Abstract
A single-nucleotide polymorphism (rs2395029) in the HCP5 gene associated with HLA-B*5701 is correlated with lower HIV-1 viral set point. The two allelic forms of coding region were ectopically expressed in TZM-bl cells for an effect on HIV-1 replication. No significant HIV-1 restriction was observed in the cells with infectivity assays throughout HIV-1 life cycle, suggesting that the association of HCP5 variant with viral control is likely due to HLA-B*5701-related effect or other functional variants in the haplotype or both.
A single-nucleotide polymorphism (SNP), rs2395029 in the HLA complex protein P5 (HCP5) gene, is highly associated with HIV-1 viral load at set point [1–3]. HCP5, mainly expressed in lymphocytes [4], is a human endogenous retroviral element (HERV) that has been suggested to control retroviruses [5, 6]. The minor allele, associated with improved viral control, is a coding change (Val112Gly) in the putative polypeptide and is in almost perfect linkage disequilibrium with HLA-B*5701 [1, 2, 7]. The HLA-B*5701 allele has been repeatedly associated with improved viral control, delayed progression to AIDS and has been shown to present HIV-1 epitopes evoking strong CD8+ T cell responses [8–10], suggesting that the observed control associated with the HCP5 SNP is at least partially a reflection of the known controlling effect of HLA-B*5701. Here, we have investigated an independent role of the HCP5 variant against HIV-1 through cellular HIV-1 infectivity assays.
We utilized TZM-bl cells that express high levels of CD4 and of the HIV-1 coreceptors CCR5 and CXCR4, are highly sensitive to infection with diverse isolates of HIV-1, and contain a reporter cassette of the luciferase gene expressed under the control of an HIV-1 long terminal repeat [11] to assess viral infectivity. The cDNA of HCP5 open reading frame containing the major allele (399 bp, GI: 47087196) was isolated from peripheral blood mononuclear cells and subcloned into pcDNA3.1/mycHisA (Invitrogen, Carlsbad, California, USA) in frame with myc epitope at carboxy terminal. The minor allele was generated with a mutagenesis using the major allele. TZM-bl cells were transiently transfected with the HCP5 plasmids using Lipofectamine 2000 (Invitrogen) and subjected to HIV-1 infection with four different HIV-1 viruses (the T-cell-line-adapted MN strain, the Tier2 BG1168 primary isolate, the Tier1 SF162 primary isolate and the Tier1 SF162 Env pseudovirus). Luciferase activity as a measurement of HIV-1 infectivity was determined after 48 h as described [12].
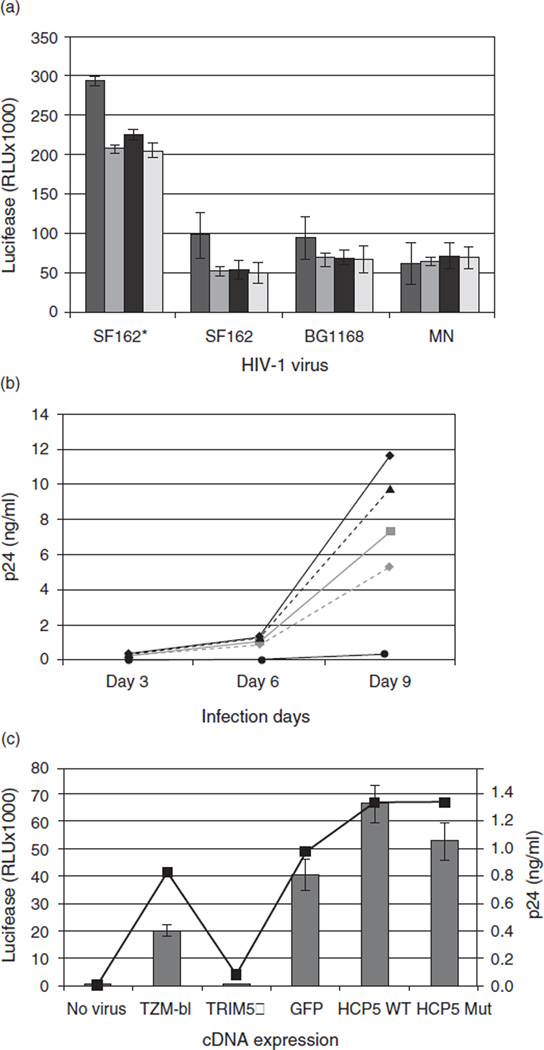
TZM-bl cells transiently transfected with HCP5 cDNAs expressed high levels of the exogenous HCP5 transcripts, over 1000-fold increase, compared with control TZM-bl cells measured by quantitative real time-PCR (TaqMan; Applied Biosystems, Foster City, California, USA). The HCP5 peptides were also confirmed by anti-myc immunoblot from the transfected cells. However, none of the HCP5 alleles showed any noticeable change in HIV-1 infectivity though the infectivity varied depending on HIV-1 strains (Fig. 1a). For each HIV-1 strain, the infectivity was independent from HCP5 allelic expression. This result demonstrates that exogenous HCP5 alleles did not affect HIV-1 infection at early stage.
Fig. 1. HCP5 alleles did not modulate HIV-1 infectivity.
(a) The major allele (HCP5 WT) or the minor allele (HCP5 mut) of HCP5 cDNA was transiently transfected into TZM-bl cells and subjected to HIV-1 infection with four different HIV-1 viruses (SF162* is the pseudovirus, SF162 and BG1168 are the primary isolates and MN is the T-cell-line-adapted virus). HIV-1 infectivity was measured for early-stage HIV-1 infection by luciferase activity as expressed relative light unit (RLU). Negative controls were TZM-bl cells with no transfection (No Tx) or with an empty vector (Vector) ( , no Tx;
, no Tx;  , vector;
, vector;  , HCPS WT;
, HCPS WT;  , HCPS mut). (b) HIV-1 antigen p24 peptide was measured from culture media in SF162-infected TZM-bl cells stably expressing HCP5 alleles, GFP or TRIM5α (from rhesus monkeys) along with control TZM-bl cells at infection days 3, 6 and 9. (
, HCPS mut). (b) HIV-1 antigen p24 peptide was measured from culture media in SF162-infected TZM-bl cells stably expressing HCP5 alleles, GFP or TRIM5α (from rhesus monkeys) along with control TZM-bl cells at infection days 3, 6 and 9. ( , TZM-bl;
, TZM-bl;  , GFP;
, GFP;  , HCP5 WT;
, HCP5 WT;  , HCP5 mut;
, HCP5 mut;  , TRIM5α) (c) Culture supernatants from the infection day 6 were added to fresh TZM-bl cells for a second round of infectivity for 48 h (
, TRIM5α) (c) Culture supernatants from the infection day 6 were added to fresh TZM-bl cells for a second round of infectivity for 48 h ( , p24;
, p24;  , infectivity). Shown are the results of a representative experiment from three independent experiments.
, infectivity). Shown are the results of a representative experiment from three independent experiments.
We also attempted to deplete host HCP5 allele by siRNAs (Dharmacon, Lafayette, Colorado, USA) targeting 3′ untranslated region of endogenous HCP5 transcripts during the overexpressing. We observed that the siRNAs reduced not only endogenous HCP5 transcript but also HIV-1 infectivity, but this effect is likely due to ‘off-target effect’ of siRNAs [13] because coexpression of the siRNA resistant-HCP5 cDNA (coding region) did not rescue the phenotype.
To assess the potential role of HCP5 alleles at later stage of HIV infection, we generated stable TZM-bl cells expressing the HCP5 alleles through retroviral transduction using a retroviral vector, pLPCX (Clontech, Mountain View, California, USA) and packaging vectors, pVPack-GP and pVPack-VSV-G (Stratagene, La Jolla, California, USA) according to the manufacturer’s instructions. Transduced TZM-bl cells were then selected with 0.5 µg/ml puromycin for 1 week, and the HCP5 expression was verified by a quantitative real time-PCR. To validate our infectivity assay, the rhesus monkey TRIM5α gene, a well known HIV-1 restriction factor [14], was used as a positive control inhibiting HIV-1 infection. First, we determined whether the high expression of HCP5 alleles affects viral productivity in HIV-1-infected cells. Production of virus assessed by p24 ELISA (PerkinElmer, Boston, Massachusetts, USA) in the culture supernatant was dramatically increased at day 9 after infection (Fig. 1b). However, the amount of HIV-1 virions from either HCP5 allele-expressing cells was indistinguishable from each other and higher than control TZM-bl cells or GFP-expressing cells (Fig. 1b). Therefore, the HCP5 alleles did not restrict viral production in our observations. As expected, the positive control cells expressing rhesus TRIM5α displayed a very limited amount of p24 production close to background level, demonstrating the accuracy of our assay system.
The culture supernatants from HIV-1-infected cells at day 6 after infection were added to fresh TZM-bl cells as a newly produced viral stock to assess its de-novo viral infectivity. The infectivity in this second round infection was mainly corresponding to the levels of p24 in the culture media and was not significantly affected by HCP5 alleles (Fig. 1c). Two other independent experiments of p24 production and second round infection showed similar results. These results indicate that later stage of HIV-1 infection was not influenced by the HCP5 expression.
The in-vitro infectivity results reported in the present study demonstrate that HCP5 does not restrict HIV-1 in early stage or in late stage of infection, at least in our cellular system. Therefore, the results reported here and in other studies [15, 16] strongly argue against a direct antiviral effect of the HCP5 gene, though we do not entirely rule out that different HCP5 alleles could elicit different immune responses against HIV-1-infected cells in vivo. Thus, the strong functional data implicating HLA-B*5701 in HIV-1 control make it likely that the immune response triggered by this HLA class I allele is at least partially responsible for the protection. However, it is important to note that multiple functional variants could be present on the same haplotype. In particular, a common polymorphism (rs9264942) located in the upstream region of the HLA-C gene, 35 kb away from the transcription start site, has been shown to be independently associated with HIV-1 control [1]. HLA-B*5701 allele is almost always found on the HLA-C SNP-containing haplotype: the effect of this HLA-C variant is therefore systematically included in the association signal attributed to HLA-B*5701. Here, we utilized in-vitro HIV-1 infectivity assays to evaluate a host genetic candidate, a HCP5 SNP (rs2395029) against HIV-1, demonstrating that the polymorphism had no influence on cellular HIV-1 infection throughout the HIV-1 life cycle.
Acknowledgements
This work was supported by the NIAID Center for HIV/AIDS Vaccine Immunology grant AI067854. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from Dr John C. Kappes, Dr Xiaoyun Wu and Tranzyme, Inc., and pLPCX-TRIM5arh-HA (rhesus monkey) from Drs Joseph Sodroski and Matt Stremlau.
W.Y. carried out the study design, molecular biology studies and infection assays, and drafted the manuscript. B.-J.M. carried out the study design, infection assays and data analysis. J.F. participated in the study design and coordination and helped to draft the manuscript. W.H. carried out the immunoassays and molecular biology studies. S.-M.X. and R.Z. carried out the infection assays. K.V.S. and H.-X.L. participated in the study design, data analysis and coordination. B.F.H. and D.B.G. conceived of the study and participated in its design and coordination. All authors read and approved the final manuscript.
References
- 1.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MA, van’t Wout AB, Schuitemaker H. Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS. 2009;23:19–28. doi: 10.1097/QAD.0b013e32831db247. [DOI] [PubMed] [Google Scholar]
- 3.Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, Delaneau O, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 4.Vernet C, Ribouchon MT, Chimini G, Jouanolle AM, Sidibe I, Pontarotti P. A novel coding sequence belonging to a new multicopy gene family mapping within the human MHC class I region. Immunogenetics. 1993;38:47–53. doi: 10.1007/BF00216390. [DOI] [PubMed] [Google Scholar]
- 5.Kulski JK, Dawkins RL. The P5 multicopy gene family in the MHC is related in sequence to human endogenous retroviruses HERV-L and HERV-16. Immunogenetics. 1999;49:404–412. doi: 10.1007/s002510050513. [DOI] [PubMed] [Google Scholar]
- 6.Garrison KE, Jones RB, Meiklejohn DA, Anwar N, Ndhlovu LC, Chapman JM, et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 9.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie GM, Kaul R, Dong T, Yang HB, Rostron T, Bwayo JJ, et al. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS. 2002;16:961–972. doi: 10.1097/00002030-200205030-00002. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 12.Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 13.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 14.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 15.Han Y, Lai J, Barditch-Crovo P, Gallant JE, Williams TM, Siliciano RF, Blankson JN. The role of protective HCP5 and HLA-C associated polymorphisms in the control of HIV-1 replication in a subset of elite suppressors. AIDS. 2008;22:541–544. doi: 10.1097/QAD.0b013e3282f470e4. [DOI] [PubMed] [Google Scholar]
- 16.Shrestha S, Aissani B, Song W, Wilson CM, Kaslow RA, Tang J. Host genetics and HIV-1 viral load set-point in African- Americans. AIDS. 2009;23:673–677. doi: 10.1097/QAD.0b013e328325d414. [DOI] [PMC free article] [PubMed] [Google Scholar]