Abstract
Approximately 2% of the world’s population is chronically infected with hepatitis C virus (HCV). Chronic hepatitis C can culminate in end stage liver disease and liver cancer if the infection is untreated. Current therapy is only partially effective and a vaccine for HCV does not exist. Since the discovery of HCV as the etiologic agent causing hepatitis C several experimental tools have been developed which have improved our understanding of the viral life cycle and the interaction of HCV with human cells. However, it remains challenging to study HCV infection in its native liver environment given its narrow species tropism, limited to humans and chimpanzees. Mice can be rendered susceptible to HCV infection by transplanting human hepatocytes into immunocompromized liver injury strains. Such human liver chimeric mice are useful as a challenge model for human hepatotropic pathogens but their utility is hampered by their inability to mount functional immune responses and practical aspects including high costs, low throughput, and donor-to-donor variability. The barriers that restrict HCV species tropism are incompletely understood. We have previously shown that expression of human CD81 and human OCLN is required for HCV uptake into mouse cells. This led to the construction of a genetically humanized mouse model for HCV infection. Here, we provide a detailed protocol for the generation of these animals and highlight some of its applications for studying HCV biology and preclinical testing of drug and vaccine candidates.
Keywords: Hepatitis C virus, animal models, viral entry, gene delivery
1. Introduction
At least 130 million individuals are chronically infected with hepatitis C virus (HCV). Chronic HCV carriers are at risk of developing severe liver disease including fibrosis, cirrhosis and hepatocellular carcinoma. HCV is a positive-sense, single stranded RNA virus of the Flaviviridae family. Current standard of care, consisting of pegylated interferon alpha (peg-IFN), ribavirin (RBV) and one of two inhibitors of the HCV RNA NS3/4A protease results in sustained virological response rates in 60–70% of patients in clinical trial cohorts[1,2]. However, this therapy is poorly tolerated and not equally effective in all patient cohorts and against all HCV genotypes. New treatment modalities, which are currently being tested in clinical trials have raised the hope that HCV can be cured in the majority of patients.
Since the discovery of HCV in 1989[3] much has been learned about the functions of HCV proteins and RNA elements and host factor requirement using cell culture systems (reviewed in[4]). However, analysis of systemic host responses to viral infection has been hampered by the lack of suitable small animal models. HCV has narrow host range, infecting only humans and chimpanzees. Chimpanzees are susceptible to patient-derived virus and infectious clones, and recapitulate the natural course of infection[5]. The ability to experimentally infect these animals has shed light on the role of the host immune response and aided the preclinical assessment of drug and vaccine candidates. However, studies in chimpanzees are hampered by limited availability, high costs, and are not considered ethically justifiable in many jurisdictions. Several distinct and potentially complementary strategies have been proposed to bridge this gap and to create more accessible small animal models[6]. One approach focuses on adapting HCV to use essential host factors from non-permissive species, such as rodents or smaller non-human primates. These efforts are based on the remarkable genetic plasticity of HCV, which fuelled by its high replicative capacity and error prone replication machinery allowing the virus to evade selection pressure exerted by the host immune system or drug regimens. This, however, can also be exploited to adapt viral genomes to use essential host molecules of other species. For example, it was previously demonstrated that certain mutations in the viral envelope proteins E1 and E2 result in a gain of function allowing HCV to efficiently engage murine CD81 and occludin (OCLN)[7] which otherwise do not facilitate efficient viral uptake[8]. While a fully mouse adapted HCV genome has not been constructed these studies provided important proof-of-concept for the feasibility of this approach.
Alternatively, the murine host can be manipulated to provide an environment that is more conducive to HCV infection. Transplantation of human hepatocytes into highly immunocompromized liver injury mouse strains can result in a high degree of human hepatocyte engraftment. Such liver chimeric mice are susceptible to human hepatotropic pathogens, including hepatitits B virus[9,10], HCV [9–11] and human malaria parasites[12,13]. However, humanized xenotransplantation models are low in throughput, costly, vary from donor-to-donor and pathogenesis studies are hampered due to the severe immunodeficiency of the recipient. The latter can be potentially overcome humanizing both the liver and the immune system in a single recipient. Recently, it was demonstrated that mice dually engrafted with human fetal hepatoblasts and components of a human immune system support HCV replication at low levels and are capable of mounting antigen-specific human immune responses which eventually result in onset of fibrosis[14], a hallmark of advancing hepatitis C. This study established important proof-of-concept for the approach but the system requires additional refinements to improve human cell chimerism, levels of HCV viremia and the functionality of the human immune system.
A genetically-engineered, inbred mouse model with inheritable susceptibility to HCV would overcome the technical difficulties of xenotransplantation. However, the determinants limiting HCV species tropism remain poorly defined. In mouse cells, the HCV life-cycle is blocked at multiple steps. HCV utilizes numerous cellular factors to enter its primary target cell, the human hepatocyte. Those include the scavenger receptor class B type I (SCARB1)[15], the tetraspanin CD81 [16], tight junction proteins, claudin-1 (CLDN1) [17] and OCLN [8,18] and the receptor tyrosine kinases epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2) [19] and the cholesterol uptake receptor Niemann Pick C1 like 1 (NPC1L1)[20]. We have previously shown that CD81, SCARB1, CLDN1 and OCLN are all required for uptake into rodent cells but only CD81 and OCLN need to be of human origin[8]. This discovery opened the door for constructing a genetically humanized mouse model. Indeed, we recently demonstrated that adenoviral delivery of human CD81 and OCLN renders mice susceptible to HCV infection with diverse HCV genotypes[21]. In order to visualize HCV uptake into murine hepatocytes, which generally do not readily support HCV RNA replication, we constructed a highly sensitive detection system where an in vivo reporter is activated by CRE recombinase expressed in the context of the incoming recombinant HCV genome. We have shown that viral uptake can be blocked by passive immunization strategies and that inoculation of these animals with a vectored vaccine induces humoral immunity and confers partial protection to heterologous challenge. We have also demonstrated proof-of-principle for combining this system with gene knockout analysis to begin to dissect viral entry in vivo.
Unfortunately, this current model supports only HCV uptake and genome translation. The ability to recapitulate the entire viral life-cycle in mice would facilitate studies on immune responses and pathogenesis. There is evidence, that can HCV replicate in murine cells, albeit to low levels, using recombinant genomes expressing a dominant selectable marker[22,23]. This suggests that all essential host factors for RNA replication are expressed in mouse cells but the rodent orthologues may interact less efficiently with the viral proteins. HCV replication is substantially increased in mouse embryonic fibroblasts derived from PKR and IRF3-deficient mice[24,25]. Thus, the inability of HCV to overcome murine defenses may also impair replication.
Adenoviral delivery to express exogenous human entry factors in mice is high-throughput and allows rapid evaluation of mutant genes but induces strongly antiviral interferon stimulated genes (ISGs), creating an environment which is not conducive for viral replication. It is conceivable that stable expression of entry factors in the form of transgenic or knock-in mice would overcome this caveat and may allow identification of a mouse background, which supports RNA replication. HCV assembly and egress in the HCV life cycle are likely to be supported in mice, as it was recently reported that infectious particles can assemble in murine hepatoma cell lines [26].
Here, we provide a detailed protocol for the construction of genetically humanized mice and highlight some of their applications in studying HCV entry and preclinical assessment of drug and vaccine candidates.
2. Genetic humanization
2.1. Theory
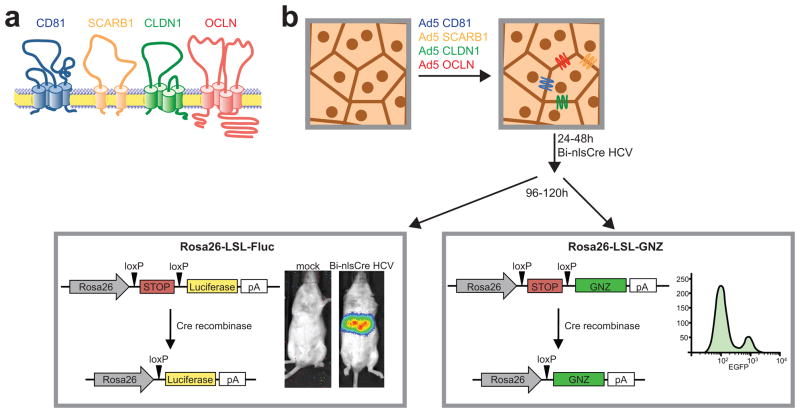
The detection of HCV entry into hepatocytes of wild-type mice[21] is based on the adenoviral delivery of the human HCV entry factors that are required for promoting viral uptake into mouse cells [8] (Figure 1). Adenoviral delivery of high particle numbers ensures that most hepatocytes will be infected with at least one adenovirus, thus maximizing the possibility that a larger number cells expresses all critical HCV entry factors. We have previously determined that an average of 5–10% of hepatocytes express CD81, SCARB1, CLDN1 and OCLN following adenoviral infection with 1×1011 of each vector. Even though incompatibilities between mouse and human CD81 and OCLN primarily limit HCV species tropism [8], we observed that overexpression of CLDN1 and SCARB1 irrespective of mouse or human origin, leads to more robust HCV entry.
Figure 1. Mechanism of studying HCV entry in vivo.
(a) The four HCV entry factors CD81, scavenger receptor BI (SCARB1), claudin-1 (CLDN1) and occludin (OCLN) (b) are delivered to the hepatocytes of reporter mice via adenoviral delivery. 24–48 hours post intravenous adenovirus delivery, 2×107 TCID50 of a Cre recombinase (Cre)-expressing HCV genome are injected i.v. and entry is analyzed either by bioluminescence imaging, histology or flow cytometry 72 hours post injection of HCV-Cre.
The inability of HCV to replicate in mouse cells [8] led to the development of Cre-lox system to detect HCV entry events in vivo [27]. Cells expressing the essential human HCV entry factors can take up intravenously injected recombinant HCV encoding for CRE recombinase. Upon initial translation of the HCV polyprotein, Cre recombinase shuttles to the nucleus and excises a transcriptional stop cassette which blocks expression of a reporter gene, such as lacZ [28], firefly luciferase [27] or fluorescent proteins [29,30]. Low levels of Cre recombinase are sufficient to permanently mark cells, which have taken up HCV. Principally, the Cre-activable reporter can be combined with any viable mouse mutant strain to assess the impact of a specific gene deletion on HCV entry, as long as the specific gene does not affect adenoviral uptake.
2.2. Preparation of reagents
The following sections described detailed step-by-step protocols for preparing the reagents needed for the construction of genetically humanized mice. All experiments involving recombinant adenoviruses and HCV should be performed under appropriate institutionally approved biosafety conditions.
2.2.1. Preparation of adenoviruses
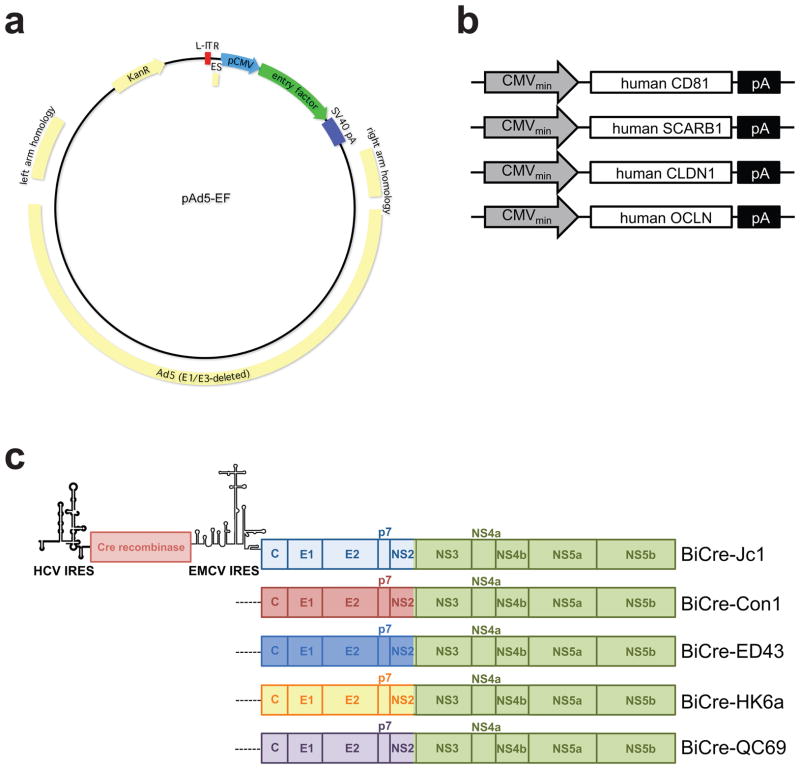
Adenoviral vectors are produced using the pAdEasy AV5 System (Agilent) according to the manufacturer’s instruction. The HCV entry factors expressing human CD81, SCARB1, CLDN1 and OCLN have been cloned into individual pAdEasy Adenoviral vectors to produce individual stocks of adenovirus [31].
Digest 20mg of each adenoviral vector expressing human CD81, SCARB1, CLDN1 and OCLN using PacI for 2 hours at 37°C. Heat-inactivate the reaction for 20 minutes at 75°C.
One day prior to transfection, plate HEK293 cells at 15% confluency in T25 flasks.
6 hours prior to transfection, replace the medium of the HEK293 cells with fresh medium containing 1.5% fetal bovine serum (FBS).
Add 125mL filter-sterilized 0.5M CaCl2 to the digested adenoviral vector and mix by vortexing. Add 250mL 2× Hank’s buffered saline (HBS) to a sterile Eppendorf tube and dropwise add the CaCl2/plasmid mixture while vortexing. Incubate the mixture at room temperature for 10 minutes before adding it dropwise to the HEK293 cells. Replace the medium on the HEK293 cells with complete medium containing 7.5% FBS 18–24 hours post transfection.
Expand the cells into a T175 flask and harvest the cells after they exhibit clear signs of cytopathic effect. Freeze-thaw cells and medium three times before centrifuging and storing the seed stock supernatants at −20°C.
Expand untransfected HEK293 cells to at least 15 P150 plates at 50–70% confluency per adenoviral construct the day before large-scale adenoviral infections.
Thaw the seed stock and adjust its volume to 75mL using Dulbecco’s Modified Eagle Medium (DMEM) without FBS. Add 5mL to each of the 15 P150 plates and incubate for 48–72 hours.
Following the onset of full cytopathic effect collect medium and cells, centrifuge and discard supernatants. Resuspend the cell pellet in 7.5mL 0.01M sodium phosphate buffer (pH 7.2). Add 2mL 5% v/w deoxycholate and incubate at 37°C for 30 minutes. Add 120mL DNAse I and 1mL 1M MgCl2 and incubate at 37°C for 30 minutes. Centrifuge for 30 minutes at 4500×g and collect the supernatant.
Separate the supernatants on two consecutive 1.2 – 1.45 g/mL cesium chloride gradients by centrifuging at 100.000×g for 12–16 hours.
Dialyze the concentrated cesium chloride-purified adenovirus preparation for 12 hours using two consecutive dialysis steps of 300mL 4% sucrose in 50mM Tris pH 8.0 at 4°C each (Slide-A-lyser dialysis cassettes, Thermo Scientific, 10.000 MWCO). Determine the particle count of the adenovirus preparation by measuring the optical density at 260 and 280nm. The particle density of the virus preparation is 20 × OD260 × 1012 particles/mL. Store the adenoviral stock at −80°C.
2.2.2. Preparation of recombinant HCV
The advent of the infectious cell culture system for HCV[32–34] has opened unprecedented opportunities to study the entire viral life-cycle in vitro. The system is based on the unique features of an HCV genome isolated from a Japanese patient with fulminant hepatitis, termed JFH1, to replicate and produce infectious particles in vitro. JFH1 belongs to HCV genotype 2a, which has limited clinical relevance. In order to extend neutralization and entry studies to other HCV genotypes, intergenotypic chimeras have been generated consisting of the core-NS2 regions of representative genomes of all seven HCV genotypes and NS3-NS5B of JFH1 [35–40]. To facilitate simpler read-out of infection and reporter genomes have been generated. Heterologous proteins have been expressed in bicistronic genomes or inserted as fusion proteins with the viral proteins [41–43]. We have taken advantage of these tools to generate recombinant HCV intergenotypic chimeras expressing Cre recombinase. Here, the 5′ untranslated region (UTR) of HCV drives expression of the first 12 codons of the core protein fused to the CRE coding sequence, while a heterologous encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES) drives expression of the HCV full-length HCV polyprotein C through NS5B (figure 3); the genome ends with the native HCV 3′ NTR.
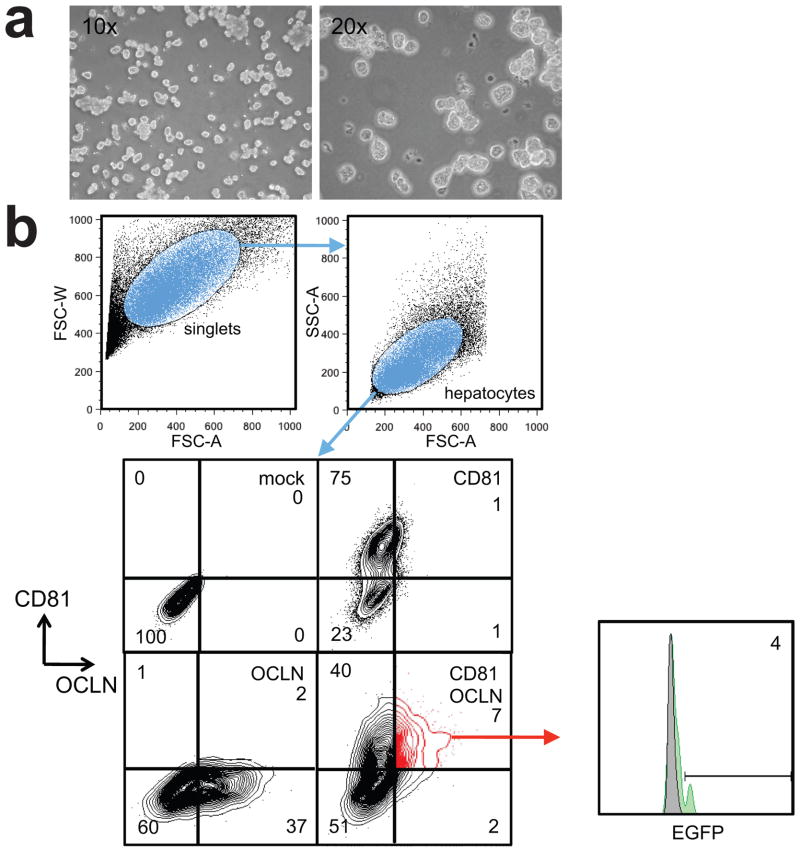
Figure 3. Detection of HCV entry by flow cytometry.
(a) Isolated hepatocytes following isolation by perfusion of the murine liver with Collagenase. (b) Gating scheme of murine hepatocytes following injection of fluorescently labeled entry factor-expressing adenovirus and HCV-Cre.
The following large scale protocol (adapted from [32]) describes the production CRE-expressing HCV stocks:
Linearize 10μg HCV-containing plasmid (i.e. pBi-nlsCre-Jc1) with XbaI for 3 hours at 37°C. Purify the reaction using MinElute Spin columns (Qiagen) according to the manufacturer’s instructions. Determine purity and completeness of digest by agarose gel electrophoresis.
HCV RNA is transcribed in vitro using T7 RiboMAX™ Express Large Scale RNA transcription kit according to the manufacturer’s instructions. Briefly, for each in vitro transcription, 1μg linearized HCV-containing plasmid are needed. For large-scale electroporations, 200 μg RNA are required (40 electroporations, 5 μg HCV RNA per electroporation). Purity and integrity are determined by RNA agarose electrophoresis. HCV RNA is frozen at −80°C in 5μg aliquots
Huh-7.5.1 cells[34], which are maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented to contain 10% fetal bovine serum and 100 mM non-essential amino acids (all supplements from Gibco) are expanded to obtain a total of 40 T600 Flasks (Millicell HY, Millipore) per large scale electroporation.
Huh-7.5.1 cells are washed twice with phosphate-buffered saline (PBS) without CaCl2/MgCl2 and trypsinized using 0.05% Trypsin-EDTA (Gibco). Cells are centrifuged at 300 × g for 5 minutes at 4 °C and washed three times with ice-cold PBS w/o CaCl2/MgCl2. After the final washing step the cells are resuspended in PBS w/o CaCl2/MgCl2 at a concentration of 2.5 × 107 cells / mL and kept on ice.
400 μL of the cell suspension (1.0 × 107 cells) are mixed with 5 μg HCV RNA, immediately transferred into a sterile 2 mm gap cuvette (BTX) and electroporated using a BTX ECM 830 electroporator (Harvard Apparatus, 860V, 5 pulses, 99 msec pulse length, 1.1s pulse interval). Following electroporation, the cells are incubated in the cuvettes for 10 minutes at room temperature. Cells are then transferred into complete DMEM (30mL per electroporation) and cuvettes are flushed with 1 mL complete DMEM. Cells are plated in 150 mm plates (6.7 × 106 cells / plate). More than 80% of cells should adhere 4 hours post electroporation in order to guarantee sufficient virus yield.
24 hours post electroporation, the medium on the cells is replaced by fresh complete DMEM. At this point, cells should be ~ 60 – 70% confluent.
48 – 60 hours post electroporation, the medium on the cells is exchanged with 15 mL pre-warmed DMEM without serum supplemented with 100 mM non-essential amino acids per 150 mm plate. From this time, supernatants are harvested and stored at 4 °C every 3–6 hours until 120 hours post electroporation (yielding a total of 9 – 18 L viral supernatants).
Viral supernatants are concentrated using a stirred cell with a 100 kDa nominal molecular weight limit to a final volume of 45 – 90 mL (200-fold concentration), aliquoted and frozen at −80 °C.
24 hours following freezing of the concentrated viral stocks, HCV 50% tissue culture infectious dose (TCID50) are determined as previously described [32]. Titers of a successfully concentrated large scale HCV production should be 4 – 8 × 107 TCID50 / mL.
Titering of the virus stocks on both Huh-7.5 and Huh-7.5 CALNL cells, a cell line engineered to express a CRE activatable GFP reporter, is performed to ensure that CRE recombinase is expressed correctly following infection of a cell. Both cell lines should yield approximately the same titers.
2.2.3. Cre responsive reporter mice
Currently available Cre responsive reporter mouse lines cover a wide range of readily detectable markers, for example lacZ[28], firefly luciferase[27] or fluorescent proteins[29,30] (Table 1).
Table 1.
Selection of CRE reporter mouse strains
| Strain name | Reporter | Read-out | Ref. |
|---|---|---|---|
| B6;129P2-Gt(ROSA)26Sortm1(CAG-ALPP)Fawa/J | ALPP | Histology | [49] |
| B6; 129-Gt(ROSA)26Sortm2Sho/J | EGFP | Flow Cytometry | [50] |
| FVB/N-Tg(CAG-EGFP,-ALPP)2.6Ggc/J | EGFP, ALPP | Flow Cytometry, Histology | [51] |
| B6.FVB-Tg(CAG-EGFP,-ALPP)2.6Ggc/J | EGFP, ALPP | Flow Cytometry, Histology | [51] |
| FVB.Cg-Gt(ROSA)26Sortm1(CAG-lacZ,-EGFP)Glh/J | EGFP, LacZ | Flow cytometry, Histology | [52] |
| B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J | EYFP | Flow Cytometry | [53] |
| B6.129S4-Gt(ROSA)26Sortm1Sor/J | LacZ | Histology | [23] |
| B6; 129S4-Gt(ROSA)26Sortm1Sor/J | LacZ | Histology | [28] |
| B6.129(Cg)-Tg(CAG-Bgeo/GFP)21Lbe/J | LacZ, GFP | Flow Cytometry, Histology | [54] |
| FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J | Luciferase | Bioluminescence | [27] |
| B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | tdTomato | Flow Cytometry, Histology | [53] |
| B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato, EGFP)Luo/J | tdTomato, EGFP | Flow Cytometry, Histology | [55] |
| B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J | ZsGreen | Flow Cytometry | [53] |
ALPP: alkaline phosphatase
A loxP site-flanked transcriptional stop cassette suppresses reporter gene expression under steady-state conditions. Following expression of Cre recombinase, the loxP site-flanked neomycin resistance cassette is excised leading to the initiation of reporter gene expression to constitutively high levels.
Convenient reporter strains for easy and high throughput read-out are FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael (called R26-LSL-FLuc) [27] and FVB.Cg-Gt(ROSA)26Sortm1(CAG-lacZ,-EGFP)Glh (called R26-LSL-GNZ)[29] mice which, upon activation by Cre recombinase, drive expression of firefly luciferase or a fusion protein of nuclear EGFP and β-galactosidase, respectively.
These mice can be readily intercrossed with a variety of transgenic, knock-out or knock-in mice to study HCV entry and usually the presence of a heterozygous reporter allele is sufficient for maximal reporter activity. Another important aspect of the Rosa26-based reporter mice is their availability on various different backgrounds (i.e. C57BL/6, Balb/C, FVB,129) thus allowing for flexibility when performing experiments requiring the intercrossing of different mouse strains. Any proposed vivo work should be reviewed and approved by appropriate institutional animal care and ethics committees.
2.3. Read-outs
Reporter gene activation following productive HCV can be visualized by microscopy in tissue sections, flowcytometrically at the single cell level (2.3.1), or using bioluminescent imaging (2.3.2).
2.3.1. Single cell analysis of HCV infection by multicolor flow cytometry
Single cell analysis of HCV infected murine hepatocytes requires the perfusion of the liver of live mice in order to ensure efficient harvest of hepatocytes (Fig. 2). In order to simplify detection of ectopically expressed HCV entry factors, we have constructed adenoviruses expressing fluorescent protein-entry factor fusions (Table 2). We have previously determined that N-terminal fusions to CD81, SCARB1, CLDN1 or OCLN do not impair their ability to support HCV glycoprotein mediated uptake [8,21]. Depending on the laser and filter set-up of the available flowcytometer different fluorescent proteins may be preferable, but the set-up in table 1 allows for unambiguous distinction, which also offers possibility for single cell sorting of HCV infected hepatocytes.
Figure 2. Constructs used for the detection of HCV entry in vivo.
(a) Genome structure of the adenoviral Ad5 constructs used to deliver the four HCV entry factors and (b) structure of the CD81, SCARB1, CLDN1 and OCLN expressing adenoviral constructs. (c) Structure of the Cre recombinase-expressing HCV genomes. Bicistronic vectors expressing Cre recombinase driven by the HCV IRES are followed by different intergenotypic chimeras of HCV genotypes 1 (Con1), 2 (Jc1), 4 (ED43), 6 (HK6a) or 7 (QC69) driven by an EMCV IRES.
Table 2.
Detection of HCV infection and HCV entry factor expression by flowcytometry
Inject Rosa26-LSL-GNZ mice with an equal mixture of adenoviruses encoding all four human entry factors in a final volume of 250 μL intravenously (1011 adenoviral particles for each entry factor)
24 – 48 hours following adenoviral entry factor delivery, inject intravenously mice with 2 × 107 TCID50 Bi-nlsCre HCV in a total volume of 250 μL.
72 – 96 hours following injection of Bi-nlsCre HCV, prepare the animal for live liver perfusion. For this purpose, mice are anaesthetized (e.g. using ketamine and xylazine). The animal is restrained using flexible bands and the skin is sprayed with 70% isopropanol. A medial axial incision is made through the skin on the ventral side across the lower abdomen up to the mediastinum and the skin spread to reveal the peritoneum. Then, the peritoneum is cut open revealing the liver and other inner organs. The gut is moved aside to expose the inferior vena cava and the portal vein. A 24-gage catheter (24 G × 0.75 in. BD Saf-T-Intima IV catheter) is inserted into the inferior vena cava and secured by suturing it using 5-0 nylon suture. After ensuring that the catheter is secured in the inferior vena cava, the portal vein is cut and perfusion with chelating solution (0.05M HEPES pH7.2, 10mM EGTA in HBSS w/o CaCl2/MgCl2) is started at a flow rate of 5 mL / minute for 5 minutes. This exsanguination under anesthesia results in the death of the animal. The chelating solution is then switched to collagenase solution (0.05M HEPES pH7.2, 4.7 mM CaCl2, 0.05% v/v collagenase type IV, Sigma C5138) at a flow rate of 5 mL / minute for 5 – 10 minutes. The liver is then removed and passed through a 100 mm cell strainer with 10 mL ice-cold PBS w/o CaCl2/MgCl2. The resulting single cell suspension containing ~80% hepatocytes is centrifuged at 50 × g for 5 minutes at 4 °C and washed twice with ice-cold PBS. The viability of the hepatocytes as determined by trypan blue exclusion should be above 70% and the yield from one mouse liver is 4 – 8 × 109 hepatocytes.
Hepatocytes are fixed using 4% paraformaldehyde in PBS pH 7.2 for 20 minutes. Following washing of the hepatocytes three times with PBS, hepatocytes are permeabilized using 0.1 % Triton-X 100 in PBS for 15 minutes at room temperature followed by three times washing with PBS. In order to ensure correct estimation of HCV-infected hepatocytes, 5 × 106 hepatocytes are stained with an antibody detecting murine albumin to distinguish hepatocytes from contaminating non-parenchymal cells (i.e. goat anti-mouse albumin, Cedarlane, Burlington, NC). A second sample is then stained for human CD81 for the analysis of infection frequency.
Cells are analyzed on a standard flow cytometer (i.e. BD LSR II) using a nozzle size of 100 μm or more. Acquisition of a minimum of 106 total events ensures sufficient statistical strength.
We have previously determined that following HCV-CRE infection up to 20% of cells expressing human CD81, SCARB1, CLDN1 and OCLN become GFP positive [21].
2.3.2 Analysis of HCV entry by in vivo bioluminescence imaging (IVIS)
In vivo bioluminescence imaging allows for rapid, minimally invasive, longitudinal quantification of reporter gene activation following HCV infection. While some imaging systems allow the detection of fluorescent protein expression in vivo, luminescence measurements generally ensure greater sensitivity through deep tissues and the skin.
Inject Rosa26-LSL-Fluc mice with an equal mixture of adenovirus encoding all four human entry factors in a final volume of 250 μL intravenously (1011 adenoviral particles for each entry factor)
24 – 48 hours following adenoviral entry factor delivery, inject intravenously mice with 2 × 107 TCID50 Bi-nlsCre HCV in a total volume of 250 μL
72 – 96 hours following injection of Bi-nlsCre HCV, prepare the animal for in vivo bioluminescence imaging. For this purpose, the animal is anaesthetized and injected with 100 μL D-Luciferin (30 mg / mL, Caliper LifeSciences) intraperitoneally.
Starting approximately 10 minutes following injection of D-Luciferin, acquisition of the resulting bioluminescence is recorded using an IVIS Lumina II in vivo bioimaging system until maximal luciferase activity is reached (Binning medium, F/stop 1, exposure time 300s).
For the analysis of results, background bioluminescence has to be recorded which is especially important when using different mouse strains on the Rosa26-LSL-Fluc background possessing distinct coat colors. Signals of 1–2 × 105 photons / second / mm2 are generally detected in positive controls whereas negative controls exhibit background signals of ~ 2 × 104 photons / second / mm2.
3. Technical trouble-shooting
Multiple steps during the process of determining HCV entry in vivo can lead to negative results of different magnitude.
3.1. Animal mortality following adenoviral infection
Due to the high adenovirus dose mice die occasionally within the first 24 hours of the experiment. This is most likely due to either contaminating CsCl or other contaminants in the adenoviral preparations. Re-dialysis of the adenoviral preparations followed by filtration through a 0.45 um syringe filter usually prevents mortality.
3.2. Variability of results
In order to assure reproducibility of results, all adenovirus and HCV preparations should be quality controlled. For optimal results, all adenovirus and HCV stocks should be stored in ready to inject aliquots preventing freeze-thaw cycles. HCV-Cre should be titered using both, Huh-7.5 and Huh-7.5 CALNL cells to ensure presence of Cre recombinase in all viral genomes. Titers determined on Huh-7.5 should be equal to those determined by flow cytometry of Huh7.5 CALNL cells.
Another important factor for success is accurate dosing and delivery of all viral preparations. All injections have to be performed by tail-vein injection ensuring delivery of virus to the liver. Other routes of inoculation (i.e. intraperitoneal injection) do not lead to efficient hepatocyte transduction.
3.3. Difficulties isolating hepatocytes for flow cytometry
Viable and complete hepatocyte isolation is important for accurate detection of HCV entry by flow cytometry. Viability should be determined prior to fixation. The average viability should exceed 75% and the yield of hepatocytes from a single animal can exceed 5×108 cells. Critical factors for the isolation of viable hepatocytes are the temperature and pH of the perfusion solutions.
Furthermore, if the isolated hepatocytes are used for flow cytometry, ensure that the antibodies used for staining are effectively staining fixed cells and adjust the forward and side-scatter detector voltages in order to correctly visualize hepatocytes (good starting values are 50 volts (FSC) and 120V (SSC) on an LSR2 flow cytometer).
4. Applications
4.1. Genetic dissection of HCV entry in vivo
Current cell culture systems for HCV, i.e. human hepatoma cells lines, do not adequately mimic in vivo human hepatocytes [44], which are polarized cells displaying the tight junction HCV entry factors (OCLN and CLDN1) in highly organized arrays. While in vitro cultures of primary human hepatocytes potentially provide a more clinically relevant system, they have not been widely adopted due to the difficulty in obtaining human liver tissue, donor variability, and the fastidious nature of hepatocytes maintained in culture. Genetic manipulation of primary cells also posesses significant challenges. Combining mouse knockout technology with our genetically humanized mouse models therefore offers a unique opportunity for HCV research. Indeed, we have provided proof-of-concept that genetically humanized can be used to genetically dissect molecular pathways involved in HCV entry [21]. The adenoviral delivery system provides considerable flexibility to express mutant variants of HCV entry factor to assess how specific mutation affect uptake efficiency [21]. Similarly, the role of host factors of interest can be assessed using mouse mutant strains harboring targeted gene disruptions. The genetically humanized mouse model allows determine the impact of loss of function for many genes to be analyzed simply by producing Rosa26 reporter mice with the homozygous mutant allele(s). However, some limitations do apply: i) the knock-out mouse has to be viable and fertile. This limitation can be potentially circumvented by employing conditional knock-out mice and ii) the molecule of interest cannot be involved in adenovirus entry. This is of crucial importance since the current experimental setup does not control for the efficiency of adenovirus infection.
4.2. Preclinical assessment of entry inhibitors
Chronic HCV infection can progress to chronic hepatitis, cirrhosis, hepatocellular carcinoma, and end stage liver disease. Currently, HCV is treated with a combination of pegylated interferon, ribavirin and one of two blockers of the viral NS3-4A protease, resulting in sustained virological responses rates of up to 70% in some patient cohorts. However, this treatment is plagued with considerable side effect frequently resulting in discontinuation of therapy. With numerous second-generation inhibitors populating pharmaceutical pipelines, a broadly effective treatment regimen is a distant but tangible goal. Until then, severely diseased patients who fail treatment face liver transplantation as the final available option. Surgery does not provide a cure and the donor organ universally becomes re-infected. Entry inhibitors might be particularly useful in this context, as blocking viral uptake by the healthy organ could transform liver transplantation from a palliative into a curative procedure. Entry inhibitors might also be valuable in chronic disease, for example to slow the spread of resistant variants in patients undergoing treatment with other DAAs. Currently, only one entry blocker has been reported in clinical development [45]. While the efficacy of entry inhibitors can be estimated in cell culture, safety and efficacy ultimately needs to be tested in vivo. Human liver chimeric mice have been used to demonstrate that antibodies blocking the entry factors CD81[46] or SCARB1[47] or the viral envelope proteins [48] can prevent HCV infection. The technical complexity in generating such humanized mice diminishes throughput limiting the number of candidate inhibitors, which can only be tested in small cohorts. We have recently demonstrated that blocking of CD81 or HCV E2 also efficiently blocks viral uptake in genetically humanized mice [21]. Thus, genetically humanized mice may serve as an additional platform to prioritize entry inhibitor candidates prior to testing in more complex systems or even clinical trials.
4.3. Vaccination
HCV therapy is predicted to improve in coming years with the approval of more potent directly acting antivirals with different modes of action. However, it remains unclear how highly potent antiviral drug cocktails will affect the global HCV disease burden. High costs, lack of adequate infrastructure for distribution and medical supervision may lessen the impact of future therapies in resource poor environments. Thus, development of (a) potent, pan-genotypic cost-effective therapeutic and or prophylactic vaccine(s) remains a priority to conquer the global HCV health problem. Current technology does not allow us to reliably predict immunogenicity, efficacy and safety of novel vaccination approaches without empirical testing in animals. Until recently chimpanzees had been the only immunocompetent animal model for HCV infection. However, high cost and limited availability of these large apes for biomedical research consequently translates in small cohort sizes for testing novel vaccination approaches. Data interpretation is further hampered by considerable genetic heterogeneity within an outbred species. Consequently, few HCV vaccine candidates have entered into clinical trials. The recently described genetically humanized mouse model [21] is fully immunocompetent. Immunization of Rosa26-LSL-Fluc mice with an recombinant vaccinia virus expressing HCV structural proteins elicited a humoral immune response directed against the viral envelope, which reduced HCV entry efficiency following genetic humanization and HCV challenge [21]. Although, correlates of protection are currently limited to neutralizing antibody responses, these data suggest that genetically humanized mice may serve as a cost-effective small animal model for prescreening experimental vaccine candidates.
5. Concluding remarks
Over the past few years HCV animal models have significantly improved. While not a single one recapitulates yet all aspects of clinical hepatitis C, each platform has unique features and can be used address important questions in HCV biology. Genetic humanization has gained traction as a complementary or alternative approach to xenotransplantation. Adenoviral gene delivery facilitates rapid and simple expression of wild-type and mutant versions of HCV entry factors in various mouse lines. HCV uptake can be readily quantified by flowcytometry or bioluminescence imaging taken advantage of a highly sensitive reporter system. Genetically humanized mice can be used to study HCV entry in vivo and to assess preclinical efficacy of entry inhibitors and vaccine candidates. Current versions of genetically humanized mouse models only support viral entry but not later stages of the viral life-cycle. To overcome the vector-induced induction of innate and adaptive immunity stable expression of HCV entry factors in form of transgenic of knock-in mice will be required.
Acknowledgments
This study was supported in part by award number RC1DK087193 (to C.M.R. and A.P.) from the National Institute of Diabetes and Digestive and Kidney Diseases, R01AI072613 (to C.M.R.) from the National Institute for Allergy and Infectious Disease, The Starr Foundation and the Greenberg Medical Institute. M.D. was supported by postdoctoral fellowship from the German Research Foundation (Deutsche Forschungsgesellschaft) and A.P. is a recipient of the Infectious Disease Society of America Astella Young Investigator Award.
References
- 1.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 2.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Murray CL, Rice CM. Turning hepatitis C into a real virus. Annu Rev Microbiol. 2011;65:307–327. doi: 10.1146/annurev-micro-090110-102954. [DOI] [PubMed] [Google Scholar]
- 5.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 6.Ploss A, Rice CM. Towards a small animal model for hepatitis C. EMBO Rep. 2009 doi: 10.1038/embor.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meuleman P, Libbrecht L, De Vos R, de Hemptinne B, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- 10.Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 12.VanBuskirk KM, O’Neill MT, De La Vega P, Maier AG, Krzych U, Williams J, Dowler MG, Sacci JB, Jr, Kangwanrangsan N, Tsuboi T, et al. Preerythrocytic, live-attenuated Plasmodium falciparum vaccine candidates by design. Proc Natl Acad Sci U S A. 2009;106:13004–13009. doi: 10.1073/pnas.0906387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacci JB, Jr, Alam U, Douglas D, Lewis J, Tyrrell DL, Azad AF, Kneteman NM. Plasmodium falciparum infection and exoerythrocytic development in mice with chimeric human livers. Int J Parasitol. 2006;36:353–360. doi: 10.1016/j.ijpara.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, Barry W, Ploss A, Rice CM, Su L. A Humanized Mouse Model to Study Hepatitis C Virus Infection, Immune Response, and Liver Disease. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO Journal. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 17.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012 doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uprichard SL, Chung J, Chisari FV, Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin LT, Noyce RS, Pham TN, Wilson JA, Sisson GR, Michalak TI, Mossman KL, Richardson CD. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J Virol. 2010;84:9170–9180. doi: 10.1128/JVI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J Virol. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG., Jr Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 28.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 29.Stoller JZ, Degenhardt KR, Huang L, Zhou DD, Lu MM, Epstein JA. Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis. 2008;46:200–204. doi: 10.1002/dvg.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Current opinion in biotechnology. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 32.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 33.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, Bukh J. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology. 2007;133:1614–1626. doi: 10.1053/j.gastro.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, Eugen-Olsen J, Bukh J. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A. 2008;105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen TB, Gottwein JM, Scheel TK, Hoegh AM, Eugen-Olsen J, Bukh J. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J Infect Dis. 2008;198:1756–1765. doi: 10.1086/593021. [DOI] [PubMed] [Google Scholar]
- 38.Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 39.Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009;5:e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaller T, Appel N, Koutsoudakis G, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol. 2007;81:4591–4603. doi: 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottwein JM, Jensen TB, Mathiesen CK, Meuleman P, Serre SB, Lademann JB, Ghanem L, Scheel TK, Leroux-Roels G, Bukh J. Development and Application of Hepatitis C Reporter Viruses with Genotype 1 to 7 Core-Nonstructural Protein 2 (NS2) Expressing Fluorescent Proteins or Luciferase in Modified JFH1 NS5A. J Virol. 2011;85:8913–8928. doi: 10.1128/JVI.00049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheahan T, Jones CT, Ploss A. Advances and challenges in studying hepatitis C virus in its native environment. Expert Rev Gastroenterol Hepatol. 2010;4:541–550. doi: 10.1586/egh.10.53. [DOI] [PubMed] [Google Scholar]
- 45.Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA, et al. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol. 2011;54:48–55. doi: 10.1016/j.jhep.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 46.Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, Desombere I, Reiser H, Leroux-Roels G. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 47.Meuleman P, Catanese MT, Verhoye L, Desombere I, Farhoudi A, Jones CT, Sheahan T, Grzyb K, Cortese R, Rice CM, et al. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology. 2012;55:364–372. doi: 10.1002/hep.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 49.Que J, Wilm B, Hasegawa H, Wang F, Bader D, Hogan BL. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci U S A. 2008;105:16626–16630. doi: 10.1073/pnas.0808649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 51.Badaloni A, Bonanomi D, Albieri I, Givogri I, Bongarzone E, Valtorta F, Consalez GG. Transgenic mice expressing a dual, CRE-inducible reporter for the analysis of axon guidance and synaptogenesis. Genesis. 2007;45:405–412. doi: 10.1002/dvg.20307. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 55.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 56.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 57.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 58.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]