all the analyses. After the above exclusions, a total of 22 176 unique mutation carriers (14 123 BRCA1 and 8053 BRCA2) from 42 studies had an observed genotype for at least one of the SNPs and were therefore included in the primary analysis (Supplementary Material, Table S1).
Statistical analysis
The aim of the primary analysis was to evaluate the association between each genotype and the risk of breast cancer. The phenotype of each individual was therefore defined by their age at diagnosis of breast cancer or their age at the last follow-up. For this purpose, individuals were censored at the age of the first breast cancer diagnosis, ovarian cancer diagnosis or bilateral prophylactic mastectomy or the age at the last observation. Mutation carriers censored at ovarian cancer diagnosis were considered unaffected. Since mutation carriers were not sampled randomly with respect to their disease status, standard methods of survival analysis (such as Cox regression) may lead to biased estimates of the hazard ratios (HRs) (37). We therefore conducted the analysis by modelling the retrospective likelihood of the observed genotypes conditional on the disease phenotypes as previously described (25). The effect of each SNP was modelled either as a per-allele HR (multiplicative model) or as separate HRs for heterozygotes and homozygotes, and these were estimated on the logarithmic scale. The HRs were assumed to be independent of age (i.e. we used a Cox proportional-hazards model). The assumption of proportional hazards was tested by adding a ‘genotype × age' interaction term to the model in order to fit models in which the HR changed with age. Where there was significant evidence of a ‘genotype × age' interaction, we fitted models that allowed for age-specific HRs. These allowed for age-specific HRs to be estimated simultaneously in 10-year intervals (20–29, 30–39, … , 70–79). Thus, these models included six log-HR parameters. We examined between-study heterogeneity by comparing the models that allowed for study-specific log-HRs against models in which the same log-HR was assumed to apply to all studies. Analyses were carried out with the pedigree analysis software MENDEL (38), and details of this approach have been described previously (25). Under the retrospective likelihood approach, the baseline age-specific incidence rates in the Cox proportional-hazards model were chosen such that the overall breast cancer incidence rates, averaged over all genotypic categories, agree with external estimates of incidence for BRCA1 and BRCA2 mutation carriers. All analyses were stratified by study group and country of residence and used calendar-year- and cohort-specific breast cancer incidence rates for BRCA1 and BRCA2 (39).
To evaluate the combined effects of the ESR1 SNPs on the risk of breast cancer, we fit retrospective likelihood models where the breast cancer incidence 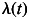 was assumed to be of the form
was assumed to be of the form 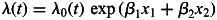 , where
, where 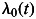 is the baseline incidence, β1 is the per-allele log-HR for SNP1, β2 is the per-allele log-HR for SNP2, and x1 and x2 represent the number of minor alleles at SNP 1 and 2, respectively (0,1,2), while allowing for linkage disequilibrium between the loci. To test whether the fit of the model is significantly improved by the inclusion of a locus into the model, we tested for the significance of parameters β1 and β2.
is the baseline incidence, β1 is the per-allele log-HR for SNP1, β2 is the per-allele log-HR for SNP2, and x1 and x2 represent the number of minor alleles at SNP 1 and 2, respectively (0,1,2), while allowing for linkage disequilibrium between the loci. To test whether the fit of the model is significantly improved by the inclusion of a locus into the model, we tested for the significance of parameters β1 and β2.
To investigate whether our results were influenced by any of our assumptions, we performed additional sensitivity analyses. If any of the SNPs were associated with disease survival, the inclusion of prevalent cases may influence the HR estimates. Current data indicate that 5-year survival after a breast cancer diagnosis is now over 80% (Cancer Research—UK, Breast cancer survival statistics). We therefore repeated our analysis by excluding mutation carriers diagnosed more than 5 years prior to the age at recruitment into the study. To examine whether SNP associations differed by type of mutations, we classified BRCA1 mutations according to their potential functional effect (40–42). Class 1 mutations comprised loss-of-function mutations, expected to result in a reduced transcript or protein level due to mRNA nonsense-mediated decay and/or degradation or instability of truncated proteins, translation re-initiation but no production of stable protein, or the absence of expression because of the deletion of transcription regulatory regions. Class 2 mutations were those likely to generate potentially stable mutant proteins that might have dominant negative action, partially preserved normal function or loss of function. Class 2 mutations include missense substitutions, in-frame deletions and insertions, as well as truncating mutations with premature stop codons occurring in the last exon. Mutations whose consequences at the transcript or protein level could not be inferred were not considered for this classification. These were mainly mutations located in splice sites but not characterized for their effect at the transcript level, or large deletions or insertions with undetermined boundaries.
We further evaluated the associations of these SNPs with the risk of ovarian cancer within a competing risk analysis framework (12,43), by estimating HRs simultaneously for breast and ovarian cancers. In this model, each individual was at risk of developing either breast or ovarian cancer, by assuming that the probabilities of developing each disease were independent conditional on the underlying genotype. A different censoring process was used in this case, whereby individuals were followed up to the age of the first breast or ovarian cancer diagnosis and were considered to have developed the corresponding disease. No follow-up was considered after the first cancer diagnosis. Individuals were censored for breast cancer at the age of bilateral prophylactic mastectomy and for ovarian cancer at the age of bilateral oophorectomy and were assumed to be unaffected for the corresponding disease. The remaining individuals were censored at the age at the last observation and were assumed to be unaffected for both diseases.
All analyses were stratified by study group and country of residence and used calendar-year- and cohort-specific cancer incidences for BRCA1 and BRCA2 (39). For sensitivity analyses, strata with a small number of mutation carriers were grouped. We used a robust variance-estimation approach to allow for the non-independence among related carriers (44). Data on the two completely correlated SNPs (rs999737 and rs10483813) were combined and treated as a single locus in the analysis of associations.
WEB RESOURCES
Breast Cancer Information Core: http://research.nhgri.nih.gov/bic/.
Cancer Research—UK, Breast cancer—survival statistics: http://info.cancerresearchuk.org/cancerstats/types/breast/survival/.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Cancer Research UK grants C12292/A11174 and C1287/A10118. The research leading to these results has received funding from the European Community's Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F22009-223175).
ACKNOWLEDGEMENTS
A.C.A. is a CR-UK Senior Cancer Research Fellow, D.F.E. is CR-UK Principal Research Fellow and G.C.T. is a NHMRC Senior Principal Research Fellow.
Study-specific acknowledgments:
The Baltic Familial Breast and Ovarian Cancer Consortium (BFBOCC)
Dr Laima Tihomirova, the Genome Database of the Latvian Population, Latvian Biomedical Research and Study Centre provided data and DNA samples for BFBOCC. This work is supported by the Research Council of Lithuania grant LIG-19/2010 to Ramunas Janavicius.
BRCA-gene mutations and breast cancer in South African women (BMBSA)
The study is supported by grants from the Cancer Association of South Africa to Elizabeth J. van Rensburg.
Breast Cancer Family Registry (BCFR)
This work was supported by the National Cancer Institute, National Institutes of Health under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Columbia University (U01 CA69398), Fox Chase Cancer Center (U01 CA69631), Huntsman Cancer Institute (U01 CA69446), Cancer Prevention Institute of California (formerly the Northern California Cancer Center) (U01 CA69417), University of Melbourne (U01 CA69638), and Research Triangle Institute Informatics Support Center (RFP No. N02PC45022-46). Samples from the FCCC, HCI and CPIC were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR.
Copenhagen Breast Cancer Study (CBCS)
We thank Susanne Kjaergaard and Mette Klarskov for clinical data. The study was supported by the NEYE Foundation.
CONSIT TEAM
CONSIT TEAM is supported by grants from Ministero della Salute (Extraordinary National Cancer Program 2006 ‘Alleanza contro il Cancro' to LV and PR and ‘Progetto Tumori Femminili' to PR), Ministero dell'Universita’ e Ricerca (RBLAO3-BETH to PR), Fondazione Italiana per la Ricerca sul Cancro (Special Project ‘Hereditary tumors' to PR), Associazione Italiana per la Ricerca sul Cancro (4017) to PP and by funds from Italian citizens who allocated the 5 ×1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5×1000') to P.P.
CONSIT TEAM thanks Marco A. Pierotti, Carla B. Ripamonti and Marilena Morganti of the Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy; Bernardo Bonanni of the Istituto Europeo di Oncologia, Milan, Italy; Alessandra Viel of the Centro di Riferimento Oncologico, Aviano, Italy and Antonella Savarese of the Istituto Regina Elena, Rome, Italy.
Epidemiological study of BRCA1 & BRCA2 mutation carriers (EMBRACE)
Douglas Easton is the PI of the study. EMBRACE Collaborating Centers are: Coordinating Centre, Cambridge: Susan Peock, Margaret Cook, Debra Frost, Radka Platte, Jean Leyland. North of Scotland Regional Genetics Service, Aberdeen: Zosia Miedzybrodzka, Helen Gregory. Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison, Lisa Jeffers. West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Carole McKeown, Kai-ren Ong, Jonathan Hoffman. South West Regional Genetics Service, Bristol: Alan Donaldson. East Anglian Regional Genetics Service, Cambridge: Joan Paterson, Sarah Downing, Amy Taylor. Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark T. Rogers, Emma McCann. St James's Hospital, Dublin & National Centre for Medical Genetics, Dublin: M. John Kennedy, David Barton. South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous, Sarah Drummond. Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman, Kathryn Hill. West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson, Eshika Haque, Ed Tobias, Alexis Duncan. South East Thames Regional Genetics Service, Guy's Hospital London: Louise Izatt, Chris Jacobs, Caroline Langman, Anna Whaite. North West Thames Regional Genetics Service, Harrow: Huw Dorkins. Leicestershire Clinical Genetics Service, Leicester: Julian Barwell. Yorkshire Regional Genetics Service, Leeds: Julian Adlard, Carol Chu, Julie Miller. Merseyside & Cheshire Clinical Genetics Service, Liverpool: Ian Ellis, Catherine Houghton. Manchester Regional Genetics Service, Manchester: D Gareth Evans, Fiona Lalloo, Jane Taylor. North East Thames Regional Genetics Service, NE Thames, London: Lucy Side, Alison Male, Cheryl Berlin. Nottingham Centre for Medical Genetics, Nottingham: Jacqueline Eason, Rebecca Collier. Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber, Irene Jobson. Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durell, Barbara Stayner. The Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Lucia D'Mello, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins, Elena Castro, Anita Mitra, Lisa Robertson. North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley. South West Thames Regional Genetics Service, London: Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester, Charlotte Eddy, Vishakha Tripathi, Virginia Attard. Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Donna McBride, Sarah Smalley. EMBRACE is supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles, Elizabeth Bancroft and Lucia D'Mello are also supported by Cancer Research UK Grant C5047/A8385.
Deutsches Krebsforschungszentrum (DKFZ) study
The DKFZ study was supported by the DKFZ.
The German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC)
GC-HBOC is supported by a grant of the German Cancer Aid (grant 107054). We thank Christian Sutter (Center Heidelberg) Sabine Preisler-Adams (Center Münster), Britta Fiebig (Center Regensburg), Wolfram Heinritz (Center Leipzig) and Dieter Schäfer (Center Frankfurt) for providing samples and Juliane Köhler for her excellent technical assistance.
Genetic Modifiers of cancer risk in BRCA1/2 mutation carriers (GEMO) study: Cancer Genetics Network ‘Groupe Génétique et Cancer', Fédération Nationale des Centres de Lutte Contre le Cancer, France
The study is supported by the Ligue National Contre le Cancer; Association for International Cancer Research Grant (AICR-07-0454); and the Association ‘Le cancer du sein, parlons-en!' Award. We wish to thank all the GEMO collaborating groups for their contribution to this study. GEMO Collaborating Centers are: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Centre Hospitalier Universitaire de Lyon/Centre Léon Bérard, & UMR5201 CNRS, Université de Lyon, Lyon: Olga Sinilnikova, Laure Barjhoux, Carole Verny-Pierre, Sophie Giraud, Mélanie Léone, Sylvie Mazoyer; and Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Carole Tirapo, Antoine de Pauw. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Audrey Remenieras, Véronique Byrde, Olivier Caron, Gilbert Lenoir. Centre Jean Perrin, Clermont-Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, François Eisinger. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier. CHU de Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis. Hôpital René Huguenin/Institut Curie, St Cloud: Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Michel Longy, Nicolas Sevenet. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU de Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung. CHU de Dijon: Fanny Coron, Laurence Faivre. CHU de St-Etienne: Fabienne Prieur, Marine Lebrun. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU de Limoges: Laurence Vénat-Bouvet. CHU de Nantes: Capucine Delnatte. CHU Bretonneau, Tours: Isabelle Mortemousque. Creighton University, Omaha, USA: Henry T.Lynch, Carrie L.Snyder.
Gynecologic Oncology Group (GOG)
GOG's participation was supported through funding provided by both intramural (Clinical Genetics Branch, DCEG) and extramural (Community Oncology and Prevention Trials Program—COPTRG) NCI programs.
Hospital Clinico San Carlos
This work was supported by the ISCIII grant RD06/0020/0021.
Helsinki Breast Cancer Study (HEBCS)
The HEBCS study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society and the Sigrid Juselius Foundation. We thank Tuomas Heikkinen and RN Irja Erkkilä for their help with the patient data and study samples.
The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON)
HEBON Collaborating Centers: Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: F.B.L. Hogervorst, S. Verhoef, M. Verheus, L.J. van ‘t Veer, F.E. van Leeuwen, M.A. Rookus; Erasmus Medical Center, Rotterdam, NL: M. Collée, A.M.W. van den Ouweland, A. Jager, M.J. Hooning, M.M.A. Tilanus-Linthorst, C. Seynaeve; Leiden University Medical Center, NL, Leiden: C.J. van Asperen, J.T. Wijnen, M.P. Vreeswijk, R.A. Tollenaar, P. Devilee; Radboud University Nijmegen Medical Center, Nijmegen, NL: M.J. Ligtenberg, N. Hoogerbrugge; University Medical Center Utrecht, Utrecht, NL: M.G. Ausems, R.B. van der Luijt; Amsterdam Medical Center, NL: C.M. Aalfs, T.A. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, Maastricht, NL: E.B. Gomez-Garcia, C.E. van Roozendaal, Marinus J. Blok, B. Caanen; University Medical Center Groningen University, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H.F. Vasen. The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088 and NKI2007-3756.
Hungarian Breast and Ovarian Cancer Study (HUNBOCS)
This work was supported by Hungarian Research Grant NKTH-OTKA CK-80745 and the Norwegian EEA Financial Mechanism Hu0115/NA/2008-3/ÖP-9.
INHERIT
Jacques Simard, Francine Durocher, Rachel Laframboise, Marie Plante, Centre Hospitalier Universitaire de Quebec & Laval University, Quebec, Canada ; Peter Bridge, Jilian Parboosingh, Molecular Diagnostic Laboratory, Alberta Children's Hospital, Calgary, Canada; Jocelyne Chiquette, Hôpital du Saint-Sacrement, Quebec, Canada ; Bernard Lesperance, Hôpital du Sacré-Cœur de Montréal, Montréal, Canada. Jacques Simard—J.S. is Chairholder of the Canada Research Chair in Oncogenetics. This work was supported by the Canadian Institutes of Health Research for the ‘CIHR Team in Familial Risks of Breast Cancer' program and by the Canadian Breast Cancer Research Alliance-grant #019511.
ILUH
The study was supported by the Icelandic Association ‘Walking for Breast Cancer Research' and by the Landspitali University Hospital Research Fund.
Istituto Oncologico Veneto—Hereditary Breast Ovarian Cancer Study (IOVHBOCS)
This study was supported by ‘Ministero della Salute' (grant numbers RFPS 2006-5-341353, ACC2/R6.9 and ‘Progetto Tumori Femminili').
Kathleen Cuningham Consortium for Research into Familial Breast Cancer (kConFab)
We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow-Up Study (funded by NHMRC grants 145684, 288704 and 454508) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. ABS is supported by an NHMRC Senior Research Fellowship.
McGill
This work was supported by the Jewish General Hospital Weekend to End Breast Cancer. MT holds a Fonds de la Recherche en Santé du Québec clinician-scientist award.
National Cancer Institute (NCI)
The research of Drs PL Mai and MH Greene was supported by the Intramural Research Program of the US National Cancer Institute, and by support services contracts NO2-CP-11019-50 and N02-CP-65504 with Westat, Inc, Rockville, MD.
Mayo Clinic (MAYO)
This work was supported by grants from the Breast Cancer Research Foundation (BCRF), Komen Foundation for the Cure, Department of Defense ovarian cancer research award (W81XWH-10-1-0341) and US National Cancer Institute, National Institutes of Health grant CA128978.
N.N. Petrov Institute of Oncology (NNPIO)
The work is supported by the Russian Foundation for Basic Research (grants 08-04-00369-a, 09-04-90402 and 10-04-92110-a), the Commission of the European Communities (grant PITN-GA-2009-238132) and through a Royal Society International Joint grant (JP090615).
Ontario Cancer Genetics Network (OCGN)
This work was supported by Cancer Care Ontario and the US National Cancer Institute, National Institutes of Health under RFA # CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR. We wish to thank Teresa Selander, Nayana Weerasooriya and members of the Ontario Cancer Genetics Network for their contributions to the study.
The Ohio State University Clinical Cancer Genetics (OSU CCG)
This work was supported by the Ohio State University Comprehensive Cancer Center. We thank Leigha Senter and Kevin Sweet for patient accrual and data management, the Human Genetics Sample bank for sample preparation and the OSU CCC Nucleic Acids Shared Resource for genotyping plate reads.
Pisa Breast Cancer Study (PBCS)
MAC from University Hospital of Pisa was supported by Istituto Toscano Tumori grant
SEABASS
SEABASS is a collaborative effort between Cancer Research Initiatives Foundation (Malaysia), University Malaya (Malaysia), National University Hospital (Singapore), University Kebangsaan Malaysia (Malaysia), Hospital Kuala Lumpur (Malaysia) and Putrajaya Hospital (Malaysia). The research has received funding from CARIF and University Malaya.
Swedish Breast Cancer Study (SWE-BRCA)
SWE-BRCA collaborators: Per Karlsson, Margareta Nordling, Annika Bergman and Zakaria Einbeigi, Gothenburg, Sahlgrenska University Hospital; Marie Stenmark-Askmalm and Sigrun Liedgren, Linköping University Hospital; Åke Borg, Niklas Loman, Håkan Olsson, Ulf Kristoffersson, Helena Jernström, Katja Harbst and Karin Henriksson, Lund University Hospital; Annika Lindblom, Brita Arver, Anna von Wachenfeldt, Annelie Liljegren, Gisela Barbany-Bustinza and Johanna Rantala, Stockholm, Karolinska University Hospital; Beatrice Melin, Henrik Grönberg, Eva-Lena Stattin and Monica Emanuelsson, Umeå University Hospital; Hans Ehrencrona, Richard Rosenquist Brandell and Niklas Dahl, Uppsala University Hospital.
University of California Irvine Study (UCI)
Now at the Beckman Research Institute of the City of Hope. This work was supported by NIH grant R01-CA74415 (to SLN). SLN is partially supported by the Morris and Horowitz Families Endowed Professorship.
UK and Gilda Radner Familial Ovarian Cancer Registries (UKGRFOCR)
UKFOCR was supported by a project grant from CR-UK to Paul Pharoah. We thank Simon Gayther, Susan Ramus, Carole Pye, Patricia Harrington and Eva Wozniak for their contributions towards the UKFOCR. We would like to acknowledge the Roswell Park Alliance Foundation for their continued support of the Gilda Radner Ovarian Family Cancer Registry. GRFOCR would like to acknowledge Kirsten Moysich (Department of Cancer Prevention and Control) and Kunle Odunsi (Departments Gynecologic Oncology and Immunology).
University California San Francisco study (USCF)
Dr Beattie was supported by a grant from the National Institutes of Health, National Cancer Institute, Bay Area Breast SPORE (P50 CA058207)
UPENN
SMD receives funding from the MacDonald Family Foundation, and KLN from Breast Cancer Research Foundation. TR is funded by NIH grants R01-CA102776 and R01-CA083855.
Cedars-Sinai Medical Center (WCRI)
This work was supported by the American Cancer Society Early Detection Professorship and Entertainment Industry Foundation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. doi:10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. doi:10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. doi:10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 5.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. doi:10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. doi:10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. doi:10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoniou A.C., Spurdle A.B., Sinilnikova O.M., Healey S., Pooley K.A., Schmutzler R.K., Versmold B., Engel C., Meindl A., Arnold N., et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am. J. Hum. Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. doi:10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniou A.C., Sinilnikova O.M., McGuffog L., Healey S., Nevanlinna H., Heikkinen T., Simard J., Spurdle A.B., Beesley J., Chen X., et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. doi:10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou A.C., Beesley J., McGuffog L., Sinilnikova O.M., Healey S., Neuhausen S.L., Ding Y.C., Rebbeck T.R., Weitzel J.N., Lynch H.T., et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. doi:10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Closas M., Hall P., Nevanlinna H., Pooley K., Morrison J., Richesson D.A., Bojesen S.E., Nordestgaard B.G., Axelsson C.K., Arias J.I., et al. Heterogeneity of breast cancer associations with five susceptibility Loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. doi:10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. doi:10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudet M.M., Kirchhoff T., Green T., Vijai J., Korn J.M., Guiducci C., Segre A.V., McGee K., McGuffog L., Kartsonaki C., et al. Common Genetic Variants and Modification of Penetrance of BRCA2-Associated Breast Cancer. PLoS Genet. 2010;6:e1001183. doi: 10.1371/journal.pgen.1001183. doi:10.1371/journal.pgen.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel C., Versmold B., Wappenschmidt B., Simard J., Easton D.F., Peock S., Cook M., Oliver C., Frost D., Mayes R., et al. Association of the Variants CASP8 D302H and CASP10 V410I with Breast and Ovarian Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. Cancer Epidemiol. Biomarkers Prev. 2010;19:2859–2868. doi: 10.1158/1055-9965.EPI-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eccles D.M. Identification of personal risk of breast cancer: genetics. Breast Cancer Res. 2008;10(Suppl. 4):S12. doi: 10.1186/bcr2172. doi:10.1186/bcr2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stacey S.N., Sulem P., Zanon C., Gudjonsson S.A., Thorleifsson G., Helgason A., Jonasdottir A., Besenbacher S., Kostic J.P., Fackenthal J.D., et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. doi:10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Q., Wen W., Qu S., Li G., Egan K.M., Chen K., Deming S.L., Shen H., Shen C.Y., Gammon M.D., et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71:1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. doi:10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakhani S.R., van de Vijver M.J., Jacquemier J., Anderson T.J., Osin P.P., McGuffog L., Easton D.F. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin. Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. doi:10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 19.Ali S., Coombes R.C. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer. 2002;2:101–112. doi: 10.1038/nrc721. doi:10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 20.Key T., Appleby P., Barnes I., Reeves G. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J. Natl. Cancer Inst. 2002;94:606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 21.Asselin-Labat M.L., Vaillant F., Sheridan J.M., Pal B., Wu D., Simpson E.R., Yasuda H., Smyth G.K., Martin T.J., Lindeman G.J., Visvader J.E. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. doi:10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 22.Joshi P.A., Jackson H.W., Beristain A.G., Di Grappa M.A., Mote P.A., Clarke C.L., Stingl J., Waterhouse P.D., Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. doi:10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 23.Lim E., Vaillant F., Wu D., Forrest N.C., Pal B., Hart A.H., Asselin-Labat M.L., Gyorki D.E., Ward T., Partanen A., et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. doi:10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 24.Cittelly D.M., Richer J.K., Sartorius C.A. Ovarian steroid hormones: what's hot in the stem cell pool? Breast Cancer Res. 2010;12:309. doi: 10.1186/bcr2627. doi:10.1186/bcr2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoniou A.C., Sinilnikova O.M., Simard J., Leone M., Dumont M., Neuhausen S.L., Struewing J.P., Stoppa-Lyonnet D., Barjhoux L., Hughes D.J., et al. RAD51 135G–>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am. J. Hum. Genet. 2007;81:1186–1200. doi: 10.1086/522611. doi:10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domchek S.M., Friebel T.M., Singer C.F., Evans D.G., Lynch H.T., Isaacs C., Garber J.E., Neuhausen S.L., Matloff E., Eeles R., et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. J. Am. Med. Assoc. 2010;304:967–975. doi: 10.1001/jama.2010.1237. doi:10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisen A., Lubinski J., Klijn J., Moller P., Lynch H.T., Offit K., Weber B., Rebbeck T., Neuhausen S.L., Ghadirian P., et al. Breast cancer risk following bilateral oophorectomy in BRCA1 and BRCA2 mutation carriers: an international case-control study. J. Clin. Oncol. 2005;23:7491–7496. doi: 10.1200/JCO.2004.00.7138. doi:10.1200/JCO.2004.00.7138. [DOI] [PubMed] [Google Scholar]
- 28.Gronwald J., Tung N., Foulkes W.D., Offit K., Gershoni R., Daly M., Kim-Sing C., Olsson H., Ainsworth P., Eisen A., et al. Tamoxifen and contralateral breast cancer in BRCA1 and BRCA2 carriers: an update. Int. J. Cancer. 2006;118:2281–2284. doi: 10.1002/ijc.21536. doi:10.1002/ijc.21536. [DOI] [PubMed] [Google Scholar]
- 29.Dunbier A.K., Anderson H., Ghazoui Z., Lopez-Knowles E., Pancholi S., Ribas R., Drury S., Sidhu K., Leary A., Martin L., Dowsett M. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. doi:10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoniou A.C., Chenevix-Trench G. Common genetic variants and cancer risk in Mendelian cancer syndromes. Curr. Opin. Genet. Dev. 2010;20:299–307. doi: 10.1016/j.gde.2010.03.010. doi:10.1016/j.gde.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Catucci I., Verderio P., Pizzamiglio S., Manoukian S., Peissel B., Zaffaroni D., Roversi G., Ripamonti C.B., Pasini B., Barile M., et al. The CASP8 rs3834129 polymorphism and breast cancer risk in BRCA1 mutation carriers. Breast Cancer Res. Treat. 2011;125:855–860. doi: 10.1007/s10549-010-1068-8. [DOI] [PubMed] [Google Scholar]
- 32.Neuhausen S.L., Brummel S., Ding Y.C., Singer C.F., Pfeiler G., Lynch H.T., Nathanson K.L., Rebbeck T.R., Garber J.E., Couch F., et al. Genetic variation in insulin-like growth factor signaling genes and breast cancer risk among BRCA1 and BRCA2 carriers. Breast Cancer Res. 2009;11:R76. doi: 10.1186/bcr2414. doi:10.1186/bcr2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuzick J., Decensi A., Arun B., Brown P.H., Castiglione M., Dunn B., Forbes J.F., Glaus A., Howell A., von M.G., et al. Preventive therapy for breast cancer: a consensus statement. Lancet Oncol. 2011;12:496–503. doi: 10.1016/S1470-2045(11)70030-4. doi:10.1016/S1470-2045(11)70030-4. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell G., Antoniou A.C., Warren R., Peock S., Brown J., Davies R., Mattison J., Cook M., Warsi I., Evans D.G., et al. Mammographic density and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Res. 2006;66:1866–1872. doi: 10.1158/0008-5472.CAN-05-3368. doi:10.1158/0008-5472.CAN-05-3368. [DOI] [PubMed] [Google Scholar]
- 35.Milne R.L., Antoniou A.C. Genetic modifiers of cancer risk for BRCA1 and BRCA2 mutation carriers. Ann. Oncol. 2011;22(Suppl. 1):i11–i17. doi: 10.1093/annonc/mdq660. doi:10.1093/annonc/mdq660. [DOI] [PubMed] [Google Scholar]
- 36.Chenevix-Trench G., Milne R.L., Antoniou A.C., Couch F.J., Easton D.F., Goldgar D.E. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. doi:10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antoniou A.C., Goldgar D.E., Andrieu N., Chang-Claude J., Brohet R., Rookus M.A., Easton D.F. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet. Epidemiol. 2005;29:1–11. doi: 10.1002/gepi.20074. doi:10.1002/gepi.20074. [DOI] [PubMed] [Google Scholar]
- 38.Lange K., Weeks D., Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet. Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. doi:10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 39.Antoniou A.C., Cunningham A.P., Peto J., Evans D.G., Lalloo F., Narod S.A., Risch H.A., Eyfjord J.E., Hopper J.L., Southey M.C., et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br. J. Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. doi:10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buisson M., Anczukow O., Zetoune A.B., Ware M.D., Mazoyer S. The 185delAG mutation (c.68_69delAG) in the BRCA1 gene triggers translation reinitiation at a downstream AUG codon. Hum. Mutat. 2006;27:1024–1029. doi: 10.1002/humu.20384. doi:10.1002/humu.20384. [DOI] [PubMed] [Google Scholar]
- 41.Mazoyer S., Puget N., Perrin-Vidoz L., Lynch H.T., Serova-Sinilnikova O.M., Lenoir G.M. A BRCA1 nonsense mutation causes exon skipping. Am. J. Hum. Genet. 1998;62:713–715. doi: 10.1086/301768. doi:10.1086/301768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrin-Vidoz L., Sinilnikova O.M., Stoppa-Lyonnet D., Lenoir G.M., Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum. Mol. Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. doi:10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 43.Ramus S.J., Kartsonaki C., Gayther S.A., Pharoah P.D., Sinilnikova O.M., Beesley J., Chen X., McGuffog L., Healey S., Couch F.J., et al. Genetic Variation at 9p22.2 and Ovarian Cancer Risk for BRCA1 and BRCA2 Mutation Carriers. J. Natl. Cancer Inst. 2011;103:105–116. doi: 10.1093/jnci/djq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boos D.D. On Generalised score tests. Am. Stat. 1992;46:327–333. doi:10.2307/2685328. [Google Scholar]