Abstract
Transcription of long noncoding RNAs (lncRNAs) within gene regulatory elements can modulate gene activity in response to external stimuli, but the scope and functions of such activity are not known. Here we use an ultra-high density array that tiles the promoters of 56 cell cycle genes to interrogate 108 samples representing diverse perturbations. We identify 216 transcribed regions that encode putative lncRNAs--many with RT-PCR-validated periodic expression during the cell cycle, show altered expression in human cancers, and are regulated in expression by specific oncogenic stimuli, stem cell differentiation, or DNA damage. DNA damage induces five lncRNAs from the CDKN1A promoter, and one such lncRNA, named PANDA, is induced in a p53- dependent manner. PANDA interacts with the transcription factor NF-YA to limit expression of pro-apoptotic genes; PANDA depletion markedly sensitized human fibroblasts to apoptosis by doxorubicin. These findings suggest potentially widespread roles for promoter lncRNAs in cell growth control.
INTRODUCTION
Mammalian genomes are more pervasively transcribed than previously expected1-4. In addition to protein coding genes, many types of non-coding RNAs (ncRNAs) are transcribed. Small regulatory ncRNAs, including small interfering RNAs (siRNAs), microRNAs, and Piwi-associated RNAs (piRNAs), function in genome defense and post-transcriptional regulation5-7. Near transcriptional start sites (TSS), divergent transcription by RNA polymerase can generate small ncRNAs ranging from 20 to 200 nucleotides, which have been variously named promoter associated sRNAs (PASRs), transcription-initiation RNAs (tiRNAs), and TSS-associated RNAs (TSSa-RNAs)8-11. However, it remains uncertain if these ncRNAs are functional or just represent byproducts of RNA polymerase infidelity12,13. Long ncRNAs (lncRNAs) vary in length from several hundred bases to tens of kilobases; they may be located in isolation from protein coding genes (long intergenic ncRNAs, or lincRNAs), or they may be interspersed nearby and within protein coding genes14,15. Moreover, recent evidence suggest that active enhancer elements are also transcribed to produce ncRNAs16,17.
Although evidence for function of ncRNAs as a group is lacking, several lncRNAs have been implicated in transcriptional regulation. Two prime examples are in the genomic loci of cell cycle genes. In the cyclin D1 (CCND1) promoter, an ncRNA transcribed two kilobases upstream of the CCND1 gene is induced by ionizing radiation and regulates transcription of CCND1 in cis by forming a ribonucleoprotein repressor complex18. This ncRNA binds to and allosterically activates the RNA-binding protein TLS (translated in liposarcoma), which inhibits histone acetyltransferases, resulting in repression of CCND1 transcription. A second example is an antisense ncRNA gene CDKN2B-AS1 (also known as p15AS or ANRIL) that overlaps with the p15 coding sequence, and CDKN2B-AS1 expression is increased in human leukemias with an inverse correlation with p15 expression19,20. CDKN2B-AS1 can transcriptionally silence p15 directly as well as through induction of heterochromatin formation. Many well-studied lncRNAs, such as those involved in dosage compensation and imprinting, regulate gene expression in cis21, but other lincRNAs, such as HOTAIR and linc-p21, can regulate the activity of distantly located genes in trans22-24. Inspired by these examples, we hypothesized that the genomic loci of cell cycle genes may harbor other functional ncRNAs that have yet to be discovered.
In this study, we create an ultrahigh-resolution tiling microarray to interrogate the transcriptional and epigenetic landscape around the TSSs of 56 cell cycle genes, including all cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (CDKIs). We analyze a diverse collection of cells and tissues samples that interrogate distinct perturbations in cell growth control. Our results reveal a map of extensive and choreographed noncoding transcription, and identify a specific set of lncRNAs that function in the DNA damage response.
RESULTS
Extensive and regulated noncoding transcription near cell cycle genes
To systematically discover functional ncRNAs in the regulatory region of human cell cycle genes, we created a tiling array that interrogates at 5-nucleotide resolution across 25kb of the 9p21 locus [which encompasses CDKN2A (p16), p14ARF, and CDKN2B (p15)], as well as from 10kb upstream to 2kb downstream of each TSS from 53 cell cycle genes to include all known cyclins, CDKs, and CDKIs (Fig 1a, Supplementary Table 1. These genes are also critical for fundamental biological processes such as senescence, self renewal, DNA damage response, and tumor formation25-27. Thus, we hybridized 54 pairs of polyadenylated RNAs from various human cells that were altered or perturbed through cell cycle synchronization, DNA damage, differentiation stimuli, oncogenic stimuli, or carcinogenesis (Supplementary Table 2).
Figure 1. Identification of ncRNAs near and within cell cycle genes.
(a) Flow chart of strategy for systematic discovery of cell cycle ncRNAs.
(b) Representative tiling array data. RNA hybridization intensity and H3K36me3 and H3K4me3 ChIP-chip signal relative to input at the CCNE1 locus in human fetal lung fibroblasts. Predicted transcripts shown in red boxes. Known mRNA exons in black boxes. Peak Calling: Each bar represents a significant peak from one of the 108 array channels.
(c) Chromatin state at transcribed regions. Average ChIP-chip signal relative to input calculated across transcriptional peaks expressed in human fetal lung fibroblasts +/- doxorubicin treatment.
(d) Codon substitution frequency (CSF) analysis. Graph of average evolutionary CSF of exons of coding genes and predicted transcripts. CSF<10 represents no protein coding potential.
(e) Transcriptional landscape of cell cycle promoters. All 56 cell cycle promoters were aligned at the TSS and average RNA hybridization signal was calculated across the 12 kilobase window. This process was repeated with all 49 samples. Output represents a 150 basepair running window of average transcription signal across all 56 promoters and all 54 arrays.
See also: Supplementary Table S1 and Supplementary Figure S1.
A peak calling algorithm searched for statistically significant signals above background and detected contiguous regions (peaks) of at least 50 basepairs. We then compiled statistically significant transcripts from all 108 channels of the 54 arrays, clustered all transcripts that overlapped by a minimum of 50 bases, and identified clusters that were present in at least 10% of the samples. Averaging the signal intensity across all probes in a peak produced a quantitative estimate of transcript abundance. Despite possible 3′ bias due to poly-adenylated RNA selection, our procedure detected exon 1 transcription from the majority of cell cycle coding genes (41 of the 56), demonstrating that this custom tiling array can detect previously reported transcribed regions. In each individual sample, we detected an average of 73 of the 216 transcribed regions (range 14-189) that did not overlap with known exons of the 56 cell cycle genes (Supplementary Figure 1; example of the CCNE1 locus in human fetal lung fibroblasts shown in Fig 1b). Across all 108 samples, we identified a total of 216 discrete transcribed regions (Supplementary Table 3). The average transcript length was 234 nucleotides (range 50- 1494). 171 of the 216 (79%) novel transcribed regions were located 5′ of the TSS of the cell cycle genes (“upstream”), 40 of the 216 (19%) were located within introns (“intronic”), and 5 of the 216 (2%) were located downstream of the 3′ end of CDKN2A.
Genes actively transcribed by RNA polymerase II are marked by trimethylation of histone H3 on lysine 4 (H3K4me3) and lysine 36 of histone H3 (H3K36me3), which reflect gene starts and bodies, respectively28. These chromatin marks can be used to identify noncoding transcription14. In a subset of our samples, we determined whether the 216 transcribed regions were similarly marked for active transcription by performing chromatin immunoprecipitation followed by hybridization to our custom tiling array (ChIP-chip). This analysis confirmed that the chromatin state at a majority of the newly defined transcripts was enriched in both H3K4me3 and H3K36me3 (Fig 1b and 1c). Using EpiGRAPH analysis to query our transcripts against approximately 900 published genomic attributes29, the 216 putative transcribed regions are enriched for H3K4me3 (p<10−9) and RNA polymerase II binding (p<10−7), providing further evidence that these genomic regions are actively transcribed.
To determine whether the 216 transcripts may encode previously unknown protein-coding exons or non-coding RNAs, we used the codon substitution frequency (CSF) analysis to assess for characteristic evolutionary signatures of protein-coding sequences across 21 sequenced mammalian genomes30. As expected, the transcribed regions that coincided with annotated exons had high CSF scores. However, over 86% of the novel transcribed regions had CSF scores well below the threshold of known protein-coding genes and resemble known ncRNAs (Fig 1d and Supplementary Table 3), suggesting that most of the novel regions do not have protein coding potential. BLAST analysis confirmed that the majority of the transcripts are not known protein coding genes (Supplementary Table 3). Furthermore, none of the transcripts intersect known pre-miRNAs, C/D box small nucleolar RNAs, H/ACA box snoRNAs, and scaRNAs as annotated in the UCSC genome browser. Thereafter, we refer to these transcribed regions as long non-coding (lnc)RNAs. We aligned the RNA hybridization signals at all 56 protein-coding loci of all 108 samples relative to their TSS (Fig 1e). As expected, we found a peak immediately downstream of the TSS corresponding to exon 1 of the protein coding gene. In addition, we found enrichment of noncoding transcription in the region 4 to 8 kilobases upstream of the TSS. Thus, unlike the previously described PASRs, tiRNAs, and TSSaRNAs, which are primarily located within 100 bp of the TSS, the majority of these ncRNAs are longer and are not clustered immediately around the TSS.
Expression patterns of ncRNAs suggest specific biological functions
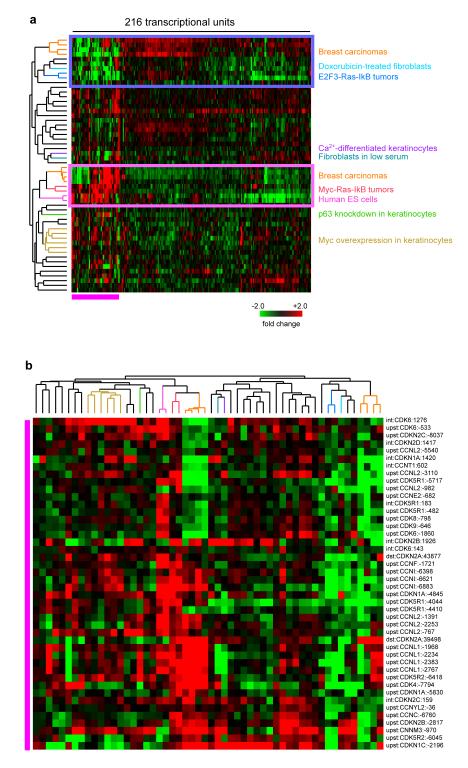
Next, we examined the biological conditions that regulate expression of these ncRNAs in order to infer possible biological functions. We assembled a matrix of the expression changes of the 216 novel transcribed regions across all 54 perturbations and hierarchically clustered the genes and samples (Fig 2a). Of the 216 novel transcribed regions, 92 (43%) had at least a 2 fold change in expression detected on the tiling array in at least one of the perturbations, suggesting that a large subset may have functional roles. The samples that had the most transcripts with at least 2 fold expression change were the embryonic stem cells (ESC) relative to day 152 fetal pancreas (40 of 216) and invasive ductal breast carcinomas relative to normal (as many as 35 of 216), suggesting that a subset of these lncRNAs may play a role in self-renewal and carcinogenesis (Fig 2a). Interestingly, lncRNA expression profiles of keratinocytes with knockdown of p63, which inhibits keratinocyte differentiation, clustered with that of ESC, suggesting that these ncRNAs may have a role in the undifferentiated state. Expression patterns from five keratinocyte samples that were transduced with the oncogene MYC alone or in combination with other oncogenes relative to controls clustered together, demonstrating that MYC has a dominant effect on ncRNA expression. MYC-RAS-IκBα transduced human keratinocytes activate an ESC-like mRNA gene expression program and acquire properties of cancer stem cells31. Notably, the lncRNA expression profile of MYC-RAS-IκBα cells clustered with that of ESC (Fig 2), suggesting a shared lncRNA signature for embryonic and cancer stem cells. In contrast, the E2F3-RAS-IκBα transduced keratinocyte, which do not express the ESC-like mRNA gene expression program, had an inverse pattern of expression for the majority of lncRNAs. In addition, 8 primary human invasive ductal breast carcinomas split into 2 different groups based on their lncRNA profiles: 4 of the cancers clustered with the ES cells and MYC-RAS-IκBα tumors and the other 4 clustered with the E2F3- RAS-IκBα tumors, suggesting that these tumor models mimic the expression pattern of not only mRNAs but also these lncRNAs in bona fide human cancers.
Figure 2. ncRNA expression across diverse cell cycle perturbations.
(a) Hierarchical clustering of 216 predicted ncRNAs across 54 arrays, representing 108 conditions. Red indicates that the cell cycle perturbation induced transcription of the ncRNA. Green indicates that the cell cycle perturbation repressed transcription of the ncRNA. Black indicates no significant expression change.
(b) Zoom in view of ncRNAs in cluster 1.
See also: Supplementary Table S2 and S3.
The 216 lncRNAs are divided into 3 main clusters based on their expression pattern across all samples (Fig 2). Notably, cluster 1 is composed of lncRNAs that are strongly induced in ES cells, keratinocytes with p63-knockdown, and Myc-Ras-IkB tumors relative to differentiated cells and GFP-Ras-IkB tumors, which we interpret to be a “stemness cluster” (Fig 2b). Interestingly, each cluster is composed of many of the ncRNAs from the same genomic locus, suggesting that multiple adjacent ncRNAs are either coordinately regulated in a shared response or are spliced together as exons of one transcript. High correlation of the dynamic expression patterns of these ncRNAs and different biological and cellular conditions suggest that these ncRNAs may be functional in the cell cycle, self renewal, and cancer.
A gene co-expression map infers trans regulatory mechanisms and biological functions
Multiple lncRNAs, including p15AS and the lncRNA upstream of CCND1, have been shown to regulate the transcription of the nearby coding gene. To determine whether gene-proximal lncRNAs are typically correlated with the expression of the nearest mRNA, we conducted whole genome expression arrays on 17 samples that were also examined on our tiling array, and calculated pair-wise Pearson correlations between the expression patterns of each cell cycle promoter lncRNAs vs. every mRNA genome-wide. Surprisingly, there was no significant correlation or anti-correlation between most of the 216 lncRNAs and the nearby protein-coding mRNA, suggesting that most of the lncRNAs may not function in cis to activate or repress nearby mRNA expression (Fig 3a). qRT-PCR analysis of lncRNAs and neighboring 5′and 3′ mRNAs in 34 additional samples confirmed these findings (Supplementary Figure 2). In contrast, we found that the median correlation between two ncRNAs of the same locus was positive, supporting our hypothesis that neighboring ncRNAs may be coordinately regulated, positively regulate each other, and/or are exons of the same transcript (Fig 3b).
Figure 3. Functional associations of ncRNAs.
(a) lncRNA expression patterns do not correlate with that of the mRNAs in cis. Histogram of Pearson correlations between each of the 216 ncRNAs and the cis mRNA across 108 samples.
(b) lncRNA expression patterns have positive correlation with neighboring lncRNA transcripts. Histogram of Pearson correlations between each of the 216 ncRNAs and nearby transcripts on the same locus across 108 samples.
(c) Genes co-expressed with lncRNAs are enriched for functional groups in cell cycle and DNA damage response. Module map of lncRNA gene sets (columns) versus Gene Ontology Biological Processes gene sets (rows) across 17 samples (p<0.05, FDR<0.05). A yellow entry indicates that the GO gene set is positively associated with the lncRNA gene set. A blue entry indicates that the GO gene set is negatively associated with the lncRNA gene set. Black entry indicates no significant association. Representative enriched GO gene sets listed.
Given that expression of the 216 ncRNAs does not generally correlate with the mRNA in cis, we further explored the genes and pathways that they may regulate, using a guilt-by-association approach14. For each lncRNA, we define a co-expression gene set as the group of mRNAs that are positively or negatively correlated with that lncRNA across the 17 samples (R>0.5 or R<0.5, respectively) (Fig S3). We then constructed a gene module map32 of the association of each lncRNA co-expression gene set vs. the Gene Ontology Biological Processes, and performed biclustering to identify lncRNAs that are associated with distinct Gene Ontology terms (Fig 3c). This analysis revealed multiple sets of lncRNAs that are associated with biological processes including cell cycle, DNA recombination, ribonucleoprotein complex biogenesis and assembly, RNA splicing, and response to DNA damage. Thus, despite having limited correlation in expression to their neighboring protein-coding gene, the expression patterns of these lncRNAs are still strongly related to the cell cycle. We constructed a similar module map with curated gene sets of metabolic and signaling pathways as well as biological and clinical states from the Molecular Signatures Database (MSigDB c2 collection)33. This module map confirmed the enrichment for cell-cycle related sets (e.g. Cell Cycle Brentani, Cell Cycle KEGG). In addition, enriched modules included several poor prognosis breast cancer gene sets (BRCA ER negative, BRCA prognosis negative, BRCA1 overexpressed up), DNA damage related gene sets (UVA/UVB), several oncogenic signatures (Ras, Myc), and stem cell gene sets (Hematopoietic stem cell, Neural Stem Cell) (Fig S4).
Validation of ncRNA expression in cell cycle, ES cell differentiation, cancer and DNA damage response
To validate these inferred functional associations, we designed quantitative RT-PCR assays for 60 of the 216 novel transcribed regions (43 upstream and 7 intronic) to obtain a more quantitative measure of these lncRNAs across different conditions. Expression in HeLa cells synchronized in cell cycle progression by double thymidine block demonstrate that most of the ncRNAs have periodic expression with peaked expression at different phases of the cell cycle (Fig 4a)34. Parallel analysis in primary human fibroblasts synchronized by serum stimulation confirmed the peak cell cycle phase of 74% of the lncRNAs with periodic expression pattern during the cell cycle (Fig 4B). Next, comparison of human ES cells and fetal pancreas at days 76 and 152 demonstrated that a majority of these ncRNAs are regulated during differentiation (Fig 4c). In addition, unsupervised clustering of lncRNA expression patterns in 5 metastatic breast cancers and 5 normal mammary tissues readily distinguished the 5 metastatic breast cancers from the normal mammary tissues (Fig 4d). Some of the lncRNAs, including upst:CCNL1::-2767 and int:CDKN1A:+885, are repressed in the metastatic breast cancers relative to normal mammary tissues, whereas others, including upst:CDKN1A:-4845, upst:CDKN2B:-2817, and int:ARF:+4517, are induced. Thus, the majority of these lncRNAs has periodic expression in the cell cycle, and is differentially expressed in different states of cell differentiation and cancer progression.
Figure 4. Validated expression of ncRNAs in cell cycle progression, ES cell differentiation, and human cancers.
Custom Taqman probes were generated and used to interrogate independent biological samples for lncRNA expression. Periodic expression of lncRNAs (blue) during synchronized cell cycle progression in HeLa cells (a) and foreskin fibroblasts (b). Cell cycle phases are confirmed by FACS and expression of genes with known periodic expression in the cell cycle (orange).
(c) Regulated expression of lncRNAs in human ES cells vs. fetal pancreas.
(d) Differential expression of lncRNAs in normal breast epithelium vs. breast cancer.
Our co-expression maps predicted associations of several ncRNAs with DNA damage response pathways (Fig 3c and Supplementary Figure 3). In support of this finding, doxorubicin-treated human fetal lung fibroblasts showed at least 2-fold change in 12 of the 216 ncRNAs on the tiling array and by qRT-PCR (Fig 2). Interestingly, 2 of those 12 ncRNAs were located 5′ of the TSS of the canonical p53 target gene CDKN1A (upst:CDKN1A: -1210 and upst:CDKN1A: -4845), and similar to the CDKN1A mRNA, were induced by doxorubicin (Fig 5a). In addition, a third lncRNA at the CDKN1A locus, upst:CDKN1A:-800, was also induced by doxorubicin, but was not included in the 216 lncRNAs because it was only expressed in one of the 108 samples, the doxorubicin-treated fibroblasts. In order to confirm whether these lncRNAs may be responsive to DNA damage, we measured expression changes of 60 lncRNAs predicted in the DNA damage pathway (as well as upst:CDKN1A:-800) by quantitative RT-PCR in human fetal lung fibroblasts treated with doxorubicin, over a 24 hour time course. Most of the lncRNAs were either significantly induced or repressed by doxorubicin, and all 5 of the tested lncRNAs surrounding the CDKN1A TSS were induced, including the 3 that were previously detected on the tiling array (Fig 5b). Notably, several ncRNAs upstream of CDKN1A are induced more rapidly and with substantially higher magnitude than CDKN1A upon DNA damage. Upst:CDKN1A:-4845 is induced up to 40 fold upon DNA damage (Fig 5c). These variations in expression patterns within the same locus suggest that the lncRNAs in the CDKN1A locus may play distinct roles in the DNA damage response from the CDKN1A protein, p21.
Figure 5. ncRNAs at CDKN1A locus are induced by DNA damage.
(a) Top: map of all detected transcripts at the CDKN1A promoter. Middle two tracks: Example of RNA hybridization intensity in control or 24 hour doxorubicin treated (200ng/ml) human fetal lung fibroblasts. Note, not all DNA damage inducible transcripts are observed in one single time point. Bottom track: p53 ChIP-chip signal relative to input confirmed the p53 binding site immediately upstream of the CDKN1A TSS upon DNA damage. RACE clone of upst:CDKN1A:-4845 closely matches predicted transcript on tiling array. See also: Supplementary Fig S7.
(b) Quantitative RT-PCR of lncRNAs shows coordinate induction or repression across a 24 hour time course of doxorubicin treatment. A cluster of lncRNAs transcribed from the CDKN1A locus are induced.
(c) Expression of transcripts from the CDKN1A locus over a 24 hour time course after doxorubicin-treatment of normal human fibroblasts (FL3). See also: Supplementary Fig S6.
(d) Northern blot of PANDA confirms transcript size of 1.5Kb.
(e) Doxorubicin induction of PANDA requires p53 but not CDKN1A. Mean + s.d. are shown, *p<0.05 relative to sictrl, student’s t-test.
(f) Expression of wild type p53 in p53-null H1299 cells restores DNA damage induction of CDKN1A and PANDA. p53(V272C) loss-of-function mutant fails to restore induction, whereas a gain-of-function Li-Fraumeni allele, p53(R273H), selectively retains the ability to induce PANDA.
PANDA: a long noncoding RNA involved in the DNA-damage response
To investigate the functional relevance of these lncRNAs at the CDKN1A locus, we selected upst:CDKN1A:-4845, hereafter termed PANDA (P21 Associated NcRNA DNA damage Activated) for further analysis. PANDA is located approximately 5 kilobases upstream of the CDKN1A TSS, coincides with a cluster of previously annotated ESTs, and is evolutionarily conserved (Supplementary Figure 5) Although the PANDA locus intersects a computationally predicted pseudogene of LAP3, quantitative RT-PCR demonstrated that PANDA was specifically induced by DNA damage, whereas LAP3 expression did not significantly change, confirming that the change in expression detected by the tiling array was not due to cross-hybridization with LAP3 (Supplementary Figure 6). Furthermore, the CSF score of PANDA, 9.3, indicated very low protein coding potential compared to LAP3 (CSF range 117-1343 for its 13 exons). Rapid amplification of 5′ and 3′ complementary DNA ends (RACE) and Northern blot analysis revealed a 1.5 kilobase transcript that is divergently transcribed from CDKN1A, antisense of the predicted LAP3 pseudogene (Fig 5d and Supplementary Figure 7). Thus, PANDA is a 5′-capped and polyadenylated non-spliced lncRNA that is transcribed antisense to CDKN1A.
Since p53 is a positive regulator of CDKN1A during the DNA damage response, we asked whether p53 also regulates PANDA expression. ChIP-chip analysis confirmed the p53 binding site immediately upstream of the CDKN1A TSS (Fig 5a)35. PANDA and CDKN1A are diametrically situated 2.5kb from this intervening p53 binding site, which supports the possibility of p53 co-regulation. Indeed, siRNA-mediated knockdown of p53 prior to DNA damage inhibited the induction of PANDA by 70% 24-hours post-DNA damage (Fig 5e and Supplementary Figure 8), similar to its effect on CDKN1A. In contrast, RNA interference of CDKN1A had no effect on PANDA expression, indicating that PANDA is not a linked transcript of CDKN1A nor is PANDA expression dependent on p21. PANDA level shows a trend of lower expression in human primary breast tumors harboring inactivating mutation in TP53 as determined by exon 2-11 DNA sequencing 36 (Supplementary Figure 9a). Further, complementation of p53-null H1299 lung carcinoma cells by wild type p53--but not loss of function p53(V272C) mutant--restored DNA damage-inducible expression of PANDA (Fig. 5f). Intriguingly, a gain-of-function p53(R273H) mutant, observed in Li-Fraumeni syndrome37, abrogated the ability to induce CDKN1A but selectively preserved the ability to induce PANDA (Fig. 5f). Selective induction of PANDA without concordant CDKN1A expression was also observed in metastatic ductal carcinomas but not normal breast tissue (Supplementary Figure 9b).
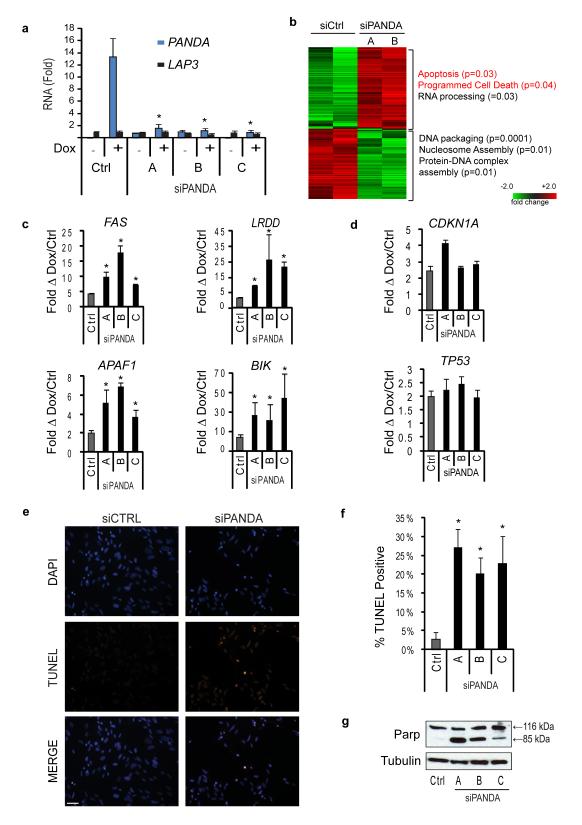
Next, we addressed whether PANDA affects the DNA damage response. We transduced human fetal fibroblasts (FL3) with custom siRNAs targeting PANDA and then applied doxorubicin for 24 hours following the knockdown (Fig 6a). Global gene expression analysis showed that 224 genes were induced and 193 genes were repressed at least 2-fold by PANDA knockdown (Fig. 6b). Genes induced by PANDA knockdown are significantly enriched for those involved in apoptosis, such as Gene Ontology terms cell death (p<0.04) and apoptosis (p<0.03) (Fig 6b). Quantitative RT-PCR confirmed that PANDA depletion induced several genes encoding canonical activators of apoptosis, including APAF1, BIK, FAS, and LRDD (Fig 6c). On the other hand, expression of neither CDKN1A itself nor TP53 was affected by PANDA depletion (Fig. 6d), suggesting that PANDA is a p53 effector that acts independently of p21CDKN1A.
Figure 6. PANDA lncRNA regulates apoptotic response to DNA damage.
(a) siRNA knockdown of PANDA in the presence of DNA damage with doxorubicin in human fibroblasts (FL3), Custom siRNAs specifically target PANDA with no discernable effect on the LAP3 mRNA. Mean + s.d. are shown in all bar graphs. * indicates p<0.05 compared to siCTRL for all panels, student’s t-test
(b) Heat map of gene expression changes with siPANDA relative to control siRNA at 24 hours of doxorubicin treatment in FL3 cells.
(c) Quantitative RT-PCR of canonical apoptosis pathway genes reveals induction with siPANDA relative to control siRNA at 28 hours of doxorubicin treatment (FL3).
(d) Quantitative RT-PCR of CDKN1A and TP53 in FL3 cells reveal no reduction in expression with siPANDA relative to control siRNA.
(e) TUNEL immunofluorescence of control and siPANDA FL3 fibroblasts at 28 hours of doxorubicin treatment. Scale bar= 20μm.
(f) Quantification of 3 independent TUNEL assays. p<0.05 for each siPANDA sample compared to siCTRL, student’s t-test.
(g) Western blot of PARP cleavage in control and PANDA siRNA FL3 fibroblasts at 24 hours of doxorubicin treatment.
DNA damage in human fibroblasts triggers p53-dependent G1 arrest, but not apoptosis38,39. Consistent with this finding, doxorubicin treatment in FL3 cells exposed to control siRNA had little to no apoptosis as measured by TUNEL. In contrast, PANDA knockdown resulted in five to seven-fold increased TUNEL-positive cells (Fig 6e, f). Immunoblot analysis of PARP, a caspase substrate and marker of apoptosis, revealed PARP cleavage only in PANDA depleted cells (Fig 6g). In contrast, six additional siRNAs targeting other transcripts within the CDKN1A promoter had no effect on apoptosis (data not shown, Supplementary Figure 10). Thus, PANDA knockdown sensitized fibroblasts to DNA damaged-induced apoptosis. Altogether, these data suggest that in parallel with p53-mediated induction of CDKN1A for cell cycle arrest, p53-mediated induction of PANDA delimit apoptosis.
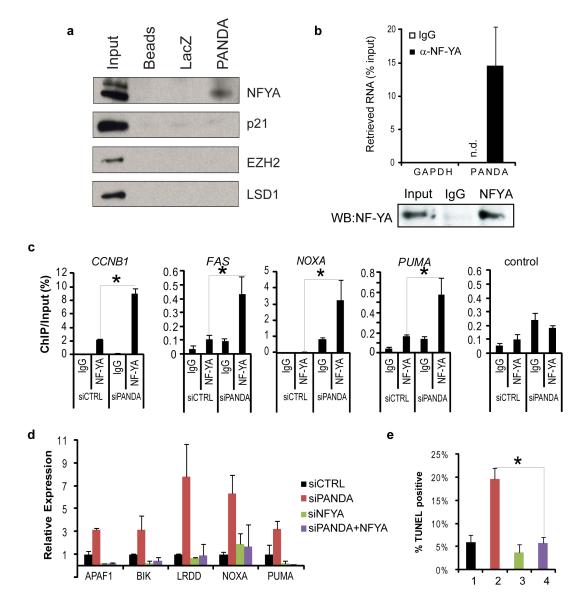
Core promoters of cell death genes downstream of p53 are distinguished from other p53 target genes by the binding site for the transcription factor NF-YA40 , and we reasoned that PANDA may affect NF-YA function. RNA chromatography41 using purified, in vitro transcribed PANDA, but not a 1.2 Kb LacZ mRNA fragment, specifically retrieved NF-YA from cellular lysates of human fibroblasts induced by DNA damage (Fig. 7a). PANDA did not retrieve other chromatin modification complexes that can bind other lncRNAs such as EZH2 or LSD142,43, nor p21, illustrating the specificity of the interaction. Immunoprecipitation of NF-YA from doxorubicin-treated primary human lung fibroblasts specifically retrieved endogenous PANDA (Fig.7b). NF-YA is a nuclear transcription factor that activates the p53-responsive promoter of FAS upon DNA damage40. Depletion of PANDA substantially increased NF-YA occupancy at target genes, including CCNB1, FAS, PUMA (also known as BBC3), and NOXA (also known as PMAIP1) (Fig. 7c). Moreover, concomitant knockdown of NF-YA and PANDA substantially attenuated induction of apoptotic genes and apoptosis as measured by TUNEL, indicating that NF-YA is required in part for cell death triggered by loss of PANDA (Fig. 7d and 7e). Thus, PANDA binding to NF-YA may evict or prevent NF-YA binding to chromatin. These data suggest that DNA damage activates p53-mediated transcription at CDKN1A and PANDA that functions synergistically to mediate cell cycle arrest and survival. CDKN1A mRNA produces p21 to mediate arrest, while PANDA impedes NF-YA activation of apoptotic gene expression program (Fig. 7f).
Figure 7. PANDA regulates transcription factor NF-YA.
(a) RNA chromatography of PANDA from doxorubicin-treated FL3 cell lysates. Retrieved proteins are visualized by immunoblot analysis.
(b) Immunoprecipitation of NF-YA from doxorubicin-treated FL3 lysates specifically retrieves PANDA as measured by qRT-PCR. Bottom: Immunoblot confirms IP of NF-YA.
(c) ChIP of NF-YA in FL3 fibroblasts nucleofected with siCTRL or siPANDA. ChIP-qPCR at known NF-YA target sites on promoters of CCNB1, FAS, NOXA, PUMA, or a control downstream region in FAS promoter lacking the NF-YA motif. Mean + s.d. are shown in all bar graphs. * indicates p<0.05 , student’s t-test
(d) Concomitant knockdown of NF-YA attenuates induction of apoptotic genes by PANDA depletion as measured by qRT-PCR. For knockdown efficiency see Fig. S11.
(e) Concomitant knockdown of NF-YA rescues apoptosis induced by PANDA depletion. Quantification of TUNEL staining is shown. Figure legend as in (D).
DISCUSSION
A regulatory network of noncoding transcription in cell cycle promoters
Recent studies have revealed that a surprisingly large fraction of mammalian genomes is transcribed. In addition to small noncoding RNAs, long noncoding RNAs can be produced from gene promoters, enhancers, as well as stand-alone intergenic loci14,15,17. New approaches are needed that not only identify ncRNAs, but also provide insight into their potential biological function. Using an ultra-high resolution tiling array, we interrogated the transcriptional landscape at cell cycle promoters in 108 samples that represent diverse perturbations. The ability to interrogate numerous and diverse biological samples in a rapid and economical fashion is advantageous for at least two reasons. First, many of the noncoding transcripts are induced only in highly specific conditions, and may have been missed if only a few conditions were surveyed. Of the 216 new noncoding transcribed regions we identified, on average only 73 of these are transcribed in any one biological sample. Second, comparison of lncRNA profiles amongst these diverse samples highlighted unexpected similarities in cell cycle promoter states among distinct perturbations. For instance, we identified a similarity of promoter states among ES cells, tumors induced by MYC, and epithelial progenitors depleted of differentiation regulator p63. Likewise, authentic human tumors can be classified based on the similarity of their promoter states to that of cells with defined oncogenic perturbation.
Noncoding transcription through regulatory elements may affect gene activity in a variety of ways. The act of transcription may open compacted chromatin over regulatory sequences, or compete with transcription factor binding (so called transcriptional interference). In addition, the ncRNA product may modulate neighboring gene expression in cis21,44, affect distantly located genes in trans22, or even serve as a target for regulation by small regulatory RNAs45,46. Because these different mechanisms predict distinct relationships between levels of ncRNAs and cognate mRNAs, we compared ncRNA and mRNA expression profiles across our samples. We found that most promoter ncRNAs are neither positively nor negatively correlated in expression with their neighboring mRNA, but are rather correlated in expression with genes located elsewhere in the genome. The genes co-expressed (and presumably co-regulated) with promoter ncRNAs function in specific biological pathways, including cell cycle, DNA damage response, stem cell differentiation, and have been associated with cancer prognosis. Quantitative RT-PCR analysis further validated that many of these ncRNAs are periodically expressed in the cell cycle, are regulated in response to DNA damage, ES cell differentiation, and are differentially expressed in human cancers. These findings suggest that cell cycle ncRNAs may participate in gene regulation in trans. In addition, noncoding transcription of cell cycle promoters may be a form of regulatory anticipation or feedback to modulate the chromatin state of cell cycle promoters.
Parallel functions of coding and noncoding RNAs driven by cell cycle promoters
Our results suggest that the human genome is organized into genomic units that code for multiple transcripts that function in the same biological pathways (Fig. 7f). 49 of 56 cell cycle protein-coding gene loci had at least one detected lncRNA and an average of four lncRNAs within 10 kilobases upstream and 2 kilobases downstream of the TSS. At the CDKN1A promoter, five lncRNAs, similar to the CDKN1A mRNA itself, are induced by DNA damage. One of these lncRNAs, which we named PANDA, is a non-spliced 1.5 kilobase ncRNA that is transcribed antisense to CDKN1A and is induced with slower kinetics than that of CDKN1A. Loss of function and complementation experiments demonstrate that PANDA induction during DNA damage is p53-dependent. In contrast, depletion of CDKN1A or depletion of PANDA had no effect on each other’s response to DNA damage, indicating that their induction by p53 occurs in parallel. PANDA inhibits the expression of apoptotic genes by sequestering the transcription factor NF-YA from occupying target gene promoters. While CDKN1A encodes a cell cycle inhibitor to mediate cell cycle arrest, PANDA promotes cell survival by impeding the apoptotic gene expression program. This linkage can be apparently exploited by tumors: The ability of Li-Fraumeni gain-of-function p53 mutant R273H to selectively retain PANDA induction instead of CDKN1A in effect uncouples cell survival from cell cycle arrest, which was similarly observed in metastatic ductal carcinomas. Thus, lncRNAs like PANDA may provide new explanations for human cancer susceptibility.
Intriguingly, Huarte et al. recently identified a distinct long intergenic noncoding RNA located 15 kilobases upstream of CDKN1A, named lincRNA-p21, that is induced by p53 and mediates p53-dependent gene repression24. Thus, the regulatory sequence upstream of the CDKN1A gene drives the expression of multiple coding and noncoding transcripts that cooperate to regulate the DNA damage response (Fig. 7f). These findings provide a vivid example that shows the blurring boundary between “genes” and “regulatory sequences”47.
Our study provides an initial catalogue of lncRNAs in cell cycle promoters that may play diverse functions. At a minimum, promoter ncRNA expression provides a convenient means of tracking the chromatin state of promoters, which may be of use in cancer biology and regenerative medicine. Future studies are needed to pinpoint the functions of these and likely other ncRNAs emanating from regulatory sequences.
METHODS
Tiling array design and RNA Hybridization
A custom tiling array (Roche Nimblegen) was designed at 5 basepair resolution across 25kb of the 9p21 region (which encompasses CDKN2a, p14ARF, and CDKN2b), as well as from 10kb upstream to 2kb downstream of each TSS from 53 other cell cycle genes including cyclins, CDKs, and CDKIs (Table S1). In addition, the HOXA and HOXD loci were placed on the array as a control. Briefly, RNA was amplified (MessageAmp Kit, Ambion), reverse transcribed (Retroscript Kit, Ambion), labeled, and hybridized according to the standard Nimblegen protocol.
Peak Calling
Robust multichip average (RMA) normalized single channel data from each array was subjected to peak calling using the Nimblescan program (Roche Nimblegen) with a window size = 50. Peaks with a peak score > 10 were considered significant transcriptional units. Peak calls from all 55 array samples were clustered using Galaxy 2,48, and only transcripts present in a minimum of 10% of the samples were considered for further analysis. Transcripts were annotated as following – “genomic location (upstream of TSS of cell cycle protein-coding gene = upst; exon of cell cycle protein-coding gene = exon; intron of cell cycle protein-coding gene = int; downstream of cell cycle protein coding gene = dst)” : “gene symbol of nearest mRNA” : “distance from TSS”.
Measuring Protein Coding Potential
To assess the coding potential of the novel transcribed regions, we evaluated the evolutionary signatures in their alignments with orthologous regions in 20 other sequenced placental mammalian genomes using the Codon Substitution Frequencies (CSF) method 30,49,50, which has also been applied to assess novel transcribed regions in mouse 14. CSF produces a score for any region in the genome considering all codon substitutions observed within its alignment, based on the relative frequency of similar substitutions in known coding and non-coding regions. Briefly, it performs a statistical comparison between two empirical codon models 51, one estimated from alignments of known coding regions and the other based on non-coding regions, and reports a likelihood ratio that quantifies whether the protein-coding model is a better explanation, while controlling for the overall level of sequence conservation30.
Module Map analysis
We generated a module map of the ncRNAs versus the protein-coding genes by computing the Pearson correlations for all pairwise combinations based on expression across 17 different samples. This map was clustered and visualized using the program Genomica (http://genomica.weizmann.ac.il/). For each ncRNA, we then defined gene sets of the protein coding genes that had a Pearson correlation that was greater than or less than 0.5 with that ncRNA. To determine functional associations, we then generated a module map of these ncRNA gene sets with Gene Ontology Biological Processes gene sets (Fig 3C) and with curated gene sets of metabolic and signaling pathways and biological and clinical states from the Molecular Signatures Database (MSigDB c2 collection) (Fig S4) 33. P-value of enrichment was determined by the hypergeometric distribution, and a false discovery rate (FDR) calculation was used to account for multiple hypothesis testing (p<0.05, FDR<0.05).
Tissue samples and cells
Informed consent was obtained for tissue donation as well as approval from institutional review boards. Human primary breast tumors from the Netherlands Cancer Institute 52, and normal breast tissues and metastatic breast tumors from the Johns Hopkins University Rapid Autopsy Program (Gupta et al., 2010) are as described. Human fetal pancreata were obtained from the Birth Defects Research Laboratory, University of Washington (Seattle, WA). Staged fetal pancreata were processed within 24 hours of receipt, minced, washed and processed for RNA isolation using standard methods. Human fetal lung fibroblasts FL3 (Coriell AG04393) or foreskin fibroblasts (ATCC CRL2091) were cultured in 10% FBS (Hyclone), 1% penicillin-streptomycin (Gibco) at 37C in 5% CO2.
PANDA Cloning and sequence analysis
3′ and 5′ RACE was performed using the FirstChoice RLM-RACE Kit (Ambion). RNA was extracted from 200ng/ml doxorubicin (Sigma) treated human fetal lung fibroblasts , polyA selected using the PolyA Purist-MAg kit (Ambion), and RLM-RACE was performed according to the standard manufacturer’s protocol.
RT-PCR
Total RNA was extracted from cells using the Trizol reagent (Invitrogen) and the RNeasy Mini Kit (Qiagen) and genomic DNA was eliminated using Turbo DNA-free (Ambion). RT-PCR using 50-250 ng of total RNA was performed using the One-Step RT-PCR Master Mix (Applied Biosystems) using Taqman Gene Expression Assays and normalized to GAPDH. Strand specific RT-PCR for PANDA was performed using the One Step RT-PCR Master Mix SYBR Green (Stratagene)).
TaqMan® custom ncRNA Assays
A panel of TaqMan® custom ncRNA assays was developed targeting 60 of the 219 novel transcribed regions using “Single-exon” design mode. The transcript specificity and genome specificity of all TaqMan assays were verified using a position specific alignment matrix to predict potential cross-reactivity between designed assays and genome-wide non-target transcripts or genomic sequences. For gene expression profiling of these ncRNAs across different conditions, cDNAs were generated from 50ng of total RNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Foster City, CA). The resulting cDNA was subjected to a 14-cycle PCR amplification followed by real-time PCR reaction using the manufacturer’s TaqMan® PreAmp Master Mix Kit Protocol (Life Technologies, Foster City, CA). Two replicates were run for each gene for each sample in a 384-well format plate on 7900HT Fast Real-Time PCR System (Life Technologies, Foster City, CA). PPIA was used as an endogenous control for normalization across different samples.
Northern Blot
5 ug of polyA RNA was obtained using RNeasy Kit (Qiagen) and PolyA Purist Mag (Ambion). Northern Blot was performed using NorthernMax Kit (Ambion) following the standard manufacturer’s protocol. Probes were generated with full length PANDA using the Prime-It RmT Random Primer Labeling Kit (Agilent).
Antibodies
The following antibodies were used for Chromatin Immunoprecipitation Assays: anti-H3K4me3 (Abcam ab8580), anti-H3K35me3 (Abcam ab9050), anti-p53 (Abcam ab28). Western blots were performed using anti-PARP (Cell Signal 9542),anti-B-tubulin (Abcam ab6046), anti LSD1 (ab17721), anti EZH2 (Cell Signal AC22), anti p21 (Santa Cruz Biotech), anti NF-YA (Santa Cruz Biotech H-209).
RNA Interference
Human fetal lung fibroblasts were transfected with 50 nM of onTargetPLUS siRNAs (Dharmacon) targeting PANDA (Supplementary Table 5). Validated siRNAs for mRNAs were obtained from Ambion (Supplementary Table 5).
TUNEL
TUNEL assays were performed using the In Situ Cell Death Detection Kit, TMR Red (Roche). Human fetal lung fibroblasts were cultured on chamber slides (Lab-Tek), treated with 200ng/ml doxorubicin (Sigma) for 24 hours, fixed with methanol at -20C for 10 minutes, and incubated with the TUNEL labeling mixture for one hour at 37C. Slides were then washed with PBS and mounted in Prolong® Gold antifade reagent with DAPI (Invitrogen) and imaged at 20x magnification.
RNA Immunoprecipitation
10 million cells were treated with 200ng/ml doxorubicin for 16 hours, trypsinized, and crosslinked with 1% formaldehyde for 10 minutes followed by the addition of .125 M Glycine for 5 minutes. After 2 PBS washes, cells were lysed with 2x volume of Buffer A (10mM HEPES pH 7.5, 1.5 mM MgCl2, 10 mM KCl, .5 mM DTT, 1mM PMSF) for 15 minutes on ice at 150 RPM. NP-40 was added to a final concentration of .25% for 5 minutes on ice. Lysates were centrifuged for 3 minutes at 2000 RPM, and the supernatant (cytosol) was collected. Next, an equal volume of Buffer C as Buffer A was added to the pellet for 20 minutes with frequent vortex (20 mM HEPES pH 7.5, 10% Glycerol, .42M KCl, 4 mM MgCl2, .5 mM DTT, 1 mM PMSF). Nuclear lysates were dounced for 5 seconds using a motorized pestle and sonicated for 7 minutes using a Diagenode Sonicator (30 seconds on, 30 seconds off, power setting H). Nuclear and cytoplasmic lysates were combined and centrifuged for 15 minutes at 13K RPM. Supernatants were transferred into Micro spin columns (Pierce 89879) and 2 ug of antibody was added and incubated overnight. 10 ul of Protein A/G Ultralink Resin (Pierce 53132) was washed 3x with RIP wash buffer (50 mM TrisHcl pH 7.9, 10% glycerol, 100mM KCl, 5mM MgCl2, 10 mM B-me, and .1% NP-40) and added to the Immunoprecipitation reaction for 1 hr at 4C. Samples were washed 4x with RIP wash buffer and 2x with 1M RIPA (50 mM Tris pH 7.4, 1M NaCl, 1 mM EDTA, .1% SDS, 1% NP-40, .5% sodium deoxycholate, .5mM DTT and 1 mM PMSF). Beads were resuspended in 200 ul 150mM RIPA (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, .1% SDS, 1% NP-40, .5% sodium deoxycholate, .5mM DTT and 1 mM PMSF) + 5 ul Proteinase K (Ambion) and incubated for 1 hr at 45C. 1 ml of Trizol was added to the sample and RNA was extracted using the RNEasy Mini Kit (Qiagen) with the on column DNAse digest (Qiagen).
RNAse mediated RNA chromatography
RNAse mediated RNA chromatography 41 was performed as previously described with the following modifications: 6 pmols of RNA (PANDA or a 1.2 KB fragment of LacZ) were used per reaction. RNA was folded (90C for 2 minutes, ice for 2 minutes, supplied with RNA structure buffer (Ambion), and shifted to room temperature for 20 minutes prior to conjugation to beads. RNAse digestion was performed with 5 ul of Rnase A/T1 cocktail (Ambion) and 2 ul of Rnase V1 (Ambion).
Cellular lysates were prepared as follows: 10 million doxorubicin treated cells (16 hrs) were incubated in 200 ul PBS, 600 ul H20, and 200 ul nuclear lysis buffer (1.28 M sucrose; 40 mM Tris-HCl pH 7.5; 20 mM MgCl2; 4% Triton X-100) on ice for 20 min. Nuclei were pelleted by centrifugation at 2,500 G for 15 min. Nuclear pellet was resuspended in 1 ml RIP buffer (150 mM KCl, 25 mM Tris pH 7.4, 0.5 mM DTT, 0.5% NP40, 1 mM PMSF and protease Inhibitor (Roche Complete Protease Inhibitor Cocktail Tablets)). Resuspended nuclei were sheared using a motorized douncer for 5 seconds. Nuclear membrane and debris were pelleted by centrifugation at 13,000 RPM for 10 min.
Chromatin Fractionation was performed as previously described53.
Chromatin Immunoprecipitation (ChIP) was performed as previously described54. Q-PCR primers for FAS and CCNB1 and FAS-control NF-YA binding sites were obtained from Morachis et al.40 Primers for PUMA and BAX were designed to surround the NF-YA consensus motif CCAAT (Supplementary Table 5).
Supplementary Material
Figure 8. Model of coding and noncoding transcripts at the CDKN1A locus coordinating the DNA damage response.
Upon DNA damage, p53 binding at the CDKN1A locus coordinately activates transcription of CDKN1A as well as noncoding transcripts PANDA and linc-p21. CDKN1A mediates cell cycle arrest, PANDA blocks apoptosis through NF-YA, and linc-p21 mediates gene silencing through recruitment of hnRPK.
Acknowledgements
We thank J. Rinn, M. Guttman and A. Regev for discussions, L. Attardi for careful reading of the manuscript, and P. Khavari for reagents. Authors Y.W., B.K., and Yu W. are employees of Life Technologies. This work was supported by grants from NIH/NIAMS (K08-AR054615 to D.J.W.), NIH/NCI (R01-CA118750 to H.Y.C.), NIH/NCI (RO1-CA130795 to M.L.W.), Juvenile Diabetes Research Foundation (S.K. K. and H.Y.C.) and American Cancer Society (H.Y.C.). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute. T.H. is supported by the Stanford Graduate Fellowship, NSF Graduate Research Fellowship, and Department of Defense (DoD) National Defense Science & Engineering Graduate Fellowship (NDSEG).
Footnotes
Data availability Tiling and microarray data are available at Gene Expression Omnibus (GSE28631). Sequence for human PANDA RNA has been deposited with GenBank under the accession number JF803844.
Contributions: H.Y.C. and D.J.W. initiated project; H.Y.C., D.J.W., and T.H. designed experiments, T.H. performed the experiments and computational analysis; Y.W., Yu W., and B.K. conducted high-throughput Taqman RT-PCRs; M.F.L. and M.K. contributed CSF analysis. These authors contributed samples or reagents: A.K., Y.K., G.G., H.M.H., N.S., C.U., P.W., A.L., S.K.K., M.V., M.V., S.S., M.L.W., and Y.X. The manuscript was prepared by H.Y.C., T.H., and D.J.W with input from all co-authors.
REFERENCES
- 1.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–6. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P. Non-coding RNA transcription: turning on neighbours. Nat Cell Biol. 2008;10:1023–4. doi: 10.1038/ncb0908-1023. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Spec No 1):R121–32. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 9.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–8. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 11.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–65. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–5. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 14.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 16.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasmant E, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–9. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 20.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–42. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 26.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- 28.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–71. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bock C, Halachev K, Buch J, Lengauer T. EpiGRAPH: user-friendly software for statistical analysis and prediction of (epi)genomic data. Genome Biol. 2009;10:R14. doi: 10.1186/gb-2009-10-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin M, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein-coding and non-coding regions. Nature Precedings. 2010 doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–8. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 36.Geisler S, et al. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 2001;61:2505–12. [PubMed] [Google Scholar]
- 37.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci U S A. 1995;92:8493–7. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–51. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 40.Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010;24:135–47. doi: 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michlewski G, Caceres JF. RNase-assisted RNA chromatography. Rna. 2010;16:1673–8. doi: 10.1261/rna.2136010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104:12422–7. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz JC, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–9. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 48.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics Chapter. 2007;10(Unit 10):5. doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin MF, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–36. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin MF, Deoras AN, Rasmussen MD, Kellis M. Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput Biol. 2008;4:e1000067. doi: 10.1371/journal.pcbi.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosiol C, Holmes I, Goldman N. An empirical codon model for protein sequence evolution. Mol Biol Evol. 2007;24:1464–79. doi: 10.1093/molbev/msm064. [DOI] [PubMed] [Google Scholar]
- 52.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 53.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–12. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.