Abstract
Th17 cells are an effector lineage of CD4 T cells that can contribute to protection against microbial pathogens and to the development of harmful autoimmune and inflammatory conditions. An increasing number of studies suggest that Th17 cells play an important protective role in mobilizing host immunity to extracellular and intracellular microbial pathogens such as Candida and Salmonella. Furthermore, the generation of Th17 cells is heavily influenced by the normal microbial flora highlighting the complex interplay between harmless microbes, pathogens, and host immunity in the regulation of pathogen-specific Th17 responses. Here, we review current understanding of microbe-induced Th17 cells in the context of infectious and inflammatory disease.
Introduction
Naïve CD4 T cells exit the thymus and, in the absence of cognate antigen, repeatedly travel through peripheral blood and lymphatic vessels (1). Initial activation of CD4 T cells occurs within secondary lymphoid organs and requires recognition of antigenic peptides presented by MHC class-II molecules on the surface of migrating or resident dendritic cells (2). Throughout the period of activation, CD4 T cells integrate signals from the T cell receptor, co-stimulatory molecules, and cytokine receptors to ultimately determine which effector capabilities will be acquired (3). Thus, antigen presenting cells and the local stimulating environment can shape the development of the effector T cell pool in an appropriate manner. In infectious disease models this plasticity allows the development of pathogen-specific effector T cells that are tailored to combat different types of microbial pathogen.
Heterogeneity within the CD4 T cell compartment was initially reported by investigators studying antibody or delayed-type hypersensitivity responses after immunization with protein or bacterial products (4–6). This led to the discovery of CD4 Th1 and Th2 populations, which represent distinct T effector lineages with differential ability to produce anti-microbial cytokines (7, 8). Th1 cells are defined by the expression of the transcription factor T-bet and secretion of IFN-γ, a cytokine that can activate macrophages to kill intracellular pathogens (9). Th2 cells are characterized by the expression of GATA-3 and production of IL-4, IL-5, and IL-13, cytokines that are critical for eradication of many extracellular parasites (10, 11). Although the definition of Th1 and Th2 provided an important conceptual framework for understanding effector CD4 T cell development, a much wider range of effector lineages is now appreciated (12). In this review we focus on the development of Th17 and how they interact with the microbes that colonize or infect the mammalian host.
Discovery and importance of Th17 cells
Th1 and Th2 differentiation can be examined by in vitro stimulation of naïve antigen-specific CD4 T cells in the presence of IL-12 and IL-4, cytokines that are normally induced by the innate immune response to microbial ligands (13). In one particular study, it was observed that if IL-12 was replaced by Borrelia burgdorferi lysate, mycobacterial lysate, or IL-6, this caused the development of effector CD4 T cells that produced IL-17 (14). Furthermore, these IL-17-producing CD4 T cells co-expressed TNF-α and GM-CSF but did not express IFN-γ or IL-4, suggesting the development of an effector subset distinct from Th1 or Th2. Further evidence that IL-17-producing cells represented a novel subset came from the observation that IL-23 could promote the development of these cells (15). IL-23 is a member of the four-chain long helix bundle family of cytokines, which includes IL-6 and IL-12. IL-23 shares the p40 subunit with IL-12 but has a unique p19 subunit (16), and the discovery that models of autoimmunity were dependent on IL-23p19 rather than IL-12p35 initiated a re-evaluation of prior studies using p40-deficient animals (17, 18). The real notoriety of Th17 cells came with the discovery that IL-17-producing T cells, driven by IL-23, are the major contributors to pathogenesis of autoimmune inflammatory diseases (19). Previously Th1 cells were thought to drive autoimmunity, but many subsequent studies in mouse models and human disease brought the realization that Th17 cells represent a new and important target for therapy of psoriasis, inflammatory bowel disease, uveitis, multiple sclerosis and arthritis (20, 21). Genetic polymorphisms or deficiencies that modulate the IL-23/Th17 axis, including IL-23R, CARD9, STAT3, and AIRE result in enhanced susceptibility to inflammatory disease, and the importance of Th17-related targets in human autoimmunity is now being validated in clinical trials targeting p40, IL-17, IL-17RA and IL-23(p19).
Initiating autoimmune disease is clearly not the raison d’etre of IL-17-producing CD4 T cells, and a more complete picture is now emerging of how Th17 cells can contribute to host defense against microbial pathogens. (22, 23). Several studies have detected IL-17-producing CD4 T cells in diverse infectious disease models and a theme that has emerged is that this lineage contributes to host defense against extracellular microbes (24, 25). The cytokines produced by Th17 cells are well suited to this role: IL-17 and TNFα can synergize to activate epithelial cell production of anti-microbial peptides, monocyte-recruiting chemokines, while G-CSF additionally drives granulopoeisis (26). IL-22 produced by Th17 cells promotes the production of anti-microbial peptides and the proliferation of epithelial cells, which can be important for repairing damage inflicted by microbial invasion (27). GM-CSF and IL-17 also activate monocytes and neutrophils to promote phagocytosis of microbes and clearance of the infection. However, it should be emphasized that Th17 cell development is not limited to extracellular infections and these cells have been observed in numerous intracellular bacterial, viral, and extracellular parasite infection models (28–30). The potent ability of Th17 cells to elicit chemokine production in tissue sites, including Th1-recruiting chemokines such as CXCL13 makes them ideally suited as first responders during re-infection (31). In addition, IL-17 can promote IL-12 production through regulation of IL-10 in dendritic cells during infection with Mycobacterium tuberculosis (32) and Francisella tularensis (33), two intracellular infections that require both IL-17A and Th1 responses for optimal pathogen control. Thus, while Th17 cells are often associated with extracellular infection, they are a CD4 lineage that is commonly elicited in response to a wide variety of pathogens. In the latter half of this review we will focus on Th17 immunity to examples of extracellular and intracellular pathogens: Candida and Salmonella.
Th17 cell differentiation
Th17 cells were officially recognized as a distinct subset of helper T cells following seminal studies demonstrating that differentiation of IL-17 producing CD4+ T cells is dependent on STAT3 and RORγt expression, but independent of putative Th1 or Th2 transcription factors (34–36). TGFβ, IL-6, and IL-21 drive the activation of STAT3, which can subsequently activate RORγt (37, 38). TGFβ together with IL-6 and IL-1 promote expression of RORγt, and IL-6 drives the expression of IL-23R (39). IL-23 subsequently acts on these early developing Th17 cells to drive effector cell differentiation and expansion (40). Some controversy persists over the precise role of TGFβ in Th17 cell differentiation in mice and humans. TGFβ and its receptor are both required for T cell intrinsic Th17 development in mouse models of colitis and encephalomyelitis (41–43). However, high concentrations of TGFβ and IL-6 stimulate production of IL-10 and inhibit the pathogenic functions of murine Th17 cells activated in vitro (44, 45). In addition, human Th17 cell differentiation does not seem to require the addition of TGFβ, which may even suppress their development (46, 47).
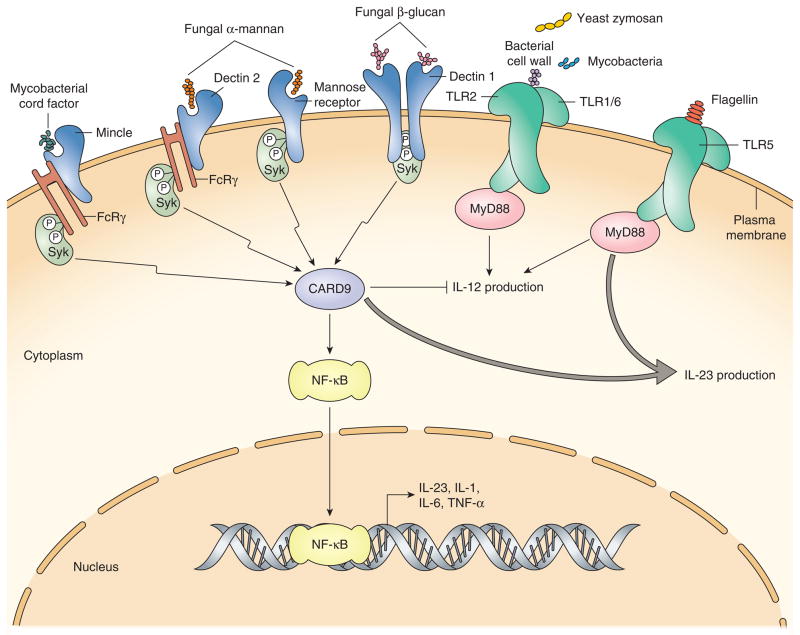
Classical Toll-like receptor (TLR) activation drives production of a variety of inflammatory cytokines, but the triggering of receptors of the C-type lectin family (CLRs) seems to provide a more specific Th17-inducing signal (Figure 1). Fungal components bind to Dectin1, Dectin2, and Mincle leading to recruitment of the tyrosine kinase Syk, activation of the adaptor CARD9 and downstream signaling via NF-KB, resulting in upregulation of IL-23, IL-1, IL-6 and TNF (48–51). Humans with loss-of-function mutations in CARD9 develop chronic mucocutaneous candidiasis and have reduced Th17 cells (52). Microbial ligands that induce Th17 responses via TLRs and CLRs have primarily been defined for Candida, but similar ligands likely exist for bacteria and viral pathogens. Mycobacterial cord factor was recently shown to bind to Mincle and activate Syk/CARD9 signaling to promote Th17 responses in a BCG vaccine model (53), and this may explain the efficacy of heat-killed Mycobacterium bovis as a Th17-inducing adjuvant. Furthermore, yeast, Mycobacterium, and heat-killed Streptococcus pneuomoniae activate TLR2 to promote Th17 development (54, 55), and flagellin expression by segmented filamentous bacteria also induces intestinal Th17 responses (56). Although CLR signaling can occur independently of TLRs, collaboration between TLRs and Dectin-1 signaling enhances the production of IL-6 and IL-23 (51, 57) and conversely it has been proposed that CLR signaling modulates TLR signaling to downregulate the production of IL-12 and favor Th17-inducing cytokines such as IL-23 (49, 57, 58).
Figure 1. Induction of Th17-promoting cytokines by microbial products.
C-type lectins Mincle, Dectin 1 and Dectin 2 as well as Mannose receptor are expressed on dendritic cells and macrophages and recognize fungal and mycobacterial components to activate Syk kinase and the CARD9 adaptor, leading to production of inflammatory cytokines that promote Th17 development in naïve T cells. Toll-like receptors (TLR) also recognize microbial products and induce production of both Th17 and Th1-promoting cytokines including IL-23 and IL-12. CLEC signaling modulates TLR signaling to downregulate Th1-promoting and favor Th17-promoting conditions, thus CLECs and TLRs co-operate to provide fine-tuning in the type of T helper response elicited.
Intestinal Th17 cells – the microbiome as regulator of tolerance verse autoimmunity
It has become increasingly clear that the resident bacterial population (the ‘microbiome’) in any given individual can profoundly impact their overall health and susceptibility to inflammatory and infectious disease (59). The microbiome is comprised of bacterial, viral, and eukaryote species that continuously colonize host mucosal and epithelial surfaces. While these microbes can be found associated with the skin, lung, genital, and oral cavity, we will focus our discussion here on the intestinal tract (60). It has long been considered that tolerance is induced to these constituent microbes, allowing them to reside in these various tissue sites without inducing inflammation (61). In fact, Inflammatory Bowel Disease (IBD) most likely arises as a consequence of developing an inappropriate immune response against host commensal flora (62). However, it is becoming increasingly clear that sub-clinical responses induced against microbes residing in the gut may also have far-reaching consequences throughout the body.
Endogenous microbial flora play a major role in the differentiation of Th17 cells and regulatory T cells in the small intestine under steady-state conditions, and antibiotic-treatment or the complete absence of bacterial flora reduces the number of Th17 cells in the gut (63). Indeed, re-colonization of the intestine with Clostridia-related commensal species, and segmented filamentous bacteria (SFB) in particular, is sufficient to induce CD4 T cells that produce IL-17 and IL-22, as well as other CD4 T helper lineages (64, 65). SFB are gram-positive anaerobes that are normally tightly attached to epithelial cells and are highly dependent on the host for many essential nutrients (66, 67). It is now apparent that intestinal flora, and especially SFB, can strongly modulate the induction of Th17 responses throughout the body, and thereby regulate the susceptibility of mice to arthritis (68), colitis (69, 70), diabetes (71), and EAE (72).
The specific bacterial products that drive homeostatic intestinal Th17 differentiation have not been well defined, however, flagellins are attractive candidates since they are expressed by SFB, recognized by TLR5-expressing intestinal phagocytes, induce IL-23 from CD103+ dendritic cells, and drive Th17 responses to enteric pathogens (56, 66, 67, 73, 74). Recent data suggest that IL-1β production by intestinal macrophages is induced by TLR recognition of microbial flora and that IL-1R signaling on intestinal T cells is required for homeostatic differentiation of intestinal Th17 cells (75). It remains to be determined how local production of IL-1β in the lamina propria directly affects naïve T cell differentiation within secondary lymphoid tissues, but it seems possible that Peyer’s patches could play a major role, since naïve CD4 T cells in this location are closely associated with SFB and lamina propria macrophages (64).
It is possible that the development of inflammatory Th17 responses to enteric pathogens differs substantially from the homeostatic development of Th17 cells in response to microbial flora. While IL-6 is reported to play a key role in Th17 development, this cytokine does not seem to be required for intestinal Th17 development under steady state conditions (75–77). Th17 cells generated in the presence of IL-23 or IL-6/TGF-beta develop different functional and pathogenic potential in autoimmune models (40, 45). Furthermore, recent data show that human Th17 cells induced by Candida albicans or Staphylococcus aureus have differential capacity to produce IFN-γ or IL-10 (78). Therefore, a degree of functional heterogeneity may exist between steady state Th17 and pathogen-specific Th17 cells, between Th17 cells induced by different classes of microbial pathogens, or between Th17 cells that are elicited at different anatomical sites. To complicate matters further, enteric pathogens such as Salmonella and Citrobacter can also modify the composition of the microbial flora (79, 80), and as a result may indirectly modulate steady state development of Th17 cells.
Th17 cells in defense against Candida
A requirement for IL-17 has been demonstrated in host resistance to extracellular pathogens such as Candida albicans (81). Mice with a deficiency in IL-17A or IL-17RA are more susceptible to intravenous Candida infection (82, 83), and IL-17RA- and IL-17RC-deficient mice are unable to clear oral infection with C. albicans (84, 85). In contrast, neither IL-17F or IL-22 are essential for host resistance to Candida infection (83, 84, 86). In the oral infection model, mice lacking IL-17RA signaling have decreased neutrophil infiltration (84), confirming a key role for IL-17A in recruiting phagocytes to the local site of infection. Candida-infected IL-23-, or IL-17RA-deficient mice also have reduced production of anti-microbial proteins, and saliva from these mice show reduced bactericidal activity (84). Thus, IL-17 plays an indirect role in defense against extracellular pathogens by recruiting phagocytes and inducing anti-microbial peptides at the site of infection. A key role for IL-17 in fungal defense has also been documented by genetic analysis of patients with increased susceptibility to chronic mucocutaneous candidiasis (CMC) (87). Patients with autosomal recessive IL-17RA or autosomal dominant IL-17F deficiency experience recurrent candida infections of the oral and genital mucosa (88). Similarly, patients with a gain of function mutation in STAT1 have increased responses to cytokines that impair the development of Th17 cells, and as a result are highly susceptible to CMC (89). Interestingly, patients with IL-17F deficiency produce a mutant IL-17F that can still form a heterodimer with IL-17A or wild-type IL-17F but with reduced functional activity (88), thus IL-17A rather than IL-17F may be critical for defense against Candida in humans. Together, these murine and human studies demonstrate that production of IL-17 at the site of fungal infection plays a key role in the resolution or susceptibility to disease. Studies with other many other microbes have established the paradigm that Th17 cytokines play a major role in mobilizing local host defense against infection with extracellular pathogens (23, 90).
Th17 cells in defense against Salmonella
Salmonella is a facultative intracellular pathogen that causes serious gastrointestinal and systemic infections (91). Like other intra-macrophage pathogens, Th1 cells are essential for protective immunity and mice with a genetic deficiency in T-bet or IFN-γ are unable to resolve Salmonella infection (92, 93). However, recent data demonstrate that Th17 cells develop during Salmonella infection and play an important role in host defense in the intestine.
Th17 cells can be induced when mice are infected with Salmonella via non-physiological routes, however, the population is typically small, and mice lacking IL-23R p19 or IL-17A experience only a minor delay in resolving primary infection (94, 95). In contrast, there is accumulating evidence that Th17 cells and associated cytokines play an important role in resistance to mucosal Salmonella infections (91, 96). Using a ligated loop model in rhesus macaques, it was noted that IL-22, IL-17, and IL-17 responsive genes were rapidly transcribed after Salmonella infection and prior depletion of CD4 T cells by SIV infection reduced this response considerably (97). SIV infection also reduced IL-17-producing CD4 T cells in the lamina propria and correlated with enhanced dissemination of Salmonella to mesenteric lymph nodes (97). This is an important finding since patients with HIV are highly susceptible to disseminated Salmonella infections and the loss of protective Th17 cells in the intestine may explain this susceptibility (96, 98). Consistent with this idea, patients with a primary genetic deficiency in Th17 development are also highly susceptible to disseminated Salmonella infections (96).
Given the rapid induction of Th17 cytokines in the ligated-loop model it is due to Salmonella-specific Th17 cells. It is more likely that intestinal Th17 cells are activated in a non-specific fashion in response to IL-1 or other inflammatory cytokines. Indeed, the early Th17 response to intestinal Salmonella infection requires expression of Myd88 and IL-1R (99). However, Nod1, Nod2, and the production of IL-6 can also cause rapid innate activation of Th17 cells (100), although IL-6 was dispensable in a different infection model (101). Non-cognate induction of effector T cells blurs the lines between innate and adaptive immunity to infection but is a common feature of immunity to Salmonella. Indeed, effector Th1 CD4 and CD8 T cells in Salmonella-infected mice rapidly secrete IFN-γ in response to innate stimuli (102, 103).
This rapid innate response does not mean that Salmonella-specific Th17 cells cannot also contribute to immunity at mucosal surfaces. A recent report documented simultaneous development of Salmonella-specific Th17 and Th1 cells in the intestine and spleen respectively after oral infection (74). These anatomically segregated Th17 and Th1 responses targeted different Salmonella antigens that were highly expressed in each tissue. The factors responsible for driving Salmonella-specific Th17 cell development in the intestine have not yet been clearly defined but previous reports show that dendritic cells conditioned with Salmonella direct Th17 and Th1 development in vitro (104, 105), and recent data show an intriguing role for B cell-derived IL-6 in the generation of Th17 cells (106). Interestingly, Salmonella-specific Th17 cells recognize Salmonella flagellin (74), an antigen that has intrinsic stimulatory capabilities and induces IL-1 and IL-6 production (107, 108). It is therefore possible that innate recognition of flagellin via TLR5 and/or NLRC4 is responsible for driving Salmonella-specific Th17 development in the intestine during infection.
As noted above, intestinal Th17 cells play a key role in early protective immunity by limiting dissemination of Salmonella to the mesenteric lymph node (97). However, it should be emphasized that there is yet no evidence for Th17 cells playing a protective role after Salmonella have spread to systemic tissues. Instead, intestinal Th17 cells prevent Salmonella dissemination using similar mechanisms implicated in immunity to extracellular bacteria. Intestinal epithelial cells can respond to local IL-17 and IL-22 in vitro by increasing production of anti-microbial proteins and chemokines during Salmonella infection (97, 109). These include anti-bacterial proteins such as, iNOS, mucin, calprotectin, RegIIIγ, and lipocalin-2, which can directly or indirectly limit bacterial growth (110). The chemokine CCL20 is also prominently produced in response to IL-17 and IL-22 and can recruit immature CCR6+ dendritic cells and presumably initiate adaptive responses (109, 111). The production of G-CSF and CXC chemokines also recruits neutrophils to the intestine and these engulf bacteria that have crossed the epithelial barrier (112).
Thus, the mechanism of Th17 defense against Salmonella penetration is similar to defense against extracellular bacteria. Indeed, while it is can be conceptually appealing to think of Th17 cells protecting against extracellular organisms and Th1 cells combating intracellular organisms, data from the Salmonella model suggest that Th17 cells can be highly active against intracellular organisms during the initial phase of trans-epithelial entry. Since most other microbial pathogens also have some degree of variation in life cycle stage or tissue tropism, a heterogenous T effector response is probably the norm rather than the exception.
Conclusions
We have discussed the role of microbes in the differentiation of Th17 cells in both autoimmune and infectious disease models and have focused on Candida and Salmonella as two examples of pathogens that induce Th17 responses. It is apparent that the differentiation of Th17 cells is surprisingly complex, involving the elicitation of multiple cytokines, most likely as a result of several microbial ligands activating receptors on DCs or other innate cells. There is also a degree of functional heterogeneity within the Th17 lineage that may mean that this lineage is adapted to fit the immune response to different pathogens or immunity at different anatomical sites. Greater understanding of how this complexity and functional heterogeneity can impact the specific effector response against different classes of pathogen is now required. Furthermore, the tripartite interactions that occur between host, microbiome, and microbial pathogen, have not been examined in any depth, and much greater understanding of how communication flows between each of these players in the regulation of Th17 cell development should be forthcoming. However, this may be challenging technically since it will likely require the definition of endogenous flora as a variable within many current experimental systems. Furthermore, the specific crosstalk that has evolved between pathogenic and non-pathogen organisms in the induction of human Th17 cells may be difficult to replicate in animal models unless greater attention is paid to the use of natural animal pathogens and routes of infection. However, greater understanding of these issues may lead to the development of vaccines and therapeutics for important infectious and inflammatory disease.
Acknowledgments
This work was supported by grants from the National Institutes of Health AI073672, AI055743, and HL112685 (to SJM).
References
- 1.Gowans JL, Knight EJ. The route of re-circulation of lymphocytes in the rat. Proc R Soc London Ser B. 1964;159:257–282. doi: 10.1098/rspb.1964.0001. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parish CR, Liew FY. Immune response to chemically modified flagellin. 3. Enhanced cell-mediated immunity during high and low zone antibody tolerance to flagellin. J Exp Med. 1972;135:298–311. doi: 10.1084/jem.135.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liew FY, Parish CR. Lack of a correlation between cell-mediated immunity to the carrier and the carrier-hapten helper effect. J Exp Med. 1974;139:779–784. doi: 10.1084/jem.139.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marrack PC, Kappler JW. Antigen-specific and nonspecific mediators of T cell/B cell cooperation. I. Evidence for their production by different T cells. J Immunol. 1975;114:1116–1125. [PubMed] [Google Scholar]
- 7.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 8.Scott P, Pearce E, Cheever AW, Coffman RL, Sher A. Role of cytokines and CD4+ T-cell subsets in the regulation of parasite immunity and disease. Immunol Rev. 1989;112:161–182. doi: 10.1111/j.1600-065x.1989.tb00557.x. [DOI] [PubMed] [Google Scholar]
- 9.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 11.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh CS, Heimberger AB, Gold JS, O’Garra A, Murphy KM. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an alpha beta T-cell-receptor transgenic system. Proc Natl Acad Sci U S A. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Infante-Duarte C, Horton HF, Byrne MC, Kamradt T. Microbial lipopeptides induce the production of IL-17 in Th cells. J Immunol. 2000;165:6107–6115. doi: 10.4049/jimmunol.165.11.6107. [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 16.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 17.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira SA, Gorman D, Kastelein RA, Sedgwick JD. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 18.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman EP, Chackerian AA, Cua DJ. Rationale and safety of anti-interleukin-23 and anti-interleukin-17A therapy. Curr Opin Infect Dis. 2006;19:245–252. doi: 10.1097/01.qco.0000224818.42729.67. [DOI] [PubMed] [Google Scholar]
- 21.Strober W, I, Fuss J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–1767. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunology. 2009;2:403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–382. doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaffen SL, Hernandez-Santos N, Peterson AC. IL-17 signaling in host defense against Candida albicans. Immunol Res. 2011;50:181–187. doi: 10.1007/s12026-011-8226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Zelante T, Iannitti R, De Luca A, Romani L. IL-22 in antifungal immunity. Eur J Immunol. 2011;41:270–275. doi: 10.1002/eji.201041246. [DOI] [PubMed] [Google Scholar]
- 28.Torrado E, Cooper AM. IL-17 and Th17 cells in tuberculosis. Cytokine Growth Factor Rev. 2010;21:455–462. doi: 10.1016/j.cytogfr.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartigan-O’Connor DJ, Hirao LA, McCune JM, Dandekar S. Th17 cells and regulatory T cells in elite control over HIV and SIV. Curr Opin HIV AIDS. 2011;6:221–227. doi: 10.1097/COH.0b013e32834577b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maizels RM, Pearce EJ, Artis D, Yazdanbakhsh M, Wynn TA. Regulation of pathogenesis and immunity in helminth infections. J Exp Med. 2009;206:2059–2066. doi: 10.1084/jem.20091903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 32.Gopal R, Lin Y, Obermajer N, Slight S, Nuthalapati N, Ahmed M, Kalinski P, Khader SA. IL-23-dependent IL-17 drives Th1-cell responses following Mycobacterium bovis BCG vaccination. Eur J Immunol. 2012;42:364–373. doi: 10.1002/eji.201141569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y, Ritchea S, Logar A, Slight S, Messmer M, Rangel-Moreno J, Guglani L, Alcorn JF, Strawbridge H, Park SM, Onishi R, Nyugen N, Walter MJ, Pociask D, Randall TD, Gaffen SL, Iwakura Y, Kolls JK, Khader SA. Interleukin-17 is required for T helper 1 cell immunity and host resistance to the intracellular pathogen Francisella tularensis. Immunity. 2009;31:799–810. doi: 10.1016/j.immuni.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 35.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 37.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 38.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, Ivanov, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 40.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 42.Acharya M, Mukhopadhyay S, Paidassi H, Jamil T, Chow C, Kissler S, Stuart LM, Hynes RO, Lacy-Hulbert A. alphav Integrin expression by DCs is required for Th17 cell differentiation and development of experimental autoimmune encephalomyelitis in mice. J Clin Invest. 2010;120:4445–4452. doi: 10.1172/JCI43796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutcher I, Donkor MK, Ma Q, Rudensky AY, Flavell RA, Li MO. Autocrine transforming growth factor-beta1 promotes in vivo Th17 cell differentiation. Immunity. 2011;34:396–408. doi: 10.1016/j.immuni.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 45.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O’Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boniface K, Blom B, Liu YJ, de Waal Malefyt R. From interleukin-23 to T-helper 17 cells: human T-helper cell differentiation revisited. Immunol Rev. 2008;226:132–146. doi: 10.1111/j.1600-065X.2008.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Annunziato F, Romagnani S. Mouse T helper 17 phenotype: not so different than in man after all. Cytokine. 2011;56:112–115. doi: 10.1016/j.cyto.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Mills KH. TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol. 2011;11:807–822. doi: 10.1038/nri3095. [DOI] [PubMed] [Google Scholar]
- 49.Lyakh L, Trinchieri G, Provezza L, Carra G, Gerosa F. Regulation of interleukin-12/interleukin-23 production and the T-helper 17 response in humans. Immunol Rev. 2008;226:112–131. doi: 10.1111/j.1600-065X.2008.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerrigan AM, Brown GD. Syk-coupled C-type lectins in immunity. Trends Immunol. 2011;32:151–156. doi: 10.1016/j.it.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis e Sousa C. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 52.Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, Jamal S, Manguiat A, Rezaei N, Amirzargar AA, Plebani A, Hannesschlager N, Gross O, Ruland J, Grimbacher B. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schoenen H, Bodendorfer B, Hitchens K, Manzanero S, Werninghaus K, Nimmerjahn F, Agger EM, Stenger S, Andersen P, Ruland J, Brown GD, Wells C, Lang R. Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J Immunol. 2010;184:2756–2760. doi: 10.4049/jimmunol.0904013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aliahmadi E, Gramlich R, Grutzkau A, Hitzler M, Kruger M, Baumgrass R, Schreiner M, Wittig B, Wanner R, Peiser M. TLR2-activated human langerhans cells promote Th17 polarization via IL-1beta, TGF-beta and IL-23. Eur J Immunol. 2009;39:1221–1230. doi: 10.1002/eji.200838742. [DOI] [PubMed] [Google Scholar]
- 55.Olliver M, Hiew J, Mellroth P, Henriques-Normark B, Bergman P. Human monocytes promote Th1 and Th17 responses to Streptococcus pneumoniae. Infect Immun. 2011;79:4210–4217. doi: 10.1128/IAI.05286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M, Kato H, Sougawa N, Matsui H, Kuwata H, Hemmi H, Coban C, Kawai T, Ishii KJ, Takeuchi O, Miyasaka M, Takeda K, Akira S. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 57.Dennehy KM, Willment JA, Williams DL, Brown GD. Reciprocal regulation of IL-23 and IL-12 following co-activation of Dectin-1 and TLR signaling pathways. Eur J Immunol. 2009;39:1379–1386. doi: 10.1002/eji.200838543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Beelen AJ, Zelinkova Z, Taanman-Kueter EW, Muller FJ, Hommes DW, Zaat SA, Kapsenberg ML, de Jong EC. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity. 2007;27:660–669. doi: 10.1016/j.immuni.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 59.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thiennimitr P, Winter SE, Baumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15:108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 62.Elson CO, Cong Y, Sundberg J. The C3H/HeJBir mouse model: a high susceptibility phenotype for colitis. Int Rev Immunol. 2000;19:63–75. doi: 10.3109/08830180009048390. [DOI] [PubMed] [Google Scholar]
- 63.Ivanov, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 65.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, Taylor TD, Ohno H, Umesaki Y, Hattori M. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 68.Wu HJ, Ivanov, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada O, Heczko P, Strus M, Bland P, Tlaskalova-Hogenova H. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 71.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SJ, McLachlan JB, Kurtz JR, Fan D, Winter SE, Baumler AJ, Jenkins MK, McSorley SJ. Temporal expression of bacterial proteins instructs host CD4 T cell expansion and Th17 development. PLoS Path. 2012;8:e1002499. doi: 10.1371/journal.ppat.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 77.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 78.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 79.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaffen SL. Recent advances in the IL-17 cytokine family. Curr Opin Immunol. 2011;23:613–619. doi: 10.1016/j.coi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 83.Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, Komatsu R, Miura N, Adachi Y, Ohno N, Shibuya K, Yamamoto N, Kawakami K, Yamasaki S, Saito T, Akira S, Iwakura Y. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 84.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ho AW, Shen F, Conti HR, Patel N, Childs EE, Peterson AC, Hernandez-Santos N, Kolls JK, Kane LP, Ouyang W, Gaffen SL. IL-17RC is required for immune signaling via an extended SEF/IL-17R signaling domain in the cytoplasmic tail. J Immunol. 2010;185:1063–1070. doi: 10.4049/jimmunol.0903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kagami S, Rizzo HL, Kurtz SE, Miller LS, Blauvelt A. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defense against Candida albicans. J Immunol. 2010;185:5453–5462. doi: 10.4049/jimmunol.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova JL. Inborn errors of mucocutaneous immunity to Candida albicans in humans: a role for IL-17 cytokines? Curr Opin Immunol. 2010;22:467–474. doi: 10.1016/j.coi.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, Gumbleton M, Toulon A, Bodemer C, El-Baghdadi J, Whitters M, Paradis T, Brooks J, Collins M, Wolfman NM, Al-Muhsen S, Galicchio M, Abel L, Picard C, Casanova JL. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulacsy V, Chernyshova L, Chernyshov V, Bondarenko A, Grimaldo RM, Blancas-Galicia L, Beas IM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel PR, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bue M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, Al-Muhsen S, Reichenbach J, Kobayashi M, Rosales FE, Lozano CT, Kilic SS, Oleastro M, Etzioni A, Traidl-Hoffmann C, Renner ED, Abel L, Picard C, Marodi L, Boisson-Dupuis S, Puel A, Casanova JL. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Griffin AJ, McSorley SJ. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal immunology. 2011;4:371–382. doi: 10.1038/mi.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 93.VanCott JL, Chatfield SN, Roberts M, Hone DM, Hohmann EL, Pascual DW, Yamamoto M, Kiyono H, McGhee JR. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med. 1998;4:1247–1252. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- 94.Schulz SM, Kohler G, Holscher C, Iwakura Y, Alber G. IL-17A is produced by Th17, gammadelta T cells and other CD4− lymphocytes during infection with Salmonella enterica serovar Enteritidis and has a mild effect in bacterial clearance. Int Immunol. 2008;20:1129–1138. doi: 10.1093/intimm/dxn069. [DOI] [PubMed] [Google Scholar]
- 95.Schulz SM, Kohler G, Schutze N, Knauer J, Straubinger RK, Chackerian AA, Witte E, Wolk K, Sabat R, Iwakura Y, Holscher C, Muller U, Kastelein RA, Alber G. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J Immunol. 2008;181:7891–7901. doi: 10.4049/jimmunol.181.11.7891. [DOI] [PubMed] [Google Scholar]
- 96.Godinez I, Keestra AM, Spees A, Baumler AJ. The IL-23 axis in Salmonella gastroenteritis. Cell Microbiol. 2011;13:1639–1647. doi: 10.1111/j.1462-5822.2011.01637.x. [DOI] [PubMed] [Google Scholar]
- 97.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, Baumler AJ. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gordon MA. Salmonella infections in immunocompromised adults. J Infection. 2008;56:413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 99.Keestra AM, Godinez I, Xavier MN, Winter MG, Winter SE, Tsolis RM, Baumler AJ. Early MyD88-dependent induction of interleukin-17A expression during Salmonella colitis. Infect Immun. 2011;79:3131–3140. doi: 10.1128/IAI.00018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Geddes K, Rubino SJ, Magalhaes JG, Streutker C, Le Bourhis L, Cho JH, Robertson SJ, Kim CJ, Kaul R, Philpott DJ, Girardin SE. Identification of an innate T helper type 17 response to intestinal bacterial pathogens. Nat Med. 2011;17:837–844. doi: 10.1038/nm.2391. [DOI] [PubMed] [Google Scholar]
- 101.Siegemund S, Schutze N, Schulz S, Wolk K, Nasilowska K, Straubinger RK, Sabat R, Alber G. Differential IL-23 requirement for IL-22 and IL-17A production during innate immunity against Salmonella enterica serovar Enteritidis. Int Immunol. 2009;21:555–565. doi: 10.1093/intimm/dxp025. [DOI] [PubMed] [Google Scholar]
- 102.Srinivasan A, Salazar-Gonzalez RM, Jarcho M, Sandau MM, Lefrancois L, McSorley SJ. Innate immune activation of CD4 T cells in salmonella-infected mice is dependent on IL-18. J Immunol. 2007;178:6342–6349. doi: 10.4049/jimmunol.178.10.6342. [DOI] [PubMed] [Google Scholar]
- 103.Kupz A, Guarda G, Gebhardt T, Sander LE, Short KR, Diavatopoulos DA, Wijburg OL, Cao H, Waithman JC, Chen W, Fernandez-Ruiz D, Whitney PG, Heath WR, Curtiss R, 3rd, Tschopp J, Strugnell RA, Bedoui S. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8 T cells. Nat Immunol. 2012;13:162–169. doi: 10.1038/ni.2195. [DOI] [PubMed] [Google Scholar]
- 104.Siegemund S, Schutze N, Freudenberg MA, Lutz MB, Straubinger RK, Alber G. Production of IL-12, IL-23 and IL-27p28 by bone marrow-derived conventional dendritic cells rather than macrophages after LPS/TLR4-dependent induction by Salmonella Enteritidis. Immunobiology. 2007;212:739–750. doi: 10.1016/j.imbio.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 105.Perona-Wright G, Jenkins SJ, O’Connor RA, Zienkiewicz D, McSorley HJ, Maizels RM, Anderton SM, MacDonald AS. A pivotal role for CD40-mediated IL-6 production by dendritic cells during IL-17 induction in vivo. J Immunol. 2009;182:2808–2815. doi: 10.4049/jimmunol.0803553. [DOI] [PubMed] [Google Scholar]
- 106.Barr TA, Brown S, Mastroeni P, Gray D. TLR and B cell receptor signals to B cells differentially program primary and memory Th1 responses to Salmonella enterica. J Immunol. 2010;185:2783–2789. doi: 10.4049/jimmunol.1001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Salazar-Gonzalez RM, McSorley SJ. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett. 2005;101:117–122. doi: 10.1016/j.imlet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 108.Mizel SB, Bates JT. Flagellin as an adjuvant: cellular mechanisms and potential. J Immunol. 2010;185:5677–5682. doi: 10.4049/jimmunol.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Salazar-Gonzalez RM, Niess JH, Zammit DJ, Ravindran R, Srinivasan A, Maxwell JR, Stoklasek T, Yadav R, Williams IR, Gu X, McCormick BA, Pazos MA, Vella AT, Lefrancois L, Reinecker HC, McSorley SJ. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity. 2006;24:623–632. doi: 10.1016/j.immuni.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, Tsolis RM, Baumler AJ. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]