Abstract
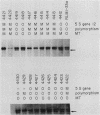
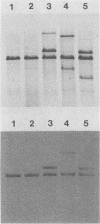
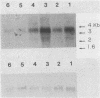
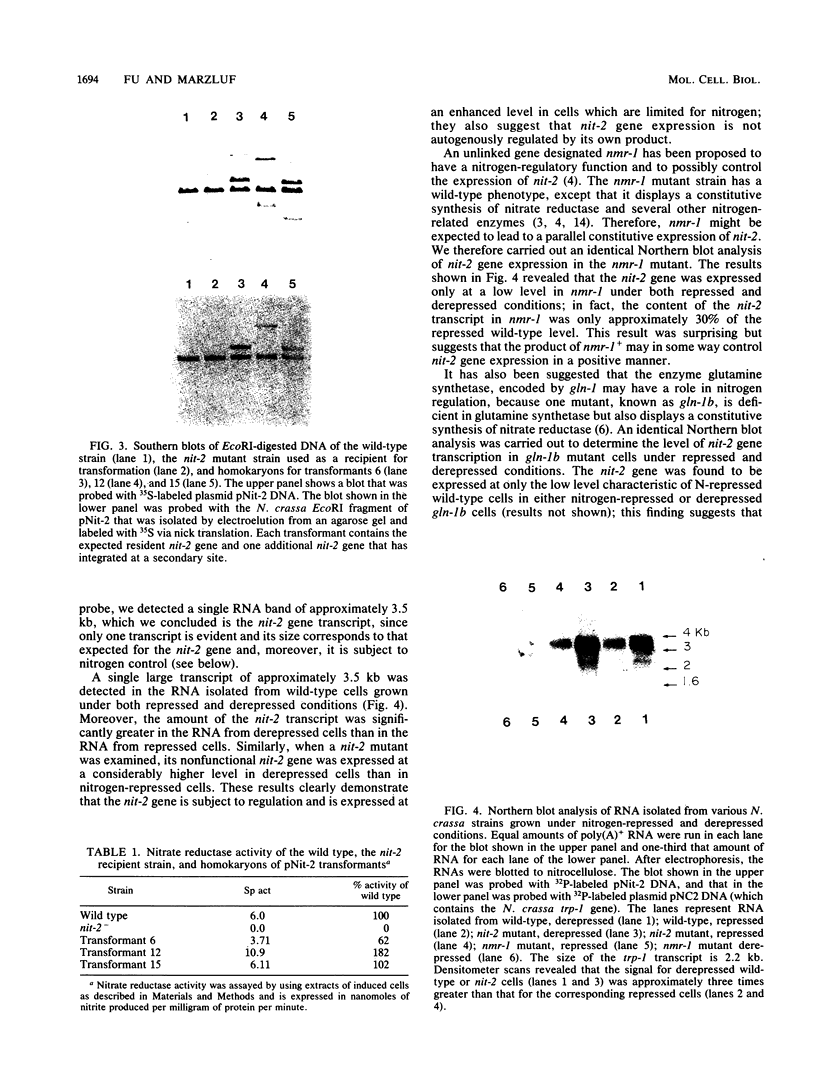
The nit-2 gene is the major nitrogen-regulatory gene of Neurospora crassa, and under conditions of nitrogen limitations, it turns on the expression of various unlinked structural genes which specify nitrogen-catabolic enzymes. The nit-2 gene was subcloned as a 6-kilobase (kb) DNA fragment from a cosmid that carried approximately a 40-kb N. crassa DNA insert. The nit-2 gene was localized in a DNA segment of approximately 3.5 kb and was shown to correspond to a unique DNA sequence located on linkage group 1. Several N. crassa nit-2 transformants were characterized and were found to possess significantly different levels of the regulated enzyme nitrate reductase. Northern blot analysis of RNA from various strains was carried out to determine whether the nit-2 gene was expressed constitutively or was itself subject to regulation. The results revealed that the nit-2 gene is transcribed to give a single large mRNA of approximately 3.5 kb. Expression of the nit-2 gene is regulated such that its transcript is present at a substantially higher level in cells which are limited for nitrogen than in cells growing under nitrogen-repressed conditions. However, the nit-2 gene is not controlled by autogenous regulation. The nit-2 gene was transcribed only at a low level in nmr-1 and in gln-1b, under both nitrogen-repressed and derepressed conditions, suggesting that these unlinked loci may exert a positive regulatory effect on nit-2.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBusk R. M., Ogilvie S. Regulation of amino acid utilization in Neurospora crassa: effect of nmr-1 and ms-5 mutations. J Bacteriol. 1984 Nov;160(2):656–661. doi: 10.1128/jb.160.2.656-661.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale S. S., Marzluf G. A. Transformation of Neurospora crassa with circular and linear DNA and analysis of the fate of the transforming DNA. Curr Genet. 1985;10(3):205–212. doi: 10.1007/BF00798750. [DOI] [PubMed] [Google Scholar]
- Dunn-Coleman N. S., Garrett R. H. The role fo glutamine synthetase and glutamine metabolism in nitrogen metabolite repression, a regulatory phenomenon in the lower eukaryote Neurospora crassa. Mol Gen Genet. 1980;179(1):25–32. doi: 10.1007/BF00268442. [DOI] [PubMed] [Google Scholar]
- Grove G., Marzluf G. A. Identification of the product of the major regulatory gene of the nitrogen control circuit of Neurospora crassa as a nuclear DNA-binding protein. J Biol Chem. 1981 Jan 10;256(1):463–470. [PubMed] [Google Scholar]
- Kinsey J. A., Rambosek J. A. Transformation of Neurospora crassa with the cloned am (glutamate dehydrogenase) gene. Mol Cell Biol. 1984 Jan;4(1):117–122. doi: 10.1128/mcb.4.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marzluf G. A. Regulation of nitrogen metabolism and gene expression in fungi. Microbiol Rev. 1981 Sep;45(3):437–461. doi: 10.1128/mr.45.3.437-461.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine K. G., Marzluf G. A. Amber nonsense mutations in regulatory and structural genes of the nitrogen control circuit of Neurospora crassa. Curr Genet. 1986;10(9):677–684. doi: 10.1007/BF00410916. [DOI] [PubMed] [Google Scholar]
- Reinert W. R., Marzluf G. A. Genetic and metabolic control of the purine catabolic enzymes of Neurospora crasse. Mol Gen Genet. 1975 Aug 5;139(1):39–55. doi: 10.1007/BF00267994. [DOI] [PubMed] [Google Scholar]
- Reinert W. R., Patel V. B., Giles N. H. Genetic regulation of the qa gene cluster of Neurospora crassa: induction of qa messenger ribonucleic acid and dependency on qa-1 function. Mol Cell Biol. 1981 Sep;1(9):829–835. doi: 10.1128/mcb.1.9.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]