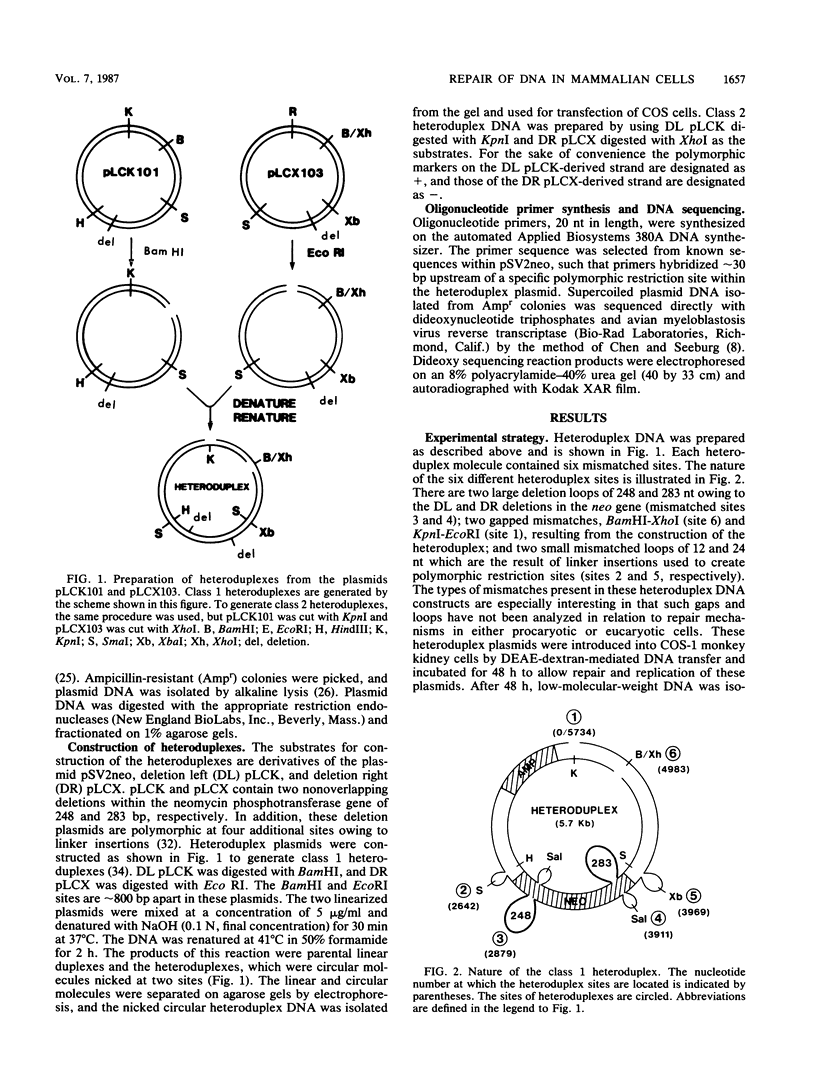
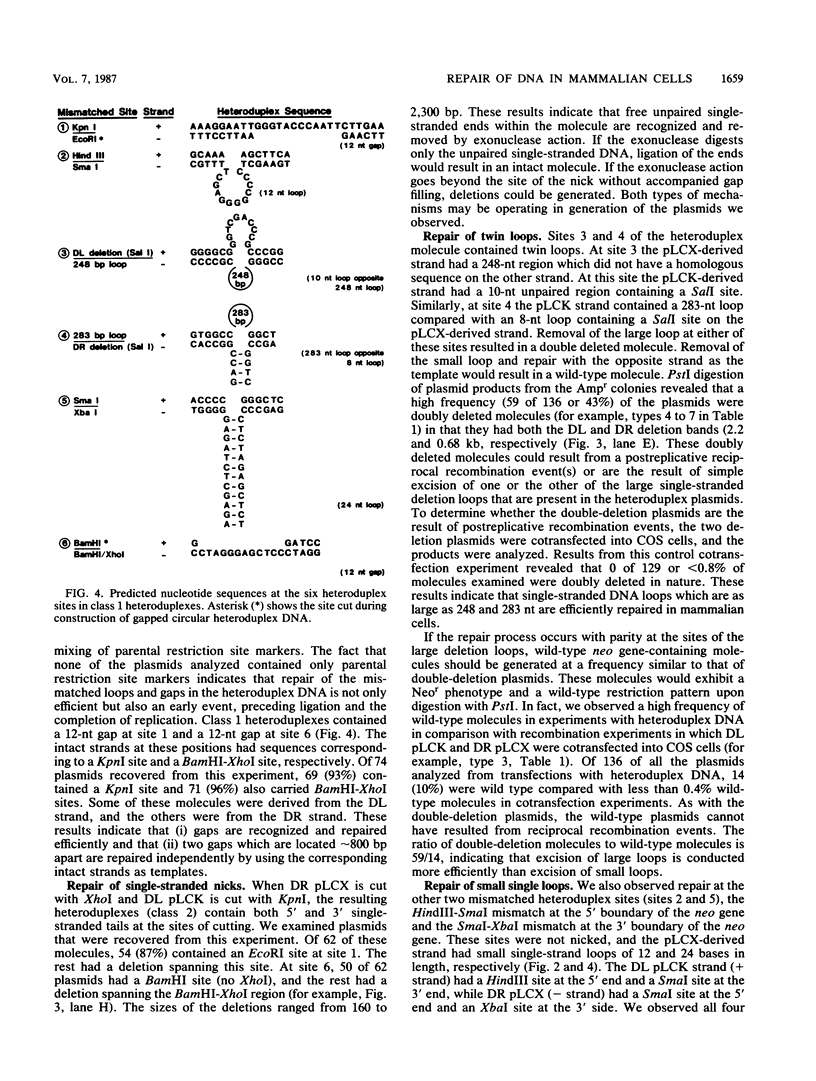
Abstract
We studied the ability of mammalian cells to repair single-stranded nicks, gaps, and loops in DNA duplexes. Heteroduplexes prepared from derivatives of the shuttle vector pSV2neo were introduced into monkey COS cells. After replication, the plasmids were recovered and used to transform Escherichia coli. Plasmid DNA from the recovered colonies was tested for repair at each of six different sites. We observed that mammalian cells are capable of repairing single-stranded gaps and free single-stranded ends most efficiently. Regions containing twin loops were recognized, and one of the loops was excised. Portions of the molecules containing small single loops were also repaired. Markers which were 58 nucleotides apart were corepaired with nearly 100% efficiency, while markers which were 1,000 nucleotides or more apart were never corepaired. The mechanisms involved in heteroduplex repair in mammalian cells seem to be similar to those involved in repairing DNA lesions caused by physical and chemical agents.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashman C. R., Davidson R. L. High spontaneous mutation frequency in shuttle vector sequences recovered from mammalian cellular DNA. Mol Cell Biol. 1984 Nov;4(11):2266–2272. doi: 10.1128/mcb.4.11.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayares D., Spencer J., Schwartz F., Morse B., Kucherlapati R. Homologous recombination between autonomously replicating plasmids in mammalian cells. Genetics. 1985 Oct;111(2):375–388. doi: 10.1093/genetics/111.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOYCE R. P., HOWARD-FLANDERS P. RELEASE OF ULTRAVIOLET LIGHT-INDUCED THYMINE DIMERS FROM DNA IN E. COLI K-12. Proc Natl Acad Sci U S A. 1964 Feb;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Asymmetric information transfer during phi X174 DNA replication. J Mol Biol. 1972 Feb 14;63(3):557–568. doi: 10.1016/0022-2836(72)90447-0. [DOI] [PubMed] [Google Scholar]
- Bauer J., Krämmer G., Knippers R. Asymmetric repair of bacteriophage T7 heteroduplex DNA. Mol Gen Genet. 1981;181(4):541–547. doi: 10.1007/BF00428750. [DOI] [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Lebkowski J. S., Botchan M. R. High mutation frequency in DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Methyl-directed repair of frameshift mutations in heteroduplex DNA. Proc Natl Acad Sci U S A. 1986 May;83(10):3395–3397. doi: 10.1073/pnas.83.10.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea V., Vovis G. F., Zinder N. D. Genetic studies with heteroduplex DNA of bacteriophage fl. Asymmetric segregation, base correction and implications for the mechanism of genetic recombination. J Mol Biol. 1975 Aug 15;96(3):495–509. doi: 10.1016/0022-2836(75)90175-8. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., Siegel E. C., Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986 Mar 20;188(2):147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Efficient correction of mismatched bases in plasmid heteroduplexes injected into cultured mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):70–74. doi: 10.1128/mcb.5.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Ayares D., Hanneken A., Noonan K., Rauth S., Spencer J. M., Wallace L., Moore P. D. Homologous recombination in monkey cells and human cell-free extracts. Cold Spring Harb Symp Quant Biol. 1984;49:191–197. doi: 10.1101/sqb.1984.049.01.022. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Cooke B. E., Fried M. Fate of mismatched base-pair regions in polyoma heteroduplex DNA during infection of mouse cells. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3073–3077. doi: 10.1073/pnas.73.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders L. H., van Kesteren A. C., Bussmann C. J., van Zeeland A. A., Natarajan A. T. Preferential repair of nuclear matrix associated DNA in xeroderma pigmentosum complementation group C. Mutat Res. 1984 Oct;141(2):75–82. doi: 10.1016/0165-7992(84)90014-9. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O., Gregg R. G., Boggs S. S., Koralewski M. A., Kucherlapati R. S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985 Sep 19;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
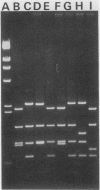
- Song K. Y., Chekuri L., Rauth S., Ehrlich S., Kucherlapati R. Effect of double-strand breaks on homologous recombination in mammalian cells and extracts. Mol Cell Biol. 1985 Dec;5(12):3331–3336. doi: 10.1128/mcb.5.12.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Smith G. R. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985 Sep 20;185(2):431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Defined oligomeric SV40 DNA: a sensitive probe of general recombination in somatic cells. Cell. 1980 Aug;21(1):141–148. doi: 10.1016/0092-8674(80)90121-x. [DOI] [PubMed] [Google Scholar]
- White J. H., Lusnak K., Fogel S. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985 May 23;315(6017):350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]
- White R. L., Fox M. S. Genetic consequences of transfection with heterduplex bacteriophage lambda DNA. Mol Gen Genet. 1975 Nov 24;141(2):163–171. doi: 10.1007/BF00267681. [DOI] [PubMed] [Google Scholar]
- Wildenberg J., Meselson M. Mismatch repair in heteroduplex DNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2202–2206. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan M. E., Cortopassi G. A., Smith C. A., Hanawalt P. C. Deficient repair of chemical adducts in alpha DNA of monkey cells. Cell. 1982 Mar;28(3):613–619. doi: 10.1016/0092-8674(82)90216-1. [DOI] [PubMed] [Google Scholar]