To the editor
At the present time, we do not have access to predictors of response to hypomethylating agents. MicroRNA (miRNAs) are noncoding RNAs of 19–25 nucleotides in length that regulate gene expression by inducing translational inhibition and cleavage of their target mRNAs through base pairing to partially or fully complementary sites. Altered miRNA expression has been documented in multiples types of leukemia [1]. Acute myeloid leukemia (AML) is a cytogenetically and molecularly heterogeneous disorder characterized by differentiation arrest and malignant proliferation of clonal myeloid precursors. Specific miRNA signatures are associated with cytogenetics and prognosis in AML [2]. Recently it has been reported that pretreatment expression levels of miR-29b are associated with clinical response in patients with AML treated with a 10-day schedule of decitabine [3]. miR-29b is involved in the regulation of DNA methylation and is down regulated in AML [3,4]. The hypothesis is that patients with higher miR-29b levels would have lower levels of DNA methyltransferase, less hypomethylation, and potentially increased sensitivity to decitabine [3]. This hypothesis is in line with previous observations associating lower levels of p15 methylation with response to decitabine based therapy [5]. The findings of Blum et al. [3] are therefore very significant as detection of miR-29b, if validated, could be used to select hypomethylating-based therapy in AML.
To follow on these observations, we have analyzed miR-29b and miR-101 expression levels in a cohort of patients with AML or high-risk myelodysplastic syndrome treated in a phase I/II study of the combination of 5-azacitidine (5-AZA), valproic acid, and ATRA [6]. We analyzed 45 patients, 28 of which had not received prior therapy. The characteristics of these patients have been previously reported [6]. Eight (29%) patients achieved a complete remission and 1 (4%) partial remission. Total cellular RNA (extracted from unselected peripheral blood mononuclear cells collected before the first cycle of treatment) was used for reverse transcription reactions using Taq-Man miRNA reverse transcription kit (Applied Biosystems). As a control, we also analyzed levels of miR-101. For real-time PCR, miR-29b and miR-101 TaqMan - miRNA assays were purchased from Applied Biosystems and analyzed with TaqMan Universal PCR Master Mix (Applied Biosystems) using an Applied Biosystems Prism 7500 Sequencing detection system. The U6 small nuclear RNA was used as internal control. Expression levels of miR-29b and miR-101 from 45 patients were compared to 6 healthy donors. Expression levels of miR-29b were significantly down-regulated in patients compared to normal controls (P = 0.001) (Fig. 1A). There were no difference in terms of miR-101 expression between patients and health donors. We then compared the miR-29b and miR-101 expression levels between patients who responded to the combination therapy versus nonresponders: no differences were observed for both miR-29b and miR-101 expression (P = 0.56) (Fig. 1B).
Figure 1.
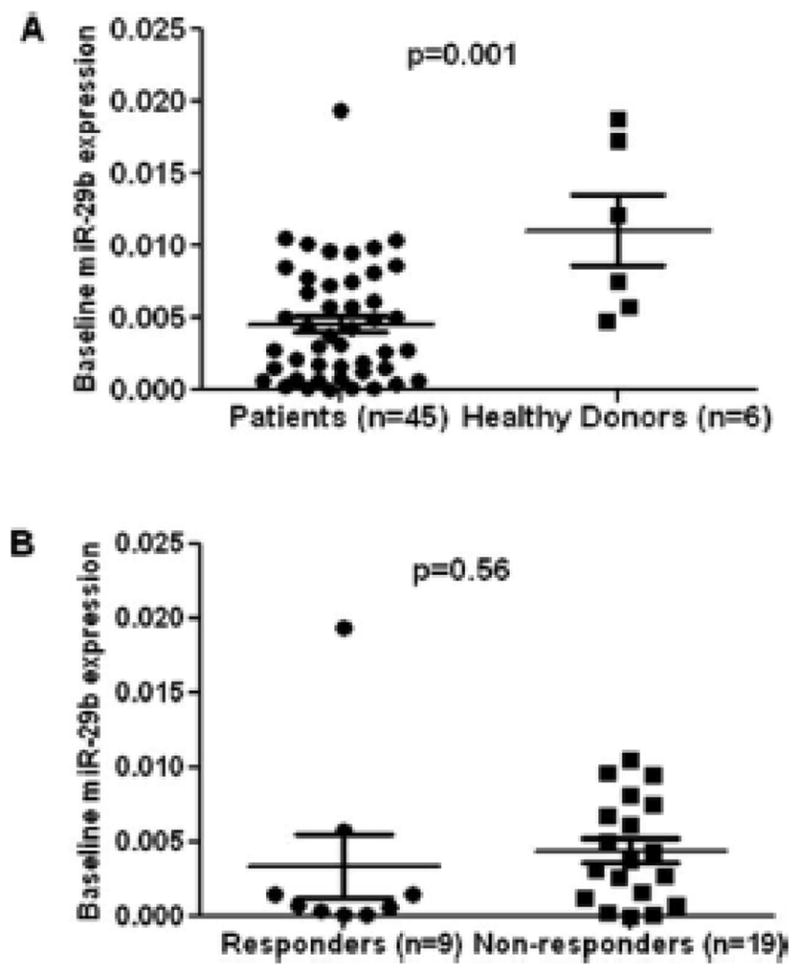
Difference in baseline miR-29b expression levels. (A) Difference in baseline miR-29b expression levels between AML patients and healthy controls. (B) Difference in baseline miR-29b expression levels between responders and nonresponders. Exact P values are shown over the columns; unpaired, two-tailed t test were performed for statistical analysis.
Our results fail to detect a relationship between miR-29b levels and response to 5-AZA based therapy. This difference could be explained by the relative small sample size, the source of RNA (peripheral blood versus bone marrow) and potentially the fact that miR-29b may predict for response to decitabine but not to 5-AZA in combination. In regards to the source of RNA, it should be noted that Blum et al. used unsorted bone marrow specimens whereas we used peripheral blood. The median blast percent in the blood of our patients was 6% (range 0–86%) whereas it was 35% (range 8–93%) in the bone marrow. In our experience, it is unlikely that the differences in blast percentage would explain the potential differences in gene expression levels. Despite these limitations, these results indicate that larger studies need to be performed to confirm the role of miRNAs in response to hypomethylating agent based therapy.
Footnotes
Conflict of interest: Nothing to report.
Author Contributions
H.Y. designed and performed research, analyzed data and wrote the manuscript; Z.F., Y.W., Y.H., G.C., and H. K. performed research, G.G.-M. designed experiment, analyzed data, wrote manuscript and funded the study.
References
- 1.Fabbri M, Garzon R, Andreeff M, et al. MicroRNAs and noncoding RNAs in hematological malignancies: Molecular, clinical and therapeutic implications. Leukemia. 2008;22:1095–1105. doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- 2.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum W, Garzon R, Klisovic RB, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci USA. 2010;107:7473–7478. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garzon R, Liu S, Fabbri M, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soriano AO, Yang H, Faderl S, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110:2302–2308. doi: 10.1182/blood-2007-03-078576. [DOI] [PubMed] [Google Scholar]


