Abstract
Background
The provirus integration site for Moloney murine leukemia virus (Pim) 1 kinase is an oncogenic serine/threonine kinase implicated in cytokine-induced cell signaling, whereas Runt-related transcription factor (Runx) has been implicated in the regulation of T-cell differentiation. The interaction of Pim1 kinase and Runx3 in the pathogenesis of peanut allergy has not been defined.
Objectives
We sought to determine the effects of Pim1 kinase modulation on Runx3 expression and TH2 and TH17 cell function in an experimental model of peanut allergy. Methods: A Pim1 kinase inhibitor was administered to peanut-sensitized and challenged wild-type and Runx3+/− mice. Symptoms, intestinal inflammation, and Pim1 kinase and Runx3 mRNA expression and protein levels were assessed. The effects of Pim1 kinase inhibition on TH1, TH2, and TH17 differentiation in vivo and in vitro were also determined.
Results
Peanut sensitization and challenge resulted in accumulation of inflammatory cells and goblet cell metaplasia and increased levels of Pim1 kinase and TH2 and TH17 cytokine production but decreased levels of Runx3 mRNA and protein in the small intestines of wild-type mice. All of these findings were normalized with Pim1 kinase inhibition. In sensitized and challenged Runx3+/− mice, inhibition of Pim1 kinase had less effect on the development of the full spectrum of intestinal allergic responses. In vitro inhibition of Pim1 kinase attenuated TH2 and TH17 cell differentiation and expansion while maintaining Runx3 expression in T-cell cultures from wild-type mice; these effects were reduced in T-cell cultures from Runx3+/− mice.
Conclusion
These data support a novel regulatory axis involving Pim1 kinase and Runx3 in the control of food-induced allergic reactions through the regulation of TH2 and TH17 differentiation.
Keywords: Pim1 kinase, Runx3, peanut, intestinal allergy, TH2, TH17
In hosts with peanut allergy, several cell types are recruited to the intestine and activated to release cytokines and chemokines, contributing to intestinal inflammation.1-4 In addition to IL-4 and IL-13, increased levels of IL-17A have been found in the small intestine and mesenteric lymph nodes (MLNs) in a mouse model of food allergy.2 The data suggest that CD4+ T cells, which produce both TH2 and TH17 cytokines, play an important role in food-associated allergic disease.
The provirus integration site for Moloney murine leukemia virus (Pim), a proto-oncogene encoding a family of serine/threonine protein kinases, has multiple cellular functions.5 Pim1 kinase has been implicated in cytokine-dependent signaling in hematopoietic cells and T lymphocytes,6,7 and kinase expression was enhanced during T-cell activation.8 Pim1 kinase increases T-cell proliferation by enhancing the activity of nuclear factor of activated T cells (NFAT) c1, increasing IL-2 production in T cells.7 Pim1 kinase expression was upregulated in the lungs of mice after sensitization and challenge with allergen.9
Pim1 kinase regulates Runt-related transcription factor (Runx) expression in vitro.6 In this family of transcription factors, Runx3 is required for epigenetic silencing in cytotoxic lineage thymocytes.10 Runx3 cooperates with T-box transcription factor (T-bet) to repress the production of IL-4 by binding to the IL-4 silencer in the TH2 cytokine locus and promotes the production of IFN-γ in TH1 cells.11-13 Loss of Runx3 results in the spontaneous development of inflammatory bowel disease, as well as allergic asthma.14,15
We investigated the role of Pim1 kinase and its relationship to Runx3 expression in an experimental model of peanut-induced intestinal allergy. Pim1 kinase was essential to the development of peanut-induced intestinal allergy. Moreover, inhibition of this kinase prevented the intestinal inflammation and attenuated TH2 and TH17 differentiation and cytokine production by regulating Runx3.
Methods
For further description of the methods used in this study, see the Methods section in this article's Online Repository at www.jacionline.org.
Mice
Five- to 6-week-old female wild-type (WT) C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, Me). Runx3 heterozygous (Runx3+/−) mice were provided by Dr James Hagman (National Jewish Health, Denver, Colo). All studies were conducted under a protocol approved by the Institutional Animal Care and Use Committee of National Jewish Health.
Preparation of peanut protein
Crude peanut extract (PE) was prepared as described in the Methods section in this article's Online Repository.
Sensitization and intragastric challenge
The experimental protocol for sensitization and challenge to peanut was previously described.2 Because C57BL/6 mice did not have peanut-induced intestinal allergy to the same extent as BALB/c mice because of their limited development of peanut-specific IgE antibody, Runx3+/− and C57BL/6 mice were passively sensitized with serum containing peanut-specific IgE.2 All systemically and passively sensitized and challenged Runx3+ − and C57BL/6 mice had diarrhea by the seventh day of challenge.
Pim1 kinase inhibitor and treatment in vivo
The small-molecule Pim1 kinase inhibitor (AR460770; Array Biopharma, Boulder, Colo) cellular inhibitory concentration of 50% was 93, 9200, and 340 nmol/L for Pim1, Pim2, and Pim3, respectively.9 PE-sensitized and challenged mice received different doses (0-100 mg/kg) of the inhibitor by means of gavage and based on earlier experiments.9 For more information, see the Methods section in this article's Online Repository.
Assessment of hypersensitivity reactions
Symptoms were evaluated as previously reported16 and described in the Methods section in this article's Online Repository.
Histology
The jejunum was processed and stained with periodic acid–Schiff, chloroacetate esterase, and anti-mouse major basic protein antibody (kindly provided by Dr J. J. Lee, Mayo Clinic, Scottsdale, Ariz) for detection of mucosal mucus-containing cells, mast cells, and eosinophils, respectively, as previously described.2,17,18 Numbers of CD4, CD8, Pim1, Pim3, and Runx3 mucosal cells were identified by means of immunohistochemical staining with anti-mouse CD4, CD8, Pim1, Pim3, and Runx3 antibodies (Abcam, Cambridge, Mass), respectively.
Cytokine levels in cell culture
Levels of the cell-culture supernatant IL-4, IL-13, IL-17A, and IFN-γ were measured by means of ELISA (eBioscience, San Diego, Calif), as described by the manufacturer.
Measurement of peanut-specific antibody levels
Serum peanut-specific IgE, IgG1, and IgG2a levels were measured by using ELISA, as described previously.16
Histamine levels in plasma
Histamine levels in plasma were measured as described in the Methods section in this article's Online Repository.
T-cell differentiation and treatment with the Pim1 kinase inhibitor in vitro
Differentiation of TH1, TH2, or TH17 cells was performed as previously described19,20 and in the Methods section in this article's Online Repository.
Western blot analysis
Cell lysates were prepared from jejunal tissue and cultured cells as previously described2,21 and in the Methods section in this article's Online Repository.
Quantitative real-time PCR
RNA was extracted from jejunal tissue homogenates or from CD4 T cells cultured in vitro with Trizol (Invitrogen, Carlsbad, Calif). cDNA was generated with the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, Calif). Quantitative real-time PCR was performed on the ABI Prism 7300 sequence detection system (Applied Biosystems, Foster City, Calif). All primers and probes used were purchased as TagMan Gene Expression Assays from Applied Biosystems. Fold change was calculated by using the ΔΔ cycle threshold method.
Anti-Runx3 antibody
Rabbit anti-human Runx3 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif) was biotinylated with the EZ-link sulfo-NHS-LC-biotin kit (Pierce, Rockford, Ill). Allophycocyanin-conjugated streptavidin (eBioscience) was used to detect biotinylated primary Runx3 antibodies.
Intracellular cytokine staining and flow cytometry
Cells from MLNs or differentiated CD4 T cells were labeled with anti-CD3 or anti-CD4 antibody (eBioscience) and stained for intracytoplasmic IL-4, IL-13, IL-17A, IFN-γ, and Runx3 using antibodies from BD Biosciences (San Jose, Calif) or as described above (Runx3 antibody).22 Cells were analyzed on a FACSCalibur (BD Biosciences) by using CellQuest software (BD Biosciences).
Cell proliferation
TH1-, TH2-, or TH17-polarized CD4 T cells were incubated with anti-CD3 and anti-CD28 (eBioscience) at 37°C for 24 hours. Tritiated thymidine (PerkinElmer, Boston, Mass) was added to the cultures for another 6 hours, and incorporation was measured in a liquid scintillation counter (Packard Bioscience Company, Meriden, Conn).
Cell viability and apoptosis
Cell viability was determined using a trypan blue dye exclusion assay. Cell apoptosis was detected by means of flow cytometry with surface staining with 7AAD and Annexin V (BD Biosciences).
Statistical analysis
ANOVA was used to determine the levels of difference among all groups. Comparisons for all pairs used the Tukey-Kramer highest significance difference test. P values for significance were set at .05. All results were expressed as means ± SEMs.
Results
Pim1 kinase levels are upregulated in the small intestines of peanut-sensitized and challenged mice
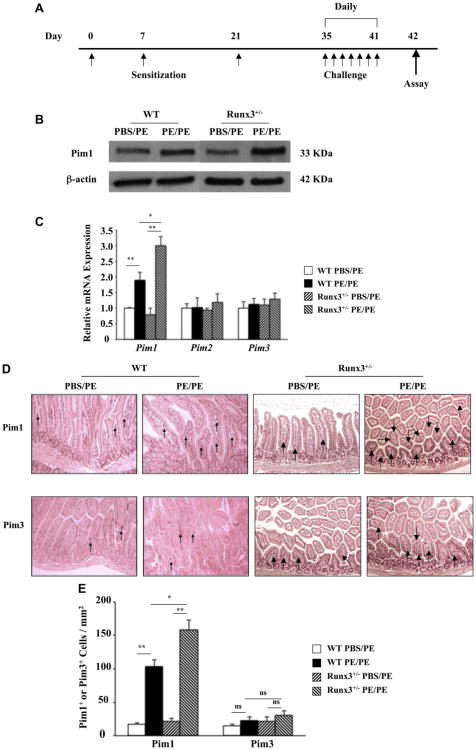
After PE sensitization and challenge (Fig 1, A), Pim1 kinase protein expression was increased in the jejunums of WT and Runx3+/− mice (Fig 1, B). Increases in Pim1 kinase protein levels were greater in the jejunums of sensitized and challenged Runx3+/− mice compared with those seen in WT mice. Pim1 kinase mRNA levels were 2- and 3-fold higher in the jejunums of PE-sensitized and challenged WT and Runx3+/− mice, respectively (Fig 1, C); Pim2 and Pim3 mRNA levels were not altered after sensitization and challenge of WT or Runx3+/− mice. Pim1 kinase was expressed predominantly in the lamina propria of the jejunum, and positive cell numbers were increased by approximately 5- and 7-fold in WT and Runx3+/− mice, respectively (Fig 1, D and E). The numbers of Pim3-positive cells were lower, with little alteration after PE sensitization and challenge.
Fig 1.
Pim1 kinase is expressed in WT and Runx3+/− mouse jejunum. A, Protocol for induction of peanut allergy. B, Western blot analysis of Pim1 kinase expression. The results are from the same experiment and the same membranes, but intervening lanes have been cut out for the photograph. C, Relative mRNA expression of Pim family members. D, Representative immunohistochemical staining for Pim1 and Pim3 kinases. E, Quantitation of mucosal Pim1 and Pim3 kinase–expressing cells. Results were from 3 independent experiments; each experiment included 4 mice per group (n = 12). *P < .05 and **P < .01. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Runx3 is downregulated in the small intestines of peanut-sensitized and challenged mice
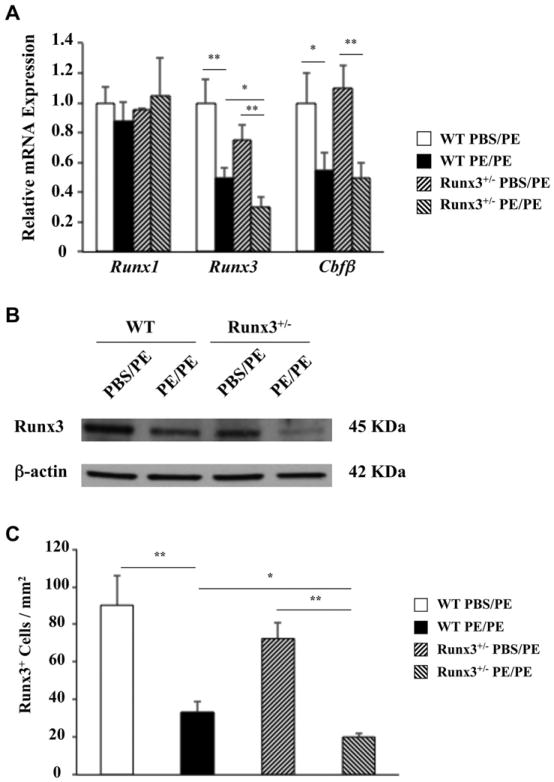
Levels of Runx3 mRNA were approximately 20% to 30% lower in sham-sensitized Runx3+/− mice than in WT mice (Fig 2, A). Runx3 and Runx/core binding factor β (Cbfβ) mRNA levels were decreased in the small intestines of PE-sensitized and challenged WT and Runx3+/− mice. Levels of Runx3 and Cbfβ mRNA, but not Runx1 mRNA, were approximately 2-fold lower in the jejunums of PE-sensitized and challenged WT and Runx3+/− mice compared with those seen in control animals. Levels of Runx3 mRNA were significantly lower in the jejunums of sensitized and challenged Runx3+/− mice than in WT mice. In parallel, Runx3 protein expression was also lower in the jejunums of sensitized and challenged mice and lower in Runx3+/− mice than in WT mice (Fig 2, B). Immunohistochemical analysis revealed that Runx3 protein was mainly expressed in the lamina propria, and levels of expression were decreased by 2- to 3-fold in sensitized and challenged WT and Runx3+/− mice (Fig 2, C). Numbers of Runx3+ cells were also significantly lower in PE-sensitized and challenged Runx3+/− mice compared with those seen in WT mice.
Fig 2.
Expression of Runx3 in the mouse jejunum. A, Relative expression levels of Runx3, Runx1, and Cbfβ mRNA in jejunums of WT and Runx3+/− mice. B, Runx3 protein levels in jejunums of WT and Runx3+/− mice. C, Quantitation of mucosal Runx3-expressing cell numbers in jejunums of WT and Runx3+/− mice. Results are from 2 independent experiments; each experiment included 4 mice per group. *P < .05 and **P < .01. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Inhibition of Pim1 kinase attenuates PE-induced intestinal responses in vivo
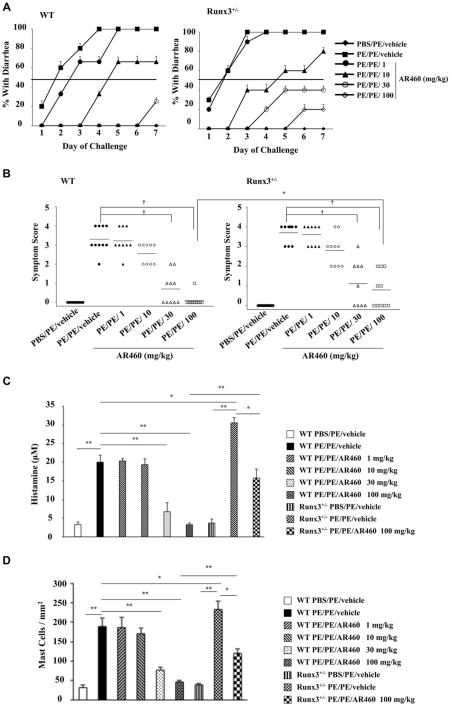
These data suggested that Pim1 kinase and Runx3 play essential roles in the control of intestinal allergy. To test this hypothesis, we investigated whether inhibition of Pim1 kinase alters the severity of PE-induced intestinal allergy using the small-molecule inhibitor AR460770. The specificity of AR460770 for Pim1 kinase was previously demonstrated.9 Administration of the inhibitor to sensitized WT mice resulted in a dose-dependent inhibitory effect on intestinal allergy induction; 30 to 100 mg/kg of the inhibitor fully prevented the development of diarrhea and symptoms in PE-sensitized and challenged WT mice. In contrast, inhibitor treatment of Runx3+/− mice resulted in reduced inhibitory effects; 30 to 100 mg/kg of the inhibitor partially inhibited diarrhea and symptoms in Runx3+/− mice. These effects were significantly lower in Runx3+/− than in WT mice (Fig 3, A and B).
Fig 3.
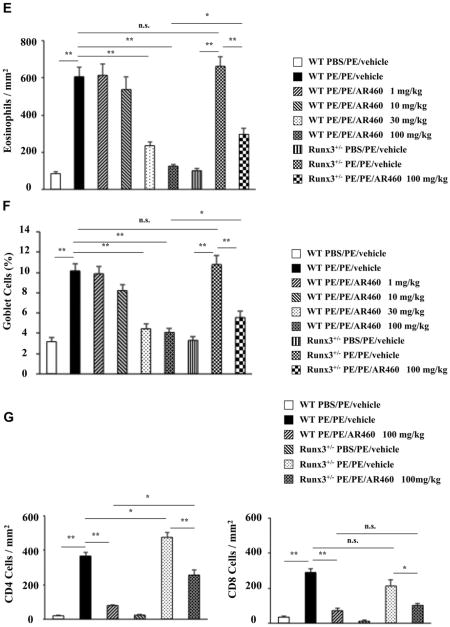
Inhibition of Pim1 kinase reduces intestinal responses. A, Kinetics of development of diarrhea. The incidence of diarrhea was significantly lower in inhibitor (100 mg/kg)-treated WT mice than in Runx3+/− mice (P< .05). B, Symptom scores were assessed 30 minutes after oral challenge. C, Plasma histamine levels were assessed within 30 minutes of the last oral challenge. D-G, Quantitation of mucosal mast cells, eosinophils, goblet cells, and CD4 and CD8 T cell numbers in the jejunum. Results were from 3 independent experiments; each experiment included 4 mice per group. *P< .05, **P< .01, and †P< .001. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Mast cells are involved in the response to PE sensitization and challenge.2 We monitored mast cell degranulation by quantitating plasma levels of histamine within 30 minutes of the last challenge. Levels of histamine were increased after sensitization and challenge, and in Runx3+/− mice levels were significantly increased over levels seen in WT mice. Levels of histamine in inhibitor (30 and 100 mg/kg)–treated WT mice were significantly decreased, almost to baseline levels, after sensitization and challenge. In Runx3+/− mice after inhibitor treatment, levels were significantly decreased, although to a smaller extent than in WT mice (Fig 3, C).
When administered after sensitization and during challenge, the inhibitor had no effect on peanut-specific IgE, IgG1, and IgG2a serum levels in WT or Runx3+/− mice (see Fig E1 in this article's Online Repository at www.jacionline.org).
PE-sensitized and challenged mice demonstrated increased numbers of mast cells, eosinophils, and periodic acid–Schiff–positive goblet cells in the mucosa of the small intestine (Fig 3, D-F, and see Figs E2-E4 in this article's Online Repository at www.jacionline.org). WT mice treated with the inhibitor at a dose of 30 to 100 mg/kg demonstrated markedly reduced numbers of these cells. In contrast, the decreases in numbers of these cells were significantly lower in inhibitor (100 mg/kg)–treated Runx3+/− mice. The lamina propria of the sham-sensitized group contained few CD4 and CD8 T cells in WT or Runx3+/− mice. These numbers were significantly increased in the untreated PE-sensitized and challenged WT group and reduced to baseline levels in the treated WT group (Fig 3, G). In contrast, CD4 T-cell numbers were significantly increased in the untreated PE-sensitized and challenged Runx3+/− mice, and after treatment, there were only modest decreases in the numbers of CD4 T cells; the effect on numbers of CD8 T cells was similar in Runx3+/− and WT mice (Fig 3, G).
Collectively, these results indicated that Pim1 kinase activation played an essential role in enhancing allergic diarrhea, intestinal inflammation, and goblet cell metaplasia and was inversely associated with Runx3 levels. In Runx3+/− mice the inhibitor exhibited reduced effects on all of these parameters, supporting the notion of an interaction (negative) between Pim1 kinase and Runx3.
Pim1 kinase regulates IL-13 and IL-17 production
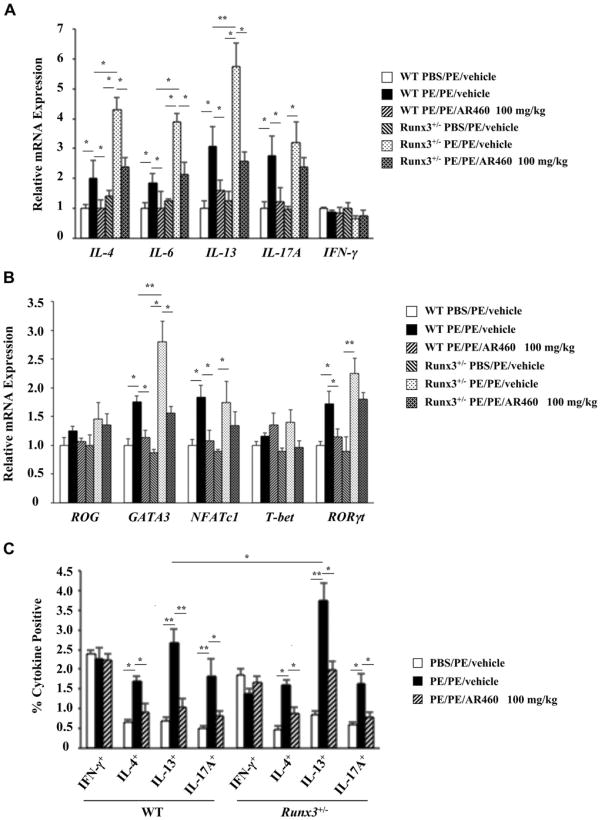
In addition to TH2 cells, TH17 cells have been implicated in allergic disease.23-25 After 7 days of PE challenges, intestinal tissue from sensitized WT and Runx3+/− mice demonstrated significant increases in IL4, IL6, IL13, and IL17A but not IFNG mRNA expression. Furthermore, IL4, IL6, and IL13 mRNA expression were significantly higher in Runx3+/− mice compared with that seen in WT mice (Fig 4, A). After treatment with AR460770, mRNA expression levels for these cytokines returned to control levels in WT but not in Runx3+/− mice. The expression levels of GATA3, NFATc1, and retinoic acid–related orphan receptor γt (RORγt) mRNA were also significantly increased in sensitized and challenged WT and Runx3+/− mice, whereas T-bet and repressor of GATA mRNA levels were not altered (Fig 4, B). After treatment with the inhibitor, these increased levels also returned to control levels in WT but not in Runx3+/− mice.
Fig 4.
Effect of Pim1 kinase inhibition on cytokine and transcription factor expression. A, TH1, TH2, and TH17 mRNA expression in jejunums of WT and Runx3+/− mice treated with AR460770 or vehicle. B, TH1, TH2, and TH17 transcription factor expression in jejunums of WT and Runx3+/− mice treated with AR460770 or vehicle. ROG, Repressor of GATA. C, Percentages of IFN-γ+–, IL-4+–, IL-13+–, and IL-17A+–producing MLN CD3+CD4+ cells of WT and Runx3+/− mice treated with the inhibitor or vehicle. Data are from 3 independent experiments (n = 12). *P < .05 and **P < .01. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
To assess the effect of Pim1 kinase inhibition on T-lymphocyte cytokine production, we isolated MLN CD4 T cells from WT and Runx3+/− mice treated with the inhibitor or vehicle and stimulated them with anti-CD3/anti-CD28. PE sensitization and challenge resulted in significant increases in the numbers of IL-4–, IL-13–, and IL-17A–producing CD4 T cells from WT and Runx3+/− mice (Fig 4, C, and see Fig E5 in this article's Online Repository at www.jacionline.org). Numbers of IL-13–producing, but not IL-4– or IL-17A–producing, CD4 T cells were significantly increased in PE-sensitized and challenged Runx3+/− compared with WT mice. Mice treated with the inhibitor exhibited 2-to 4-fold and 1.5- to 2-fold decreases in the numbers of these CD4 cytokine–producing cells in WT and Runx3+/− mice, respectively. The percentages of CD4+IFN-γ+ cells were not altered by sensitization and challenge or treatment with the inhibitor.
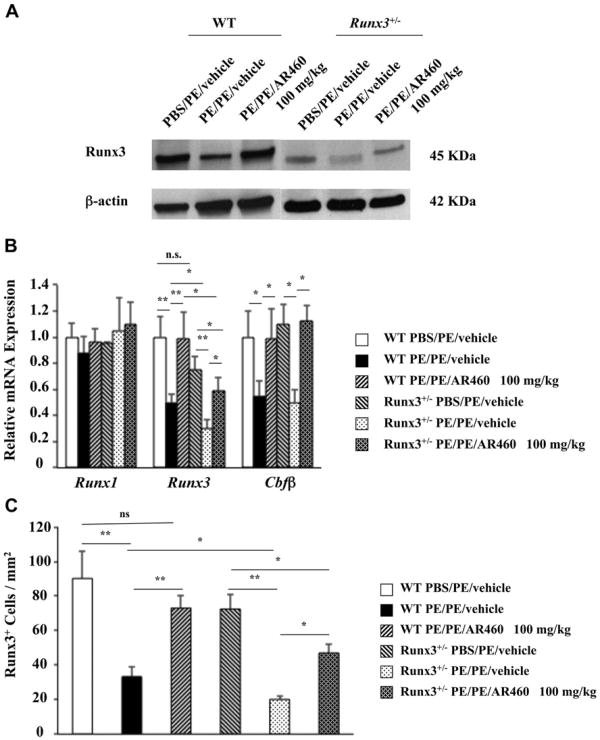
Pim1 regulates Runx3 expression
We next determined whether inhibition of Pim1 kinase affects Runx3 expression and whether Runx3 plays an important role in peanut allergy. Protein was extracted from jejunal tissue of WT and Runx3+/− mice, and Western blot analysis showed a decrease in levels of Runx3 in PE-sensitized and challenged WT and Runx3+/− mice; levels of Runx3 protein were decreased to a greater degree in Runx3+/− mice (Fig 5, A). In mice treated with AR460770, Runx3 levels were restored to baseline levels in WT but not in Runx3+/− mice. In parallel, levels of Runx3 and Cbfβ mRNA were also decreased in sensitized and challenged WT and Runx3+/− mouse tissue, with levels of Runx3 mRNA lower in the PE-sensitized and challenged Runx3+/− mice than in WT mice (Fig 5, B). Runx3 mRNA levels were restored to control values after treatment with the inhibitor in WT but not in Runx3+/− mice. Runx3 protein expression was decreased in the jejunums of PE-sensitized and challenged WT and Runx3+/− mice and was similarly restored to almost control values after treatment with the inhibitor in WT but not Runx3+/− mice (Fig 5, C, and see Fig E6 in this article's Online Repository at www.jacionline.org). Together, these data indicated that PE sensitization and challenge resulted in Pim1 activation and reductions in levels of Runx3 protein and mRNA. In WT mice treatment with the Pim1 kinase inhibitor normalized these levels to baseline, although not in Runx3+/− mice.
Fig 5.
Pim1 kinase regulates Runx3 transcription factor expression in the intestine. A, Runx3 protein levels in jejunums of Pim1 kinase inhibitor–treated PE/PE WT and Runx3+/− mice. The results are from the same experiment and the same membranes, but intervening lanes have been cut out for the photograph. B, Runx mRNA expression in the jejunums of WT and Runx3+/− mice treated with or without AR460770. C, Quantitation of mucosal Runx3+/− cell numbers in the jejunums of WT and Runx3+/− mice. Results are from 3 independent experiments with 4 mice per group. *P < .05 and **P < .01. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Effects of Pim1 kinase inhibition on TH1, TH2, and TH17 cell differentiation and Runx3 expression in vitro
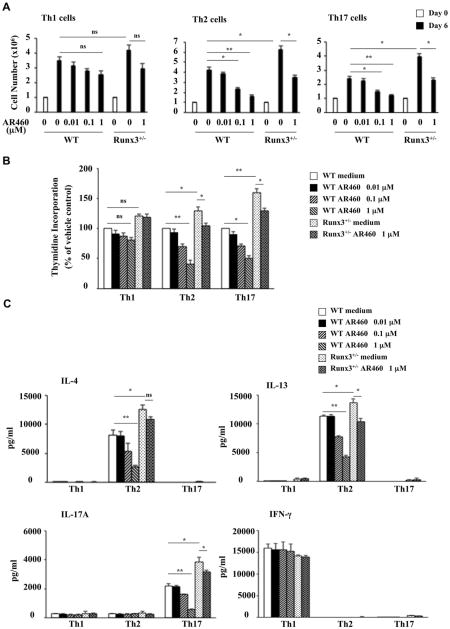
Considering the effects of Pim1 kinase inhibition on Runx3 and TH2/TH17 cytokine production in vivo, we investigated the effect of Pim1 kinase inhibition on Runx3 expression and T-cell differentiation and function in vitro. Isolated naive CD4+CD45RB+ T cells from WT and Runx3+/−- mice were cultured under TH1-, TH2-, and TH17-polarizing conditions in the presence or absence of the inhibitor for 6 days and then stimulated with the combination of anti-CD3/anti-CD28. Overall, TH2 cells expressed higher levels of Pim1 mRNA than TH1 cells in WT and Runx3+/− mice but expressed lower levels of Runx3 mRNA than TH1 cells in WT and Runx3+/− mice (see Fig E7 in this article's Online Repository at www.jacionline.org). The inhibitor suppressed TH2 and TH17 cell expansion in cells from WT mice in a dose-dependent manner; 0.1 to 1 μmol/L inhibited cell number increases (Fig 6, A), and 1 μmol/L inhibited TH2 and TH17 cell proliferation, as assessed using tritiated thymidine incorporation (Fig 6, B). TH1 cell expansion was not significantly affected. In contrast, the inhibitor (1 μmol/L) suppressed TH2 and TH17 cell expansion to a lesser extent in cells from Runx3+/− mice. The numbers of untreated Runx3+/− TH2 and TH17 cells were significantly higher than in WT cells (Fig 6, A and B). As with WT cells, Runx3+/− TH1 cell expansion was not significantly affected. In parallel, the decreases in TH2 and TH17 cytokine levels in the polarized T-cell cultures (IL-4, IL-13, and IL-17A, respectively) from Runx3+/− mice were lower than in WT cell cultures (Fig 6, C); IFN-γ levels were not affected by the inhibitor. These effects were not due to altered cell viability (see Figs E8 and E9 in this article's Online Repository at www.jacionline.org).
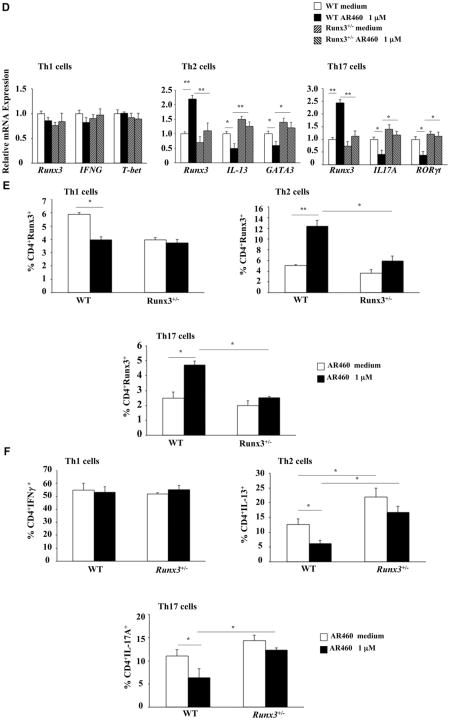
Fig 6.
Pim1 kinase inhibitor modulates Runx3 expression and suppresses the differentiation of naive CD4 T cells into TH2 and TH17 lineages in vitro. A, Cell proliferation reported as number of cells. B, Cell proliferation measured using tritiated thymidine incorporation. C, Cytokine levels in supernatants of cultured CD4 T cells under TH1-, TH2-, and TH17-polarizing conditions. D, Runx3 and cell-specific transcription factor mRNA expression in naive CD4 T cells differentiated in vitro into TH2 or TH17 cells. E and F, Intracellular staining of Runx3 and cytokines in polarized TH1, TH2, and TH17 cells. Results are from 3 independent experiments. *P < .05 and **P < .01.
In the polarized T-cell cultures we examined the effects of AR460770 on the expression of Runx3 and lineage-specific transcription factors using quantitative PCR. In polarized WT but not Runx3+/− TH2 cultures treated with the inhibitor, Runx3 mRNA expression was upregulated (Fig 6, D). Similar results were seen inTH17-polarized cells from WT and Runx3+/− mice. In parallel, levels of IL13 and GATA3 and IL17A and RORγt mRNA expression were decreased in TH2 and TH17 cells from WT but not from Runx3+/− mice (Fig 6, D). No effects were detected in TH1 cells.
We examined the effects of Pim1 kinase inhibition on Runx3 protein and cytokine levels in the polarized T-cell cultures from WT and Runx3+/−mice using intracellular staining. The inhibitor increased the percentages of CD4+Runx3+ cells in the polarized TH2 and TH17 cell cultures from WT but not in the polarized cultures from Runx3+/− mice (Fig 6, E, and Fig E10 in this article's Online Repository at www.jacionline.org). In parallel, the percentages of CD4+IL-13+ and CD4+IL-17A+ cells were significantly decreased in inhibitor-treated TH2 and TH17 cell cultures from WT but not Runx3+/− mice (Fig 6, F). Of note, the percentage of CD4+IL-13+ cells was significantly higher in untreated TH2 cell cultures from Runx3+/− than from WT mice, whereas the percentage of CD4+IFN-γ+ cells was not affected in untreated and treated TH1 cell cultures from WT or Runx3+/−mice. These data demonstrated that inhibition of Pim1 kinase affected TH2 and TH17 but not TH1 differentiation and promoted expression of Runx3. Thus Pim1 kinase functions as a positive regulator for TH2 and TH17 differentiation and expansion and as a negative regulator of Runx3 expression.
Discussion
The family of Pim protein kinases includes 3 members; Pim1 and Pim2 are primarily expressed in hematopoietic cells, whereas Pim3 is expressed in brain, kidney, and mammary tissue. Pim1 is involved in cytokine signaling and has been implicated in many signal transduction pathways.5,26 Pim2 and Pim3 have largely overlapping functions.5,27 Several studies have implicated Pim kinases in hematopoiesis, T-cell development, and differentiation.28,29 Pim gene expression is induced by several factors, particularly by cytokines, including IL-2, IL-3, IL-12, IL-15, and IFN-γ.30,31 Pim1 is expressed in cell types linked to allergic disease, such as T cells and eosinophils28,32; is upregulated in peripheral T cells after antigen activation33; and influences both mouse and human CD4 T-cell activation and differentiation.29,33
In this study we investigated the roles of Pim1 kinase and Runx3 in peanut-induced allergic intestinal responses. Because homozygous Runx3 deficiency is embryonically lethal, we used Runx3+/− mice, which expressed roughly 30% lower Runx3 mRNA and protein levels than WT mice. We first demonstrated that Pim1 kinase expression was upregulated in the jejunums of sensitized and challenged WT mice, with little change in either Pim2 or Pim3 expression. Next, we determined the role of Pim1 kinase using a selective small-molecule inhibitor. In vivo treatment with this inhibitor, which was administered after sensitization but during the oral challenge phase, reduced the incidence and severity of diarrhea and intestinal inflammation (mast cell, eosinophil, and CD4 and CD8 T-cell accumulation and goblet cell metaplasia) in WT mice accompanied by decreases in IL-13 and IL-17A levels in the intestines and MLNs. The inhibitor did not alter the development of specific antibodies, including PE-specific IgE, likely because sensitization was completed before treatment. These data identified for the first time that Pim1 kinase contributed in important ways to the development of peanut-induced allergic responses.
There are several possibilities whereby Pim1 kinase regulates the initiation of an allergic response. IL-13 is a pivotal TH2 effector cytokine in several allergic diseases.34,35 We previously demonstrated2 and confirmed here increased IL-13 levels in the intestines of sensitized and challenged mice. Furthermore, targeting IL-13 alone with a soluble IL-13 receptor 2 protein, which neutralized IL-13,36 resulted in almost complete elimination of allergic diarrhea and suppressed intestinal inflammation and goblet cell metaplasia in vivo.2 The major sources of IL-13 are activated T cells and mast cells.2,37 Inhibition of Pim1 kinase resulted in the reduction of T-cell and mast cell recruitment and activation, resulting in lower levels of IL-13 and IL-17A and an improved outcome after challenge of sensitized mice. In addition to IL-13, IL-17A might contribute to severe allergic responses in mice by enhancing IL-13 function.25 CD4 TH2 memory/effector cells contribute to IL-17 cytokine production and promote the exacerbation of allergic disease.38
Differentiation of TH1, TH2, and TH17 cells is mainly regulated by cytokines and transcription factors.29,39-42 In vivo our data showed that inhibition of Pim1 kinase significantly reduced not only TH2 expansion and cytokine production but also expression of the TH2 lineage-specific transcription factor GATA3 in WT mice. In addition, expression levels of IL17A and RORγt were inhibited, indicating that inhibition of Pim1 kinase activity suppressed TH2 and TH17 differentiation and cytokine production in WT mice and was mediated through inhibition of GATA3 and RORγt transcription factor induction, respectively. On the other hand, TH1 differentiation was unaffected.
To understand the consequences of Pim1 kinase inhibition, we focused on a potential downstream transcriptional regulator, Runx3. Runx, a novel family of transcription factors, is associated with the development of allergic responses.43 There are 3 mammalian Runx genes: Runx1, Runx2, and Runx3. Runx1 is required for hematopoiesis,44 and Runx2 is a regulator of osteogenesis.45 Runx3 resides on human chromosome 1p36.1,46 which maps to a region containing susceptibility genes for asthma,47 and on mouse chromosome 4,48 which contains a susceptibility gene for atopic dermatitis.49 Loss of Runx3 results in spontaneous development of inflammatory bowel disease,14 as well as constitutive airway hyperreactivity and eosinophilic inflammation.43 We demonstrated that Runx3 (but not Runx1 or Runx2) expression was downregulated in the jejunums of sensitized and challenged WT and Runx3+/− mice and that the increases in Pim1 kinase mRNA and protein levels were greater in the jejunums of peanut-sensitized and challenged Runx3+/− mice than in WT mice. The decreases in Runx3 expression were prevented in WT but not Runx3+/− mice after inhibition of Pim1 kinase, suggesting an important inverse relationship between Pim1 kinase and Runx3 expression. By all accounts, the effects of Pim1 kinase inhibition in vivo were lower in Runx3+/− than in WT mice. TH2 cytokine (IL4, IL6, and IL13) mRNA expression was significantly higher in the intestines of sensitized and challenged Runx3+/−mice compared with that seen in WT mice. In Runx3+/− mice inhibition of Pim1 kinase resulted in reduced inhibitory effects on TH2 and TH17 expansion and cytokine production, as well as expression of GATA3 and RORγt. As in WT mice, TH1 differentiation was unaffected.
Naive CD4 T cells from WT and Runx3+/− mice were treated with the Pim1 kinase inhibitor under TH1-, TH2-, or TH17-polarizing conditions to extend the link between Pim1 kinase and Runx3. We observed that the inhibitor suppressed TH2 and TH17 cell differentiation in cells from WT but not Runx3+/− mice in vitro, which was detected as lower expression levels of GATA3/IL13 and RORγt/IL17A. The expression levels of Runx3 mRNA and the percentages of CD4+Runx3+ cells were upregulated in CD4 T cells from WT but not Runx3+/− mice induced to differentiate into TH2 or TH17 cells in the presence of the Pim1 kinase inhibitor. In contrast, inhibition of Pim1 kinase activity had little to no effect on levels of expression of IFNG and IFNG mRNA and protein or the percentages of CD4+IFN-γ+ cells from WT and Runx3+/− mice under TH1 differentiating conditions. In addition to blocking TH2 and TH17 differentiation, the inhibitor prevented anti-CD3/anti-CD28–triggered expansion of these cells from WT mice but not the proliferation of TH1 cells. The inhibitor suppressed TH2 and TH17 cell expansion to a lesser extent in cells from Runx3+/− mice.
Although the direct interactions between Pim1 kinase and Runx3 require further definition, collectively, these results demonstrate that Pim1 kinase inhibition attenuates TH2 and TH17 differentiation in cells from WT mice by suppressing expression of TH2 and TH17 lineage-specific transcription factors, limiting expansion of TH2 and TH17 cells, and reducing TH2 and TH17 cytokine production. At the same time, Runx3 expression was maintained. In cells from Runx3+/− mice, Pim1 kinase inhibition had less to no effect on TH2 and TH17 differentiation, nor could Runx3 expression be maintained. Thus it appears that in the generation of an allergic response, as shown here for PE-induced intestinal allergy, Pim1 kinase is upregulated, Runx3 is downregulated, and TH2 and TH17 differentiation is facilitated. As a corollary, targeting Pim1 kinase results in maintenance of Runx3 expression and prevention of TH2 and TH17 differentiation, resulting in a markedly reduced allergic response. Targeting this novel regulatory axis involving Pim1 kinase and Runx3 offers new therapeutic opportunities for the control of food-induced allergic reactions.
Supplementary Material
Fig E1. Treatment with a Pim1 kinase inhibitor had no effect on serum immunoglobulin production in WT and Runx3+/− mice. Serum levels of peanut-specific IgE, IgG1, and IgG2a were assessed by means of ELISA 24 hours after the last challenge and expressed as the optical density of diluted serum, as described in the Methods section. Results were obtained from 3 individual experiments with 4 mice per group. n.s., Not significant; PBS/PE, sham sensitized but PE challenged; PE/PE, PE sensitized and challenged. *P < .05 and **P < .01.
Fig E2. Decreased mast cell infiltration in the intestinal walls of PE/PE mice treated with the Pim1 kinase inhibitor. Intestinal mucosal mast cells were quantified in the jejunum by using chloroacetate esterase staining. Representative sections of PBS/PE/vehicle WT mice (A), PE/PE/vehicle WT mice (B), PE/PE/AR460 (1 mg/kg) WT mice (C), PE/PE/AR460 (10 mg/kg) WT mice (D), PE/PE/AR460(30 mg/kg) WT mice (E), PE/PE/AR460 (100 mg/kg) WT mice (F), PBS/PE/vehicle Runx3+/− mice (G), PE/PE/vehicle Runx3+/− mice (H), and PE/PE/AR460 (100 mg/kg) Runx3+/−(I) mice are shown (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E3. Decreased eosinophil accumulation in the intestines of PE/PE mice treated with the Pim1 kinase inhibitor. Eosinophils were identified and numbers were quantitated in the jejunum by staining with anti-MBP antibody. Representative sections of PBS/PE/vehicle WT mice (A), PE/PE/vehicle WT mice (B), PE/PE/AR460 (1 mg/kg) WT mice (C), PE/PE/AR460 (10 mg/kg) WT mice (D), PE/PE/AR460 (30 mg/kg) WT mice (E), PE/PE/AR460 (100 mg/kg) WT mice (F), PBS/PE/vehicle Runx3+/− mice (G), PE/PE/vehicle Runx3+/− mice (H), and (PE/PE/AR460 (100 mg/kg) Runx3+/− mice (I) are shown (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E4. Decreased numbers of goblet cells in intestinal epithelia of sensitized and challenged mice treated with the Pim1 kinase inhibitor. Goblet cells were identified by means of periodic acid–Schiff staining 24 hours after the last challenge. Representative sections of PBS/PE/vehicle WT mice (A), PE/PE/vehicle WT mice (B), PE/PE/AR460 (1 mg/kg) WT mice (C), PE/PE/AR460 (10 mg/kg) WT mice (D), PE/PE/AR460 (30 mg/kg) WT mice (E), PE/PE/AR460 (100 mg/kg) WT mice (F), PBS/PE/vehicle Runx3+/− mice (G), PE/PE/vehicle Runx3+/− mice (H), and PE/PE/AR460 (100 mg/kg) Runx3+/− mice (I) are shown (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E5. Effect of the Pim1 kinase inhibitor on cytokine production. Flow analysis of intracellular cytokine staining of isolated MLNs from WT and Runx3+/− mice is shown. Cells were stimulated with phorbol 12-myristate 13-acetate/ionomycin/brefeldin A, as described in the Methods section. Numbers shown in the quadrants represent percentages of the CD4+ T lymphocyte–gated cell population, and numbers are representative of 3 independent experiments. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E6. Immunohistochemical analysis of Runx3 expression in the jejunal tissues. Representative pictures show Runx3 protein in jejunums of PBS/PE/vehicle WT and Runx3+/− mice and PE/PE WT and Runx3+/− mice treated with AR460770 or vehicle (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E7. Pim1 and Runx3 mRNA expression in naive CD4 T cells differentiated in vitro into TH1, TH2, or TH17 cells from WT and Runx3+/− mice determined using real-time PCR. Data are from 3 independent experiments. *P < .05.
Fig E8. Cell viability was determined by means of trypan blue dye staining of polarized TH1, TH2, and TH17 cells cultured in the presence of different concentrations of the inhibitor for 6 days. Data shown are from 3 independent experiments.
Fig E9. Cell apoptosis was determined by means of flow cytometry with Annexin V and 7AAD dual staining of polarized TH1, TH2, and TH17 cells cultured in the presence of different concentrations of inhibitor for 6 days. Numbers indicate percentages of cells in each quadrant and are representative of 3 independent experiments.
Fig E10. Effect of the Pim1 kinase inhibitor on Runx3 protein levels in the polarized TH1, TH2, and TH17 cells determined by intracellular staining. Biotinylated rabbit anti-human Runx3 polyclonal antibody was prepared as described in the Methods section. Cells were stimulated with phorbol 12-myristate 13-acetate/ionomycin/brefeldin A, as described in the Methods section. Numbers shown in the quadrants represent percentages of the CD4+ T lymphocyte–gated cell population, and numbers are representative of 3 independent experiments.
Clinical implications.
Pim1 kinase and Runx3 are potential new targets for the treatment of peanut-induced anaphylaxis and intestinal inflammation.
Acknowledgments
Supported by National Institutes of Health grants HL-36577 and AI-77609. M.W. was supported by a fellowship from the Eugene F. and Easton M. Crawford Charitable Lead Unitrust. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Abbreviations used
- Cbfβ
Core binding factor β
- MLN
Mesenteric lymph node
- NFAT
Nuclear factor of activated T cells
- PE
Peanut extract
- Pim
Provirus integration site for Moloney murine leukemia virus
- RORγt
Retinoic acid–related orphan receptor γt
- Runx
Runt-related transcription factor
- T-bet
T-box transcription factor
- WT
Wild-type
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.Kweon MN, Yamamoto M, Kajiki M, Takahashi I, Kiyono H. Systemically derived large intestinal CD4 (+) Th2 cells play a central role in STAT6-mediated allergic diarrhea. J Clin Invest. 2000;106:199–206. doi: 10.1172/JCI8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, Takeda K, Shiraishi Y, Okamoto M, Dakhama A, Joetham A, et al. Peanut-induced intestinal allergy is mediated through a mast cell-IgE-FcepsilonRI-IL-13 pathway. J Allergy Clin Immunol. 2010;126:306–16. doi: 10.1016/j.jaci.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eigenmann PA. T lymphocytes in food allergy: overview of an intricate network of circulating and organ-resident cells. Pediatr Allergy Immunol. 2002;13:162–71. doi: 10.1034/j.1399-3038.2002.01015.x. [DOI] [PubMed] [Google Scholar]
- 4.Blumchen K, Ulbricht H, Staden U, Dobberstein K, Beschorner J, de Oliveira LC, et al. Oral peanut immunotherapy in children with peanut anaphylaxis. J Allergy Clin Immunol. 2010;126:83–91. doi: 10.1016/j.jaci.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37:726–30. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Aho TL, Sandholm J, Peltola KJ, Ito Y, Koskinen PJ. Pim-1 kinase phosphorylates RUNX family transcription factors and enhances their activity. BMC Cell Biol. 2006;7:21–9. doi: 10.1186/1471-2121-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rainio EM, Sandholm J, Koskinen PJ. Transcriptional activity of NFATc1 is enhanced by the Pim-1 kinase. J Immunol. 2002;168:1524–7. doi: 10.4049/jimmunol.168.4.1524. [DOI] [PubMed] [Google Scholar]
- 8.Wingett D, Long A, Kelleher D, Magnuson NS. Pim-1 proto-oncogene expression in anti-CD3-mediated T cell activation is associated with protein kinase C activation and is independent of Raf-1. J Immunol. 1996;156:549–57. [PubMed] [Google Scholar]
- 9.Shin YS, Takeda K, Shiraishi Y, Jia Y, Wang M, Jackson L, et al. Inhibition of Pim1 kinase activation attenuates allergen-induced airway hyperresponsiveness and inflammation. Am J Respir Cell Mol Biol. 2012;46:488–97. doi: 10.1165/rcmb.2011-0190OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 11.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate IFNγ and silence IL-4 in T helper type 1 cells. Nat Immunol. 2007;8:145–53. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 12.Kohu K, Ohmori H, Wong WF, Onda D, Wakoh T, Kon S, et al. The Runx3 transcription factor augments Th1 and down-modulates Th2 phenotypes by interacting with and attenuating GATA3. J Immunol. 2009;183:7817–24. doi: 10.4049/jimmunol.0802527. [DOI] [PubMed] [Google Scholar]
- 13.Lee SH, Jeong HM, Choi JM, Cho YC, Kim TS, Lee KY, et al. Runx3 inhibits IL-4 production in T cells via physical interaction with NFAT. Biochem Biophys Res Commun. 2009;381:214–7. doi: 10.1016/j.bbrc.2009.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, Woolf E, et al. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc Natl Acad Sci U S A. 2004;101:16016–21. doi: 10.1073/pnas.0407180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–15. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, et al. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J Allergy Clin Immunol. 2000;106:150–8. doi: 10.1067/mai.2000.107395. [DOI] [PubMed] [Google Scholar]
- 17.Friend DS, Ghildyal N, Austen KF, Gurish MF, Matsumoto R, Stevens RL. Mast cells that reside at different locations in the jejunum of mice infected with Trichinella spiralis exhibit sequential changes in their granule ultrastructure and chymase phenotype. J Cell Biol. 1996;135:279–90. doi: 10.1083/jcb.135.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomkinson A, Cieslewicz G, Duez C, Larson KA, Lee JJ, Gelfand EW. Temporal association between airway hyperresponsiveness and airway eosinophilia in ovalbumin-sensitized mice. Am J Respir Crit Care Med. 2001;163:721–30. doi: 10.1164/ajrccm.163.3.2005010. [DOI] [PubMed] [Google Scholar]
- 19.Komine O, Hayashi K, Natsume W, Watanabe T, Seki Y, Seki N, et al. The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J Exp Med. 2003;198:51–61. doi: 10.1084/jem.20021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashino S, Wakita D, Shiohama Y, Iwakura Y, Chamoto K, Ohkuri T, et al. A T(h) 17-polarized cell population that has infiltrated the lung requires cells that convert to IFN-γ production in order to induce airway hyperresponsiveness. Int Immunol. 2010;22:503–13. doi: 10.1093/intimm/dxq034. [DOI] [PubMed] [Google Scholar]
- 21.Ohnishi H, Miyahara N, Dakhama A, Takeda K, Mathis S, Haribabu B, et al. Corticosteroids enhance CD8+ T cell-mediated airway hyperresponsiveness and allergic inflammation by upregulating leukotriene B4 receptor 1. J Allergy Clin Immunol. 2008;121:864–71. doi: 10.1016/j.jaci.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto M, Takeda K, Joetham A, Ohnishi H, Matsuda H, Swasey CH, et al. Essential role of Notch signaling in effector memory CD8+ T cell-mediated airway hyperresponsiveness and inflammation. J Exp Med. 2008;205:1087–97. doi: 10.1084/jem.20072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tesmer LA, Lundy SK, Sarkar S, Fox DA. Th17 cells in human disease. Immunol Rev. 2008;223:87–113. doi: 10.1111/j.1600-065X.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–76. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 25.Lajoie S, Lewkowich IP, Suzuki Y, Clark JR, Sproles AA, Dienger K, et al. Complement-mediated regulation of the IL-17A axis is a central genetic determinant of the severity of experimental allergic asthma. Nat Immunol. 2010;11:928–35. doi: 10.1038/ni.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aujla M. Targeted therapies: Pim kinase inhibition and chemoresistance. Nat Rev Clin Oncol. 2010;7:3. [Google Scholar]
- 27.Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller L. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95:1004–15. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox CJ, Hammerman PS, Thompson CB. The Pim kinases control rapamycinresistant T cell survival and activation. J Exp Med. 2005;201:259–66. doi: 10.1084/jem.20042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aho TL, Lund RJ, Ylikoski EK, Matikainen S, Lahesmaa R, Koskinen PJ. Expression of human pim family genes is selectively up-regulated by cytokines promoting T helper type 1, but not T helper type 2, cell differentiation. Immunology. 2005;116:82–8. doi: 10.1111/j.1365-2567.2005.02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dautry F, Weil D, Yu J, Dautry-Varsat A. Regulation of pim and myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J Biol Chem. 1988;263:17615–20. [PubMed] [Google Scholar]
- 31.Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I. Interferon-α activates multiple STAT proteins and up-regulates proliferation-associated IL-2Rα, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–91. [PubMed] [Google Scholar]
- 32.Stout BA, Bates ME, Liu LY, Farrington NN, Bertics PJ. IL-5 and granulocytemacrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J Immunol. 2004;173:6409–17. doi: 10.4049/jimmunol.173.10.6409. [DOI] [PubMed] [Google Scholar]
- 33.Jackson LJ, Wright D, Gross S, Robinson J, Marmsater FP, Allen S, et al. Inhibition of T cell function in vitro and in vivo using small molecule antagonists of PIM kinases. J Immunol. 2009;182:35. [Google Scholar]
- 34.Walter DM, McIntire JJ, Berry G, McKenzie AN, Donaldson DD, DeKruyff RH, et al. Critical role for IL-13 in the development of allergen induced airway hyperreactivity. J Immunol. 2001;167:4668–75. doi: 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- 35.Miyahara S, Miyahara N, Matsubara S, Takeda K, Koya T, Gelfand EW. IL-13 is essential to the late-phase response in allergic rhinitis. J Allergy Clin Immunol. 2006;118:1110–6. doi: 10.1016/j.jaci.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Donaldson DD, Whitters MJ, Fitz LJ, Neben TY, Finnerty H, Henderson SL, et al. The murine IL-13 receptor alpha 2: molecular cloning, characterization, and comparison with murine IL-13 receptor alpha 1. J Immunol. 1998;161:2317–24. [PubMed] [Google Scholar]
- 37.Taube C, Miyahara N, Ott V, Swanson B, Takeda K, Loader J, et al. The leukotriene B4 receptor (BLT1) is required for effector CD8+ T cell-mediated, mast cell-dependent airway hyperresponsiveness. J Immunol. 2006;176:3157–64. doi: 10.4049/jimmunol.176.5.3157. [DOI] [PubMed] [Google Scholar]
- 38.Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, et al. A novel subset of CD4 (+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med. 2010;207:2479–91. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 40.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–55. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–5. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 43.Fainaru O, Shseyov D, Hantisteanu S, Groner Y. Accelerated chemokine receptor 7-mediated dendritic cell migration in Runx3 knockout mice and the spontaneous development of asthma-like disease. Proc Natl Acad Sci U S A. 2005;102:10598–603. doi: 10.1073/pnas.0504787102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–30. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 45.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbf1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 46.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–32. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 47.Haagerup A, Bjerke T, Schiøtz PO, Binderup HG, Dahl R, Kruse TA. Asthma and atopy—a total genome scan for susceptibility genes. Allergy. 2002;57:680–6. doi: 10.1034/j.1398-9995.2002.23523.x. [DOI] [PubMed] [Google Scholar]
- 48.Calabi F, Rhodes M, Williamson P, Boyd Y. Identification and chromosomal mapping of a third mouse runt-like locus. Genomics. 1995;26:607–10. doi: 10.1016/0888-7543(95)80184-n. [DOI] [PubMed] [Google Scholar]
- 49.Christensen U, Møller-Larsen S, Nyegaard M, Haagerup A, Hedemand A, Brasch-Andersen C, et al. Linkage of atopic dermatitis to chromosomes 4q22, 3p24 and 3q21. Hum Genet. 2009;126:549–57. doi: 10.1007/s00439-009-0692-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig E1. Treatment with a Pim1 kinase inhibitor had no effect on serum immunoglobulin production in WT and Runx3+/− mice. Serum levels of peanut-specific IgE, IgG1, and IgG2a were assessed by means of ELISA 24 hours after the last challenge and expressed as the optical density of diluted serum, as described in the Methods section. Results were obtained from 3 individual experiments with 4 mice per group. n.s., Not significant; PBS/PE, sham sensitized but PE challenged; PE/PE, PE sensitized and challenged. *P < .05 and **P < .01.
Fig E2. Decreased mast cell infiltration in the intestinal walls of PE/PE mice treated with the Pim1 kinase inhibitor. Intestinal mucosal mast cells were quantified in the jejunum by using chloroacetate esterase staining. Representative sections of PBS/PE/vehicle WT mice (A), PE/PE/vehicle WT mice (B), PE/PE/AR460 (1 mg/kg) WT mice (C), PE/PE/AR460 (10 mg/kg) WT mice (D), PE/PE/AR460(30 mg/kg) WT mice (E), PE/PE/AR460 (100 mg/kg) WT mice (F), PBS/PE/vehicle Runx3+/− mice (G), PE/PE/vehicle Runx3+/− mice (H), and PE/PE/AR460 (100 mg/kg) Runx3+/−(I) mice are shown (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E3. Decreased eosinophil accumulation in the intestines of PE/PE mice treated with the Pim1 kinase inhibitor. Eosinophils were identified and numbers were quantitated in the jejunum by staining with anti-MBP antibody. Representative sections of PBS/PE/vehicle WT mice (A), PE/PE/vehicle WT mice (B), PE/PE/AR460 (1 mg/kg) WT mice (C), PE/PE/AR460 (10 mg/kg) WT mice (D), PE/PE/AR460 (30 mg/kg) WT mice (E), PE/PE/AR460 (100 mg/kg) WT mice (F), PBS/PE/vehicle Runx3+/− mice (G), PE/PE/vehicle Runx3+/− mice (H), and (PE/PE/AR460 (100 mg/kg) Runx3+/− mice (I) are shown (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E4. Decreased numbers of goblet cells in intestinal epithelia of sensitized and challenged mice treated with the Pim1 kinase inhibitor. Goblet cells were identified by means of periodic acid–Schiff staining 24 hours after the last challenge. Representative sections of PBS/PE/vehicle WT mice (A), PE/PE/vehicle WT mice (B), PE/PE/AR460 (1 mg/kg) WT mice (C), PE/PE/AR460 (10 mg/kg) WT mice (D), PE/PE/AR460 (30 mg/kg) WT mice (E), PE/PE/AR460 (100 mg/kg) WT mice (F), PBS/PE/vehicle Runx3+/− mice (G), PE/PE/vehicle Runx3+/− mice (H), and PE/PE/AR460 (100 mg/kg) Runx3+/− mice (I) are shown (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E5. Effect of the Pim1 kinase inhibitor on cytokine production. Flow analysis of intracellular cytokine staining of isolated MLNs from WT and Runx3+/− mice is shown. Cells were stimulated with phorbol 12-myristate 13-acetate/ionomycin/brefeldin A, as described in the Methods section. Numbers shown in the quadrants represent percentages of the CD4+ T lymphocyte–gated cell population, and numbers are representative of 3 independent experiments. PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E6. Immunohistochemical analysis of Runx3 expression in the jejunal tissues. Representative pictures show Runx3 protein in jejunums of PBS/PE/vehicle WT and Runx3+/− mice and PE/PE WT and Runx3+/− mice treated with AR460770 or vehicle (magnification ×200). PBS/PE, Sham sensitized but PE challenged; PE/PE, PE sensitized and challenged.
Fig E7. Pim1 and Runx3 mRNA expression in naive CD4 T cells differentiated in vitro into TH1, TH2, or TH17 cells from WT and Runx3+/− mice determined using real-time PCR. Data are from 3 independent experiments. *P < .05.
Fig E8. Cell viability was determined by means of trypan blue dye staining of polarized TH1, TH2, and TH17 cells cultured in the presence of different concentrations of the inhibitor for 6 days. Data shown are from 3 independent experiments.
Fig E9. Cell apoptosis was determined by means of flow cytometry with Annexin V and 7AAD dual staining of polarized TH1, TH2, and TH17 cells cultured in the presence of different concentrations of inhibitor for 6 days. Numbers indicate percentages of cells in each quadrant and are representative of 3 independent experiments.
Fig E10. Effect of the Pim1 kinase inhibitor on Runx3 protein levels in the polarized TH1, TH2, and TH17 cells determined by intracellular staining. Biotinylated rabbit anti-human Runx3 polyclonal antibody was prepared as described in the Methods section. Cells were stimulated with phorbol 12-myristate 13-acetate/ionomycin/brefeldin A, as described in the Methods section. Numbers shown in the quadrants represent percentages of the CD4+ T lymphocyte–gated cell population, and numbers are representative of 3 independent experiments.