Abstract
Impaired energy metabolism in neurons is integral to a range of neurodegenerative diseases, from Alzheimer's disease to stroke. To investigate the complex molecular changes underpinning cellular adaptation to metabolic stress, we have defined the proteomic response of the SH-SY5Y human neuroblastoma cell line after exposure to a metabolic challenge of oxygen glucose deprivation (OGD) in vitro. A total of 958 proteins across multiple subcellular compartments were detected and quantified by label-free liquid chromatography mass spectrometry. The levels of 130 proteins were significantly increased (P<0.01) after OGD and the levels of 63 proteins were significantly decreased (P<0.01) while expression of the majority of proteins (765) was not altered. Network analysis identified novel protein–protein interactomes involved with mitochondrial energy production, protein folding, and protein degradation, indicative of coherent and integrated proteomic responses to the metabolic challenge. Approximately one third (61) of the differentially expressed proteins was associated with the endoplasmic reticulum and mitochondria. Electron microscopic analysis of these subcellular structures showed morphologic changes consistent with the identified proteomic alterations. Our investigation of the global cellular response to a metabolic challenge clearly shows the considerable adaptive capacity of the proteome to a slowly evolving metabolic challenge.
Keywords: Alzheimer's, cell culture, energy metabolism, mitochondria, neurodegeneration, proteomics
Introduction
Impaired energy metabolism in neurons is integral to many central nervous system diseases. An acute, intense metabolic challenge during conditions such as stroke and brain trauma can induce irreversible neuronal damage.1 By contrast, age-related chronic metabolic impairment is thought to contribute to the evolution of neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and Huntington's disease.2, 3 Neurochemical cascades initiated by both acute and prolonged metabolic challenges can induce cell death via an elevation of proapoptotic proteins. These metabolic challenges can also activate genomic and proteomic responses promoting cell survival in the immediate phase as well as preparing the cell to deal with similar challenges in the future (conditioning).4
Subcellular organelles display complex proteomic responses to impaired energy metabolism, with these responses ultimately determining the death or survival of the cell. In mitochondria, metabolic stress can disrupt energy generation, provoke mitochondrial swelling, and initiate apoptosis, all with potentially lethal consequences.5 In response to cell stress, the endoplasmic reticulum (ER) detects and reconfigures misfolded proteins in a process known as the unfolded protein response (UPR). The UPR is integral to cell survival as excessive accumulation of these damaged proteins in the ER lumen can result in the suppression of protein synthesis and irreversible pathology.6 In the nucleus and ribosomes, metabolic stress leads to a genomic response and the synthesis of proteins such as heat-shock proteins and trophic factors, which contribute to protein repair and neuronal survival.7 The genomic response to metabolic challenge has been well characterized both in vitro and in vivo.8, 9, 10 In the present study, we provide a comprehensive cellular proteomic response and explore the extent to which proteins are altered after a metabolic challenge.
Oxygen glucose deprivation (OGD) induces metabolic stress in neurons in culture.11, 12 The intensity of the metabolic challenge can be controlled by adjusting experimental conditions such as the choice of cell line and alterations in the duration of OGD. Mass spectrometry-based proteomics provides an excellent platform to quantify OGD-dependent protein changes and to gain a fresh perspective on adaptations that occur within the proteome in response to metabolic stress.13 We have used human SH-SY5Y neuroblastoma cells and label-free liquid chromatography mass spectrometry (LC-MS) to define the integrated proteomic response to metabolic stress that induces severe mitochondrial dysfunction but only limited cell death. The majority of proteins analyzed (800 out of 1,000) were not significantly altered by OGD. Protein increases after OGD was more than twice as frequent as was decreases, consistent with an integrated cellular survival response. Protein interactomes highlight coordinated proteomic responses after OGD in relation to mitochondrial energy production, ER stress, and ribosomal dysfunction.
Materials and methods
Reagents
All standard growth media used was obtained from Invitrogen (Paisley, UK), unless otherwise stated, and all procedures were conducted in compliance with all relevant legislation in the United Kingdom. Cells were routinely maintained in standard 75 cm2 cell culture-treated flasks (Greiner Bio One, Stonehouse, UK) within a continuous flow incubator at 37°C, with 5% CO2 in fully humidified air. All growth media and solutions were prewarmed to 37°C before contact with cells, unless otherwise stated. The acetonitrile and water used in the LC-MS buffer solutions were of HPLC grade (Fisher, Loughborough, UK). Formic acid was of 98% to 100% purity (Merck, Darmstadt, Germany) and trifluoroacetic acid was 99% sequencing grade (Sigma-Aldrich, Gillingham, UK). All other chemicals used in the processing of the protein samples were of reagent grade or better. The trypsin used for protein digestion was from Promega (Southampton, UK).
Cell Culture and Oxygen Glucose Deprivation
SH-SY5Y cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F12 (DMEM/F12) supplemented with 2 mmol/L ℒ-Glutamine, 100 U/mL Penicillin, 100 mg/mL Streptomycin, and fetal bovine serum. Cells were routinely maintained at 37°C, 5% CO2, with culture medium replaced biweekly. Oxygen glucose deprivation (for 3, 6, 9, 12, and 18 hour durations) was achieved through incubation of the cells at 37°C in 95% N2:5% CO2. Aglycemia was induced by replacing standard culture medium with glucose-free DMEM supplemented with fetal bovine serum, after a glucose-free wash step. Control cells underwent the same glucose-free wash step, followed by incubation with glucose containing media (4,500 mg/L glucose). The osmolality of the DMEM solutions used was within the range of 300 to 340 mOsm/kg for the no glucose solutions, and 320 to 360 mOsm/kg for the glucose containing media. All media were equilibrated overnight before experimentation in either normoxic or anoxic chambers. Oxygen content in the media was recorded in preliminary studies using a fiber optic dual oxygen-temperature sensing probe (OxyLite 2000 E Series, Oxford Optronix, Oxford, UK). A rapid oxygen depletion phase was recorded, with oxygen partial pressure falling exponentially from 140 to 7 mm Hg over the initial 3 hours of OGD. Oxygen depletion plateaued and remained severely hypoxic between 4 and 18 hours with a recorded average partial pressure of 2 mm Hg.
Assessment of Mitochondrial Function (MTS Assay)
Cells were seeded onto poly-𝒟-lysine coated flat-bottomed 96-well plates at a density of 125,000 cells per well overnight. After OGD, glucose-free DMEM was replaced with 200 μL glucose-containing DMEM and cell viability was assessed using 20 μL of CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Madison, USA) as per the manufacturer's guidelines. Cells were incubated for 2 hours in glucose containing, normoxic media (37°C, 5% CO2) to allow residual mitochondrial function after OGD to be assessed using a Dynex MRX plate reader (Dynex Technologies Ltd., Worthing, UK) at 490λ.
Assessment of Global Cell Death (Trypan Blue)
Trypan blue staining was carried out to assess the extent of cell death occurring after each OGD time point. Cells were collected immediately at the end of each time point by trypsinization, centrifugation (200 g, 7 minutes) and resuspension in glucose-containing DMEM. Cells were incubated with 0.1% trypan blue as per the manufacturer's instructions (Invitrogen). A hemocytometer was used to count stained and unstained cells. The percentage of stained cells in each OGD and time-matched control sample was calculated.
Preparation of Protein Samples for Liquid-Chromatography Mass Spectrometry
SH-SY5Y cells were treated with 6 hours OGD (n=6 independent samples per group) and 18 hours OGD (n=6), with a time-matched control for each sample. After OGD or ‘control' incubations, SH-SY5Y cells were immediately scraped and centrifuged (200 g, 7 minutes). Cell pellets were washed in Hanks Buffered Saline Solution and recentrifuged. Washed cell pellets were resuspended in 1 ml Hanks Buffered Saline Solution and centrifuged (6,000 g, 3 minutes). Supernatants were removed and the pellets snap frozen for temporary storage at −80°C. Cell pellets were defrosted and lysed in 100 μL 8 M urea using a cordless motor pellet pestle (Sigma-Aldrich, Dorset, UK). Samples were then centrifuged (14,000 g, 15 minutes) and supernatants recovered. Total protein content was determined using the Pierce BCA Protein Assay Kit (Thermo-Fisher, Loughborough, UK). Protein extract (140 μg) from OGD and control samples were digested as previously described.14
Liquid Chromatography Mass Spectrometry
Label-free intensity-based LC-MS was used to identify proteins differentially expressed after 6 and 18 hours OGD relative to time-matched controls. Capillary LC-MS data were acquired on an online system consisting of a micro-pump (1200 binary HPLC system, Agilent, Edinburgh, UK) coupled to a hybrid LTQ-Orbitrap XL instrument controlled through Xcalibur 2.0.7 (Thermo-Fisher). Peptides were reconstituted in 20 μL of loading buffer before injection and 8 μL was loaded. The HPLC separation was provided by a gradient between buffer A (97.5% H2O, 2.5% acetonitrile, 0.1% formic acid) and buffer B (90% acetonitrile, 10% H2O, 0.025% trifluoroacetic acid, 0.1% formic acid). The peptide mixture was separated on a 140-minute gradient. The mass spectrometer was operated in ‘data-dependent mode', with a single MS scan (400 to 2,000 m/z) followed by MS/MS scans in the linear ion trap on the five most abundant ions and excluded for 120 seconds.
Protein Identification and Quantification
All LC-MS runs were analyzed using a label-free intensity-based approach with Progenesis (version 4.0 Nonlinear Dynamics, Newcastle, UK), under randomized conditions. The MS/MS data were searched using MASCOT versions 2.3 (Matrix Science Ltd, London, UK) against a human subset of the NCBI database (downloaded on 12 January 2011) with 34,281 sequences using a maximum missed-cut of 2. Variable methionine oxidation, STY phosphorylation, protein N-terminal acetylation, and fixed cysteine carbamidomethylation were used in all searches.
LC-MS label-free quantification was performed using Progenesis 4.0 (Nonlinear Dynamics). Protein conflict (peptides shared between different proteins) was solved as followed: conflict resulting from multiple sequence assignment to the same peak; we kept only the sequence having the highest score. Conflict resulting from same peptide sequences assigned to different proteins, the assignment was singly attributed to the protein that had the highest number of peptides. Regarding the label-free quantitation, the total number of Features (i.e., intensity signal at a given retention time and m/z) was reduced to MSMS peaks with charge of 2, 3, or 4+ and we only kept the five most intense MSMS spectra per ‘Feature'. The subset of multicharged ions (2+, 3+, and 4+) was extracted from each LC-MS run and the ions intensities summed for normalization. Protein quantitation was performed as follows; for a specific protein, the associated unique peptide ions were summed to generate an abundance value. The measured protein abundances were transformed using an ArcSinH function (as the method of detection can generate a significant amount of near zero measurement for which a log transform is not ideal). The within-group means were calculated to determine the fold change and the transformed data were then used to calculate the P values using one-way analysis of variance. Significant changes in protein expression (P<0.01) between control and OGD experimental groups were determined for each time point (6 and 18 hours) using Student's t-test.
Analysis of Protein Subcellular Localization
All 958 proteins identified by LC-MS by at least two or more peptides were uploaded to the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov15) to determine subcellular localization based on gene ontology (GO). Protein clusters founded on subcellular localization with DAVID scores >2 were used. Proteins significantly altered (P<0.01) after 18 hours OGD were also assessed by DAVID according to subcellular localization.
Interactome Analysis of Proteins Altered with Oxygen Glucose Deprivation
Protein identifiers of all the proteins significantly altered after 18 hours OGD (P<0.01) were uploaded with their corresponding fold change values to Ingenuity Pathway Analysis software (IPA; http://www.ingenuity.com). Interactomes were algorithmically generated based on direct relationships (physical interactions and associations) between eligible proteins. The interactomes are color coded with green nodes representing proteins that were downregulated after 18 hours OGD and red protein nodes representing proteins that were upregulated after 18 hours OGD. The shading of the interactome nodes is positively correlated with the magnitude of fold change. The IPA generates a score for each interactome, which is a putative measure of probability (see Deighton et al16 for critical analysis).
Subcellular Morphology with Electron Microscopy
SH-SY5Y cells were exposed to OGD using the same paradigm as the LC-MS study. Samples were collected immediately after 3, 9, and 18 hours of OGD and fixed for a minimum of 20 hours in aldehyde fixative (4% paraformaldehyde and 5% glutaradehyde in 0.08 M sodium cacodylate buffer, pH 7.2). Once fixed, cells were centrifuged at 100 g, the supernatant removed and replaced with a 3% solution of agar (type VII, low gelling temperature; Sigma-Aldrich, Gillingham, UK). The gels were set at 4°C for 5 minutes before being cut into 1 mm3 blocks and processed for electron microscopy (EM). Processing, embedding, and staining were performed, as described previously.17 Briefly, the blocks were cut into 70 nm thick sections, collected on 300 mesh copper grids, stained with Reynold's lead citrate and contrasted with uranyl acetate.
The EM sections were evaluated on a JEOL CX-100 II transmission electron microscope (Tokyo, Japan). The evaluator was blinded to the treatment condition of all samples. Cells were chosen from grid squares in a predetermined pattern. Fifty cells were assessed from each treatment. For each cell, the condition of mitochondria and rough ER was assessed. Mitochondria were graded as ‘normal' (cristae clearly visible across >50% of the matrix), ‘swollen' (with a proportion of the matrix lost) and ‘abnormal' (with most/ the entire matrix lost). The ER in each cell was graded similarly, i.e., ‘normal' (where ER outer membranes ran parallel to each other), ‘swollen' (where modest swelling of ER outer membrane was present), or ‘abnormal' where ER displayed extensive membrane swelling and distortions. A Chi-Squared 3 × 2 matrix was used to test the null hypothesis that OGD would have no effect on the ultrastructure of organelles.
Results
Mitochondrial Function, Cell Viability, and Ultrastructure after Oxygen Glucose Deprivation
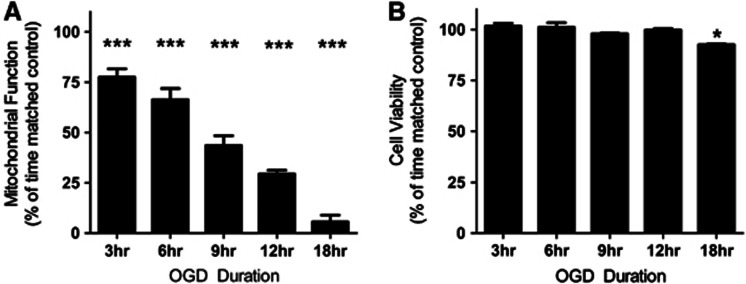
Normal mitochondrial function was significantly reduced in SH-SY5Y cells after OGD relative to time-matched control cells (P<0.0001, at each time point analyzed), with longer durations of OGD inducing more severe mitochondrial dysfunction (Figure 1A). Viability of SH-SY5Y cells treated with OGD was quantified using Trypan Blue staining (Figure 1B). Minimal cell death was detected after OGD, with 91% of cells remaining viable after 18 hours OGD.
Figure 1.
Mitochondrial function decreases after oxygen glucose deprivation (OGD), whereas cell viability is largely maintained. (A) Mitochondrial function is significantly decreased at each OGD time point compared with time-matched controls. Mitochondrial function was assessed using an MTS assay, with data presented as mean±s.d., n=8 for each time point. (B) Cell viability is maintained up to 12 hours OGD, with an increase in cell death after 18 hours OGD. Cell viability was assessed using a trypan blue dye exclusion assay. Data are presented as mean±s.d., n=3 for each time point. All data are expressed as a percentage of contemporaneous time matched controls (*P<0.05, ***P<0.0001, T-test).
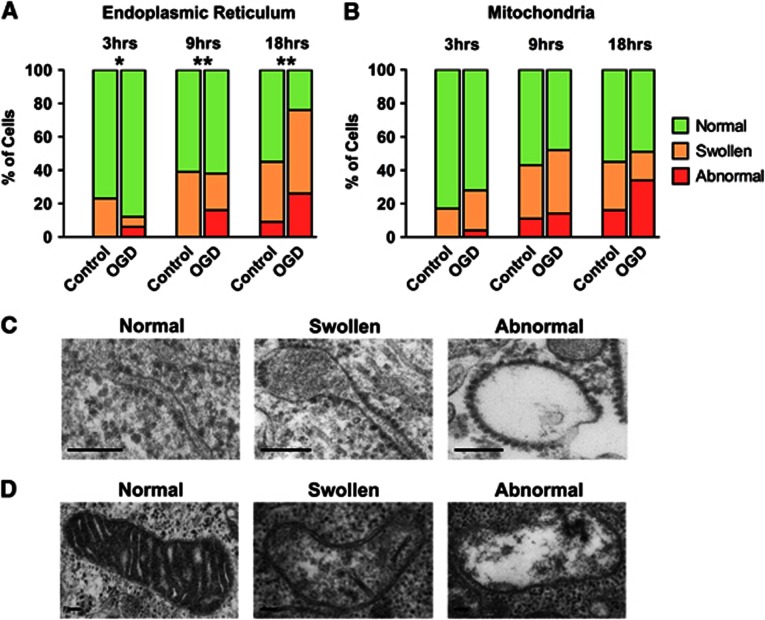
Ultrastructural morphologic changes in subcellular organelles were analyzed relative to time-matched controls by EM after OGD. With increasing OGD duration, the number of cells displaying abnormal ER was significantly increased (Chi-Square, P<0.01) (Figures 2A and 2C). SH-SY5Y cells also showed changes in mitochondrial morphology with increasing OGD duration (Figures 2B and 2D). It should be noted that even after 18 hours OGD exposure, only 26% of cells displayed abnormal ER, and only 34% had abnormal mitochondria.
Figure 2.
The endoplasmic reticulum (ER) and mitochondria display progressive morphologic changes after oxygen glucose deprivation (OGD). (A) OGD induced increases in the number of cells containing swollen and abnormal ER. Significant changes in the distribution of ‘normal', ‘swollen', and ‘abnormal' ER within cells were seen after 3, 9, and 18 hours OGD compared with the time-matched controls (*P<0.05, **P<0.01, χ2). (B) The occurrence of mitochondrial morphologic abnormality increases after longer OGD durations. (C) Representative electron micrographs of the three grades of ER morphology. Scale bar=0.1 μm. (D) Representative electron micrographs of the three grades of mitochondrial morphology. Scale bar=0.1 μm.
Global Proteomic Response after Oxygen Glucose Deprivation (6 and 18 hours)
A total of 958 proteins were identified across all samples with two or more peptides by LC-MS (Supplementary Table 1). Minimal protein changes were observed after 6 hours OGD, with only 14 proteins significantly altered in expression level (P<0.01). After 18 hours OGD, 193 proteins were significantly altered in expression level (P<0.01) with 130 proteins increased and 63 proteins decreased. For 765 proteins, no significant difference was observed between control and OGD samples after 18 hours OGD.
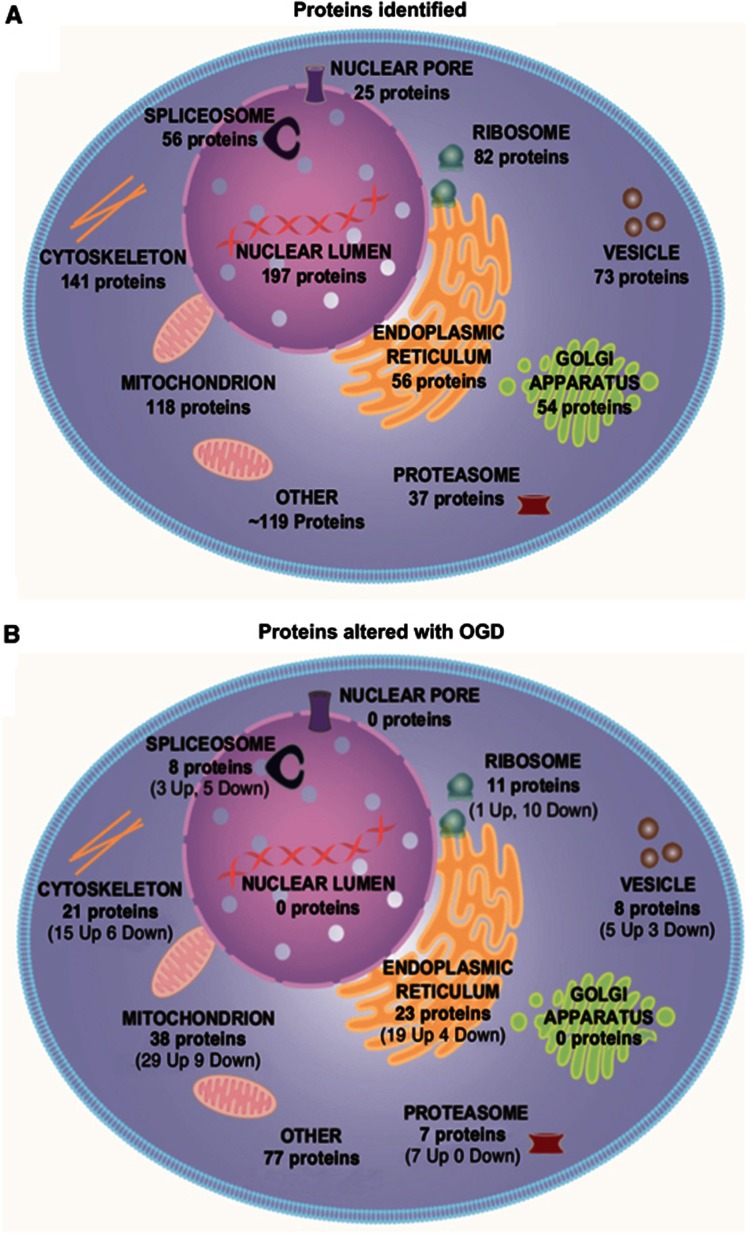
Subcellular distribution of all the 958 proteins identified by LC-MS was examined using DAVID software. Proteins were detected from across many subcellular compartments, including the nuclear lumen, cytoskeleton, mitochondria, ER, golgi apparatus, ribosomes, and vesicles (Figure 3A).
Figure 3.
Subcellular distribution of detected proteins contrasted with subcellular distribution of proteins significantly altered after oxygen glucose deprivation (OGD). Proteins were ascribed to subcellular compartment by DAVID software based on gene ontology. Proteins were significantly altered after OGD (P<0.01) relative to their original levels in time-matched control samples. (A) Liquid chromatography mass spectrometry (LC-MS) detected 958 proteins from across many subcellular compartments, showing the utility of LC-MS in providing a global overview of cellular proteomics. (B) Organelles are differentially affected by 18 hours OGD. The 193 significant proteins changed (P<0.01) were not distributed evenly across subcellular compartments: the endoplasmic reticulum and the mitochondria contained the largest proportion of significantly altered proteins, whereas no significant protein changes were detected in the golgi apparatus or nuclear pore.
The ascribed subcellular locations of the significantly altered proteins with OGD (18 hours) indicate that organelles are not uniformly affected by the metabolic challenge. None of the 197 detected nuclear lumen proteins were significantly altered after OGD, while 41% of the 56 detected ER proteins and 32% of the 118 detected mitochondrial proteins were significantly altered (Figure 3B). The significantly increased and decreased proteins after OGD (18 hours) are listed by subcellular location in Table 1 and Table 2, respectively.
Table 1. Proteins significantly increased with OGD (18 hours).
| Uniprot accn no. | Gene name | Protein name | P value | Fold change | Protein function |
|---|---|---|---|---|---|
| Mitochondria | |||||
| P30048 | PRDX3 | Peroxiredoxin III | 0.0006 | 1.57 | Cell redox homeostasis |
| P61604 | HSPE1 | 10 kDa heat-shock protein | 6.1E-06 | 1.51 | Stress response |
| P18859 | ATP5J | ATP synthase-coupling factor 6 | 0.0016 | 1.44 | ETC complex V |
| O75489 | NDUFS3 | NADH-ubiquinone oxidoreductase 30 kDa subunit | 0.0030 | 1.38 | ETC complex I |
| P24752 | ACAT1 | Acetyl-CoA acetyltransferase | 9.1E-05 | 1.38 | Ketone body metabolism |
| P09622 | DLD | Dihydrolipoyl dehydrogenase | 0.0051 | 1.36 | Gycine cleavage |
| Q9UJZ1 | STOML2 | Stomatin-like protein 2 | 0.0052 | 1.36 | Receptor binding |
| P09972 | ALDOC | Fructose-bisphosphare aldolase C | 0.0061 | 1.34 | Glycolysis |
| P38606 | ATP6V1A | V-type proton ATPase catalytic subunit A | 0.0067 | 1.33 | ATP hydrolysis transport |
| P13804 | ETFA | Electron transfer flavoprotein subunit alpha | 0.0005 | 1.33 | Respiratory ETC |
| P30084 | ECHS1 | Enoyl-CoA hydratase | 0.0043 | 1.33 | Fatty acid metabolism |
| P40926 | MDH2 | Malate dehydrogenase | 0.0009 | 1.32 | Citric acid cycle |
| P10809 | HSPD1 | 60 kDa heat-shock protein | 0.0012 | 1.31 | Protein folding |
| P62258 | YWHAE | 14-3-3 protein epsilon | 0.0056 | 1.30 | Apoptosis |
| P04181 | OAT | Orthinine aminotransferase | 0.0012 | 1.30 | Amino acid biosynthesis |
| Q13011 | ECH1 | Delta (3,5)-delta(2,4)-dienoyl-CoA isomerase | 0.0048 | 1.28 | Lipid metabolism |
| Q99714 | HSD17B10 | 3-hydroxyacyl-CoA dehydrogenase type 2 | 0.0037 | 1.27 | Lipid metabolism |
| P07954 | FH | Fumarate hydratase | 0.0038 | 1.27 | Citric acid cycle |
| P30044 | PRDX5 | Peroxiredoxin 5 | 0.0014 | 1.26 | Redox homeostasis |
| P42765 | ACAA2 | 3-ketoacyl-CoA thiolase | 0.0067 | 1.26 | Lipid metabolism |
| P25325 | MPST | 3-mercaptopyruvate sulfurtransferase | 0.0012 | 1.25 | Response to Toxins |
| P30086 | PEBP1 | Phosphatidylethanolamine binding protein 1 | 0.0067 | 1.24 | Protease inhibition |
| P25705 | ATP5A1 | ATP synthase subunit alpha | 0.0019 | 1.21 | ETC complex V |
| Q06830 | PRDX1 | Peroxiredoxin 1 | 0.0009 | 1.20 | Redox homeostasis |
| P43686 | PSMC4 | 26S protease regulatory subunit 6B | 0.0007 | 1.20 | Protein degradation |
| Q16718 | NDUFA5 | NADH-ubiquinone oxidoreductase 13 kDa subunit | 0.0022 | 1.20 | ETC complex I |
| O00154 | ACOT7 | Cytosolic acyl-coenzyme A thioester hydrolase | 0.0009 | 1.19 | Acyl-CoA hydrolysis |
| Q00610 | CLTC | Clathrin heavy chain 1 | 4.9E-05 | 1.16 | Endocytosis |
| P12277 | CKB | Creatine Kinase B type | 0.0027 | 1.16 | Creatine metabolism |
| Endoplasmic reticulum | |||||
| O60568 | PLOD3 | Procollagen-lysine 2-oxoglutarate 5-dioxygenase 3 | 7.7E-05 | 1.87 | Lysyl hydroxylation |
| P27797 | CALR | Calreticulin | 1.0E-07 | 1.56 | Protein folding |
| P11021 | HSPA5 | Heat-shock protein 5 | 3.9E-05 | 1.53 | Protein complex assembly |
| P14625 | HSP90B1 | Heat-shock Protein 90 kDa beta member 1 | 5.5E-05 | 1.49 | Protein transport |
| P30101 | PDIA3 | Protein disulfide isomerase A3 | 2.7E-05 | 1.45 | Protein folding |
| Q16799 | RTN1 | Reticulon 1 | 0.0055 | 1.41 | Neuron differentiation |
| Q8NBS9 | TXNDC5 | Thioredoxin-domain containing protein 5 | 0.0017 | 1.40 | Cell redox homeostasis |
| P13667 | PDIA4 | Protein disulfide isomerase A4 | 1.4E-05 | 1.38 | Cell redox homeostasis |
| Q9Y2B0 | CNPY2 | Protein canopy homolog 2 | 0.0051 | 1.38 | Protein binding |
| Q15084 | PDIA6 | Protein disulfide isomerase A6 | 6.1E-06 | 1.37 | Chaperone |
| P54920 | NAPA | Alpha soluble NSF attachment protein | 0.0003 | 1.35 | ER-golgi vesicle transport |
| P14314 | PRKCSH | Glucosidase 2 subunit beta | 0.0016 | 1.33 | Glycoprotein formation |
| Q9NQC3 | RTN4 | Reticulon 4 | 0.0033 | 1.32 | Neurogenesis/apoptosis |
| Q14697 | GANAB | Neutral alpha-glucosidase AB/GNAB protein | 0.0018 | 1.27 | Glycoprotein cleavage |
| P07237 | P4HB | Protein disulfide isomerase | 0.0073 | 1.25 | Redox homeostasis |
| Q9BS26 | ERP44 | Endoplasmic reticulum resident protein 44 | 0.0007 | 1.23 | ER stress response |
| Q9NYU2 | UGGT1 | UDP glucose:glycoprotein glucosyltransferase 1 | 0.0074 | 1.23 | ER protein folding |
| P30040 | ERP29 | Endoplasmic reticulum resident protein 29 | 0.0003 | 1.23 | ER protein folding |
| P55072 | VCP | Valosin-containing protein | 0.0069 | 1.15 | ER stress response |
| Ribosomes | |||||
| P69905 | HBA1 | Hemoglobin subunit alpha | 0.0003 | 1.97 | O2 transport |
| Vesicle | |||||
| O15240 | VGF | Neurosecretory protein VGF | 0.0035 | 1.39 | Cell–cell interactions |
| P08133 | ANXA6 | Annexin 6 | 0.0075 | 1.18 | Ca2+ binding |
| P21281 | ATP6V1B2 | V-type proton ATPase subunit B | 0.0098 | 1.16 | H+ ion transport |
| P07900 | HSP90AA1 | Heat Shock Protein HSP90-alpha | 0.0030 | 1.11 | Stress response |
| Q9BTT0 | ANP32E | Acidic leucine-rich nuclear phosphoprotein 32E | 0.0034 | 1.08 | Phosphatase inhibitor |
| Cytoskeleton | |||||
| P52907 | CAPZA1 | F-actin capping protein subunit alpha 1 | 0.0039 | 1.35 | Protein complex assembly |
| P16949 | STMN1 | Stathmin | 0.0029 | 1.32 | Microtubule disassembly |
| Q9Y230 | RUVBL2 | RuvB like 2 | 0.0037 | 1.31 | Transcription |
| P47756 | CAPZB | F-actin-capping protein subunit beta | 3.2E-05 | 1.25 | Protein complex assembly |
| Q03252 | LMNB2 | Lamin B2 | 0.0076 | 1.24 | Cytoskeleton regulation |
| Q16352 | INA | Alpha-internexin | 4.9E-05 | 1.24 | Neurogenesis |
| P07437 | TUBB | Tubulin beta chain | 0.0016 | 1.23 | Microtubule constituent |
| P36405 | ARL3 | ADP-ribosylation factor-like protein 3 | 0.0071 | 1.23 | Cell division |
| P61163 | ACTR1A | Alpha centractin | 0.0029 | 1.23 | Vesicle mediated transport |
| Q9UJU6 | DBNL | Debrin-like protein | 0.0095 | 1.18 | Endocytosis |
| Q16658 | FSCN1 | Fascin | 0.0018 | 1.18 | Cytoskeleton organization |
| P62158 | CALM | Calmodulin | 0.0059 | 1.17 | Calcium modulation |
| Q9BZK7 | TBL1XR1 | F-box like/WD repeat containing protein TBL1XR1 | 0.0046 | 1.17 | Protein degradation |
| P49773 | HINT1 | Histidine triad nucleotide-binding protein 1 | 0.0002 | 1.15 | Signal transduction |
| P43487 | RANBP1 | Ran-specific GTPase-activating protein | 0.0029 | 1.15 | Signal transduction |
| Spliceosome | |||||
| Q07955 | SRSF1 | Serine/Arginine-rich splicing factor 1 | 1.1E-05 | 1.42 | mRNA splicing |
| Q01081 | U2AF1 | Splicing factor U2AF 35 kDa subunit | 0.0062 | 1.23 | mRNA splicing |
| Q15029 | EFTUD2 | 116 kDa U5 small nuclear ribonucleoprotein | 0.0069 | 1.19 | mRNA splicing |
| Proteosome | |||||
| Q15008 | PSMD6 | 26s proteasome non ATPase regulatory subunit 6 | 0.0053 | 1.61 | Protein degradation |
| P17980 | PSMC3 | 26S protease regulatory subunit 6A | 0.0071 | 1.27 | Protein degradation |
| Q13200 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2 | 0.0005 | 1.22 | Protein degradation |
| P54727 | RAD23B | UV excision protein RAD23 homolog B | 0.0009 | 1.22 | Protein degradation |
| Q99460 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 0.0002 | 1.18 | Protein degradation |
| O00231 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11 | 0.0008 | 1.18 | Protein degradation |
| P28066 | PSMA5 | Proteasome subunit alpha type 5 | 0.0036 | 1.16 | Protein degradation |
ER, endoplasmic reticulum; ETC, electron transport chain; OGD, oxygen glucose deprivation.
Proteins are grouped according to subcellular compartment based on gene ontology and ranked according to magnitude of fold change. The significance threshold was set a priori at P<0.01.
Table 2. Proteins Significantly Decreased with OGD (18 hours).
| Uniprot accn no. | Gene name | Protein name | P-value | Fold change | Protein function |
|---|---|---|---|---|---|
| Mitochondria | |||||
| P21796 | VDAC1 | Voltage-dependent anion selective channel protein 1 | 0.0079 | 0.26 | Ion transport |
| P22695 | UQCRC2 | Cytochrome b-c1 complex subunit 2 | 0.0017 | 0.31 | ETC complex III |
| P31930 | UQCRC1 | Cytochrome b-c1 complex subunit 1 | 0.0097 | 0.46 | ETC complex III |
| Q9NS69 | TOM22 | Translocase of outer membrane 22 kDa subunit | 0.0027 | 0.46 | Protein transport |
| Q9Y3E5 | BIT1 | Peptidyl-tRNA hydrolase 2 | 0.0038 | 0.51 | Apoptosis |
| P02786 | TFRC | Transferrin receptor protein 1 | 0.0002 | 0.58 | Endocytosis |
| O75534 | CSDE1 | Cold shock domain containing protein E1 | 1.0E-06 | 0.62 | Transcription |
| Q9Y6H1 | CHCHD2 | Coiled-coil helix–coiled-coil helix domain protein 2 | 0.0099 | 0.70 | unknown |
| P11310 | ACADM | Medium chain specific acyl-CoA dehydrogenase | 0.0084 | 0.79 | Lipid metabolism |
| Endoplasmic reticulum | |||||
| Q81V08 | PLD3 | Phospholipase D3 | 0.0016 | 0.61 | Lipid degradation |
| Q96AG4 | LRRC59 | Leucine rich repeat-containing protein 59 | 0.0048 | 0.64 | ER membrane interactions |
| P13637 | ATP1A3 | Na+/K+ ATPase alpha (III) subunit | 0.0008 | 0.67 | ATP hydrolysis catalyst |
| P04843 | RPN1 | Ribophorin 1 | 0.0094 | 0.68 | Glycosyltransferase activity |
| Ribosomes | |||||
| P63173 | RPL38 | 60s ribosomal protein L38 | 0.0001 | 0.65 | Protein biosynthesis |
| P62249 | RPS16 | 40S ribosomal protein S16 | 7.9E-05 | 0.70 | Protein biosynthesis |
| P35544 | FAU | Ubiquitin-like protein FUB 1 | 0.0012 | 0.70 | Ubiquitination modulation |
| P62263 | RPS14 | 40S ribosomal protein S14 | 0.0004 | 0.74 | Protein biosynthesis |
| P62829 | RPL23 | 60S ribosomal protein L23 | 5.3E-05 | 0.74 | Protein biosynthesis |
| P62266 | RPS23 | 40S ribosomal protein S23 | 0.0004 | 0.76 | Protein biosynthesis |
| P46781 | RPS9 | 40S ribosomal protein S9 | 0.0002 | 0.78 | Protein biosynthesis |
| P08708 | RPS17 | 40S ribosomal protein S17 | 0.0004 | 0.80 | Protein biosynthesis |
| P42766 | RPL35 | 60S ribosomal protein L35 | 0.0018 | 0.82 | Protein biosynthesis |
| P26373 | RPL13 | 60S ribosomal protein L13 | 0.0004 | 0.84 | Protein biosynthesis |
| Vesicle | |||||
| P07355 | ANXA2 | Annexin A2 | 0.0038 | 0.49 | Ca2+ binding |
| P62491 | RAB11A | Ras-related protein Rab 11A | 0.0026 | 0.76 | Endocytic cycling |
| P07858 | CTSB | Cathepsin B | 0.0053 | 0.85 | Protein degradation |
| Cytoskeleton | |||||
| P06493 | CDK1 | Cyclin-dependent kinase 1 | 0.0022 | 0.57 | Cell-cycle control |
| P28289 | TMOD1 | Tropomodulin 1 | 0.0005 | 0.64 | Cytoskeleton organization |
| Q01518 | CAP1 | Adenylyl cyclase-associated protein 1 | 4.7E-06 | 0.70 | Signal transduction |
| Q9UQE7 | SMC3 | Structural maintenance of chromosomes protein 3 | 0.0084 | 0.71 | Cell cycle |
| P18206 | VCL | Viniculin | 0.0067 | 0.78 | Cell adhesion |
| Q01082 | SPTBN1 | Spectrin beta chain brain 1 | 0.0092 | 0.81 | Cytoskeleton movement |
| Spliceosome | |||||
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | 2.9E-06 | 0.58 | mRNA processing |
| Q9UKM9 | RALY | RNA-binding protein Raly | 0.0014 | 0.66 | mRNA splicing |
| Q86V81 | THOC4 | THO complex subunit 4 | 0.0007 | 0.75 | mRNA splicing |
| O43143 | DHX15 | Pre-mRNA splicing factor ATP-dep. RNA helicase | 5.5E-06 | 0.78 | mRNA splicing |
| P07910 | HNRNPC | Heterogeneous nuclear ribonucleoproteins Ca/C2 | 0.0005 | 0.81 | mRNA splicing |
ER, endoplasmic reticulum; ETC, electron transport chain; OGD, oxygen glucose deprivation.
Proteins are grouped according to subcellular compartment based on gene ontology and ranked according to magnitude of fold change. The significance threshold was set a priori at P<0.01.
Functional protein–protein interactions for the 193 proteins altered with 18 hours OGD were defined using IPA software. IPA-generated 10 protein interactomes (Supplementary Table 2) and three high-scoring interactomes (IPA scores >40) are discussed in context below. These interactomes highlight a shift in mitochondrial energy production, enhanced ER stress, and ribosomal dysfunction.
Mitochondrial Protein Alterations after Oxygen Glucose Deprivation (18 hours)
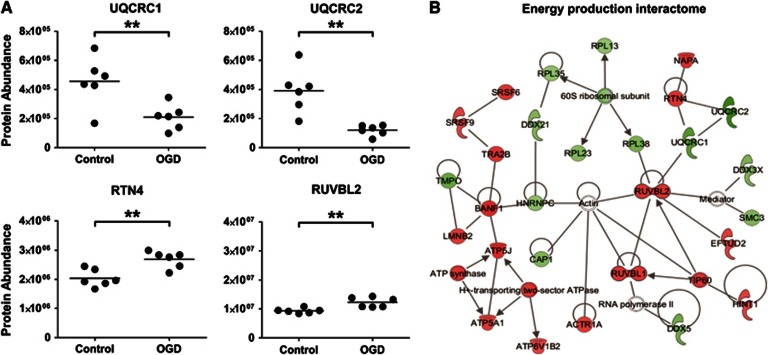
In all, 38 mitochondrial proteins (out of 118 detected; assigned using the GO term mitochondrion GO:0005739) were significantly altered after OGD (18 hours): 29 were increased and 9 were decreased. VDAC1 and TOM22, proteins involved in mediating the movement of metabolites across the mitochondrial membrane, were significantly reduced after OGD. UQCRC1 and UQCRC2, integral components of complex III of the electron transport chain (ETC), were also significantly reduced (Figure 4A). Coordinated changes in other regions of the ETC were observed after 18 hours OGD treatment: ETC complex V proteins ATP5J and ATP5A1, and ETC complex I proteins NDUFS3 and NDUFA5 were all significantly increased (P<0.01).
Figure 4.
Mitochondrial energy production proteins and associated interacting partners are affected by oxygen glucose deprivation (OGD). (A) Key mitochondrial energy production proteins (UQCRC1/UQCRC2) and their putative interacting partners (RTN4/RUVBL2) are significantly altered by OGD (18 hours). Each data point represents an independent sample with the mean abundance line shown (**P<0.01, T-test). (B) Energy Production Interactome. One of the highest scoring Ingenuity Pathway Analysis (IPA) networks gained from the 18-hour OGD proteomic data set is presented. The network contains numerous proteins involved in mitochondrial energy production, the colored nodes indicate proteins detected in our study as significantly altered by OGD. Red nodes are upregulated proteins and green nodes are downregulated. Unshaded nodes were manually inserted by IPA to augment functional coherence. Only direct protein–protein interactions were included in the analysis.
Numerous mitochondrial proteins that have a role in lipid metabolism were also found to be altered after 18 hours OGD. ACADM, the enzyme involved in the first step of fatty acid beta oxidation was significantly decreased (P<0.01); and ECH1, HSD17B10 and ACAA2, enzymes downstream in the beta-oxidation process were significantly increased. ACAT1, a mitochondrial protein involved in ketone metabolism was also significantly increased.
The highest scoring protein interactome generated by IPA features some of the alterations in energy production described above, namely increases in Complex V proteins (including ATP5J and ATP5A1) and decreases in Complex III proteins (UQCRC1 and UQCRC2) (Figure 4B). This Energy Production Interactome also highlights interactions between altered components of the ETC and other cellular machinery. Both UQCRC1 and UQCRC2 interact with RTN4, a protein predominantly associated with regulating ER function that was significantly upregulated after OGD (Figure 4A). UQCRC2 is also shown to interact with upregulated RUVBL2, a cytoskeletal protein involved in numerous cellular activities including transcription and cellular transformation (Figure 4A). The Energy Production Interactome also contains a cluster of ribosomal proteins that are distinct but intersect with the ribosomal interactome described below.
Endoplasmic Reticulum Protein Alterations after Oxygen Glucose Deprivation (18 hours)
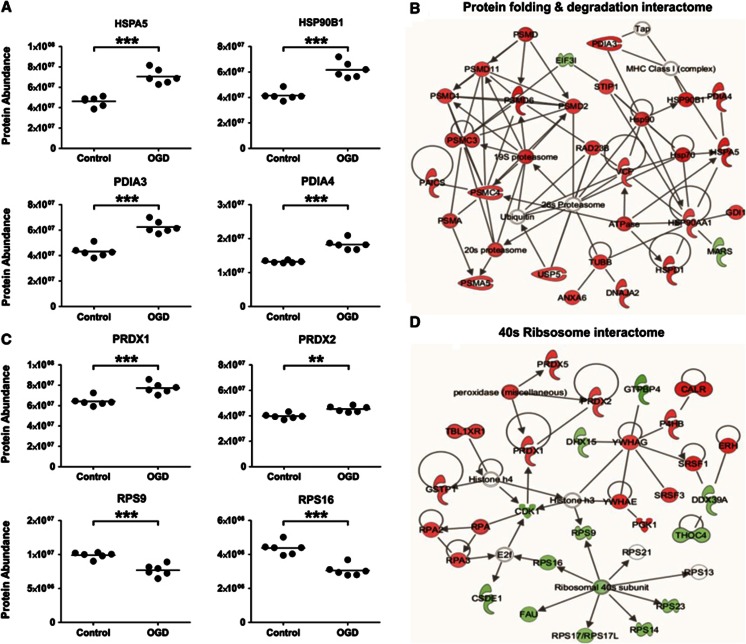
In all, 23 ER proteins (out of 56 detected; assigned using the GO term Endoplasmic reticulum GO:0005783) were significantly altered after 18 hours OGD: 19 were increased and 4 were decreased. A number of proteins centrally involved in the UPR were significantly increased, including, HSPA5, HSP90B1, PDIA3, and PDIA4 (P<0.01) (Figure 5A). Other proteins including CALR, UGGT1, ERP29, ERP44, and VCP that are integral to ER stress response and protein folding were significantly increased.
Figure 5.
Protein production, folding, and degradation pathways are affected by oxygen glucose deprivation (OGD). (A) Key proteins involved in protein folding and the endoplasmic reticulum stress response (HSPA5/HSP90B1/PDIA3/PDIA4) are significantly altered by OGD (18 hours). (B) Protein Folding and Degradation Interactome. A high scoring Ingenuity Pathway Analysis (IPA) network generated from the 18-hour OGD proteomic data set contained numerous proteins involved in protein folding and degradation is presented. The colored nodes indicate proteins detected in our study as significantly altered by OGD. Red nodes are upregulated proteins and green nodes are downregulated proteins. Unshaded nodes were manually inserted by IPA to augment functional coherence. Only direct protein–protein interactions were included in the analysis. (C) Representative ribosomal proteins (RPS9/RPS16) and the cellular stress response proteins (PRDX1/PRDX2) are significantly altered by OGD (18 hours). Each data point represents an independent sample (**P<0.01, ***P<0.0001, T-test). (D) 40S Ribosomal Interactome. A further high scoring IPA network was dominated by the downregulation of 40S ribosomal proteins and cell stress response proteins.
Multiple proteins that have a role in protein degradation were significantly altered after OGD (18 hours). Protein subunits of the 20S and 19S core structures of the 26S proteasome (PSMA5, PSMC3, PSMC4, PSMD1, PSMD2, PSMD6, and PSMD11) were increased. These proteasomal proteins, along with proteins involved in the UPR and heat-shock response after OGD dominate the high scoring, Protein Folding and Degradation Interactome generated by IPA (Figure 5B).
Ribosomal Protein Alterations after Oxygen Glucose Deprivation (18 hours)
In all, 11 ribosomal proteins (out of 82 detected; assigned using the GO term Ribosome GO:0005840) were significantly altered after 18 hours OGD: 1 was increased and 10 were decreased. The decreased ribosomal proteins were associated with both the 60S (RPL38, RPL23, RPL35, and RPL13) and 40S (RPS1, RPS14, RPS23, RPS9, and RPS17) ribosomal subunits (Figure 5C). These downregulated ribosomal proteins feature prominently in another of the high scoring protein interactomes generated by IPA (Figure 5D). An additionally important feature of this Ribosomal Interactome is the cluster of upregulated peroxidases (PRDX1, PRDX2, and PRDX5) (Figure 5C) involved in redox homeostasis and protecting neurons from oxidative insults (for a review, see Bell and Hardingham, 201118).
Discussion
This study provides the first comprehensive analysis of the proteomic and morphologic response of human SH-SY5Y cells to prolonged metabolic challenge. The robustness of SH-SY5Y cells to external stressors has allowed the evolving proteomic changes occurring in response to OGD induced metabolic stress to be defined, in the absence of cell death. The observed protein changes provide insight into the complex molecular alterations underpinning cellular adaptation to metabolic stress. The lack of acute ischemic cell death in our in vitro model of neuronal metabolic stress makes the proteomic results particularly pertinent to mild, sustained ischemia challenges and the compromised energy metabolism associated with slowly evolving neurodegenerative disorders such as Alzheimer's disease.3
Proteomics has showed alterations and interactions in mitochondrial, ER, and ribosomal proteins after OGD. The ETCproteins displayed bidirectional changes (Complex I and V proteins increased and Complex III proteins decreased) in response to OGD. Other mitochondrial proteins involved in lipid and ketone metabolism, including AD-related HSD17B10 (more commonly known as ABAD), were also increased after OGD (18 hours). The ER stress response proteins along with proteins involved in proteasomal/ubiquitination degradation also formed an important part of the upregulated proteomic response to the metabolic challenge of OGD.
A significant mitochondrial proteomic response was observed after OGD. Two of the most markedly decreased proteins after 18 hours OGD were subunits 1 and 2 (UQCRC1 and UQCRC2) of ETC complex III. Complex III is the third component of the ETC located within the inner mitochondrial membrane. Complex III mediates the transfer of electrons from ubiquinone to cytochrome C, and in doing so translocates a proton into the inner-membrane space and contributes to the generation of the electrochemical gradient used for the production of ATP.19 Complex III is also intrinsically involved in the generation of reactive oxygen species from the mitochondria during hypoxia, and these reactive oxygen species work to stabilize HIF1α and initiate protein transcription.20 A significant decrease in UQCRC1 and UQCRC2 might be an attempt by the cells to reduce the production of reactive oxygen species from the mitochondria under stress conditions. Interestingly, UQCRC1 has also been recorded as significantly decreased in the brains of AD patients.21 The similar decrease in UQCRC1 expression observed in AD patients and cells in vitro after OGD may provide a link between chronic oxidative challenge and the onset and progression of AD.
The Energy Production Interactome (Figure 4B) also highlighted putative functional protein–protein interactions between UQCRC1, UQCRC2, and RTN4 (a protein increased after 18 hours OGD). RTN4 is increased in hippocampal neurons of AD patients22 and is known to interact with BACE1, reducing AB40 and AB42 levels.23 The dysregulation of RTN4 after a metabolic challenge (18 hours OGD), its interaction with complex III proteins, and its potential role in APP processing, highlights RTN4 protein as a compelling target for future investigation.
Mitochondrial complex I and V proteins were significantly increased in expression after OGD in sharp contrast to the reduction in complex III proteins after OGD. Complex I is the first protein complex of the ETC and initiates the transfer of electrons required for oxidative phosphorylation. The process is initiated through binding and oxidation of NADH, with the concurrent transfer of two electrons from NADH to ubiquinone, and the translocation of four protons into the intermembrane space.24 An increase in Complex I proteins after OGD may reflect a compensatory mechanism to counteract the impact of decreased complex III proteins on energy production. Similarly, the increase in Complex V proteins may reflect a compensation for downregulated Complex III proteins or a general remodeling of the ETC, as has previously been shown in studies investigating ischemic preconditioning.25 Functional assays of the five complexes that constitute the ETC, in ischemic conditions will need to be carried out to confirm whether such remodeling occurs. Determining the regulatory mechanisms of these protein changes could provide valuable insight that would allow therapeutic intervention to alter the course of mitochondrial dysfunction after OGD and metabolic challenges in vivo.
Concurrent with the dysregulation of the oxidative phosphorylation pathway, OGD (18 hours) induced an increase in mitochondrial proteins involved in lipid and ketone metabolism. The increase in these proteins (ACAT1 in ketone metabolism, and ACADM, ECH1, HSD17B10, and ACAA2 in lipid metabolism) indicates a shift toward using lipid and ketone bodies as alternative sources of energy for SH-SY5Y cells faced with glucose restriction. The availability of such alternative energy sources is likely to be important in preventing cells hitting the point where energy provision is insufficient for cell survival, leading to a downward spiral of metabolic and cognitive decline. A recent study using a triple transgenic model of AD showed an induction of ketogenesis after a 2-deoxy-𝒟-glucose diet was introduced. This correlated with an increase in nonamyloidogenic APP processing pathways, reduced AD pathology and reduced oxidative stress,26 indicating a potential therapeutic role for enhanced ketone metabolism.
To use ketones as an alternative energy source in the brain, acetoacetate and β-hydroxybutyrate (the main ketone bodies used for energy) need to be synthesized from acetyl-coA. Acetyl-coA is released after beta oxidation of fatty acids, a four step process within the mitochondrial matrix requiring beta-oxidation enzymes at each stage (namely, ACAD, ECH1, HSD17B10, and ACAA2). Each of these beta-oxidation enzymes, as noted above, was significantly altered after 18 hours OGD. Of note, the third beta-oxidative enzyme HSD17B10 (ABAD) is now emerging as an important player in AD. ABAD has been shown to directly interact with Aβ in the mitochondria of AD patients and AD transgenic mouse models. Inhibition of the ABAD-Aβ interaction suppresses induced apoptosis by increasing levels of Aβ, and conversely overexpression of ABAD in transgenic mice with high levels of Aβ has been shown to cause heightened neuronal oxidative stress and memory impairments.27 The direct involvement of ABAD in AD pathophysiology makes ABAD an interesting candidate to investigate further in terms of its upregulation upon metabolic challenge.
Pore forming proteins VDAC1 and TOM22 were significantly downregulated after OGD. VDAC1 is thought to be involved in the formation of the mPTP, which in turn allows calcium and solutes <1.5 kDa to pass across the mitochondrial membranes, resulting in osmotic swelling, loss of the mitochondrial membrane potential, and damage to the electron transport respiratory chain.28 Mitochondrial swelling after OGD was clearly showed in the human cells under the electron microscope (Figure 2D). The role that these mitochondrial pore forming proteins have in mediating mitochondrial swelling after OGD needs to be defined to facilitate the discovery of novel pathways that might be manipulated to restore mitochondrial normality.
A major proteomic response in the ER was identified after OGD. The ER stress response (also known as the unfolded protein response, UPR) is a cellular reaction activated to clear misfolded proteins that accumulate upon ER stress, thus returning the cell to a state of proteostasis.29 Two central molecular chaperones, HSPA5 and HSP90B1, involved in the early and late stages of the UPR respectively were upregulated after OGD in SH-SY5Y cells (as well as numerous other UPR proteins and ER stress proteins) (Figure 5A). These data are consistent with recent results from OGD in mixed primary cortical cultures where increases in both HSPA5 and HSP90B1 were detected.30 The induction of the UPR in response to OGD was also implied in the electron microscopic data. The ER showed a progressive morphologic swelling that significantly increased after longer durations of OGD exposure compared with time-matched controls (P<0.01, Figure 2C). An expanded ER forms an important part of the UPR as it enables larger numbers of misfolded proteins to be incorporated within the ER membrane, thus increasing cellular capacity to deal with abundant protein damage.31 A larger ER volume also reduces the concentration of protein intermediates, therefore reducing the risk of protein aggregate formation.32 The morphologic observation of enlarged ER along with the significant increase in UPR proteins HSPA5 and HSP90B1 detected in our proteomic data provides telling insights into how the human neuronal cells respond to the metabolic challenge of OGD.
In accord with activation of the UPR, an upregulation of numerous proteins involved in proteasomal degradation was highlighted by the Protein Folding and Degradation Interactome. Subunits of the 20S and 19S (core and regulatory) particles that constitute the 26S proteasome were increased, suggesting 26S proteasomal degradation to be a fundamental mechanism for degrading the misfolded proteins that accumulate in response to metabolic challenge. There has been a long-standing hypothesis that protein aggregates in diseased brain impair the protein degradation function of the 26S proteasome.33, 34, 35 Our proteomic data show proteasomal proteins to be upregulated after a metabolic challenge. It is therefore possible to hypothesize that any proteasomal response to metabolic challenge might be adversely affected in the presence of protein aggregates associated with neurodegenerative diseases such as AD. In this situation, the cellular capacity to cope with metabolic stress would be impaired, potentially leading to a deterioration in cellular function.
A coordinated response after OGD (18 hours) was also observed in the ribosomal proteome. Numerous ribosomal proteins that constitute the core ribosomal machinery were downregulated, and in line with this, several eukaryotic initiation factors central to protein synthesis were also decreased. It is well established that cells undergoing severe metabolic stress experience a downregulation of protein translation, which can be reversible in more resistant brain areas, and irreversible in vulnerable regions.36 Further work involving a recovery phase would need to be undertaken to understand whether the decrease in protein synthesis machinery is a survival mechanism to conserve energy, or whether it ultimately pushes the cells down a cell death route. The proteomic data does suggest that activation of the UPR correlates with a decrease in global protein synthesis, perhaps to deal with the accumulation of misfolded proteins.37
Substrate deprivation both in vivo and in vitro has long been recognized to evoke a marked genomic response. Whether this genomic response translates into distinct alterations in protein expression has been a matter of debate. The present study clearly shows the considerable adaptive capacity of the SH-SY5Y proteome to a prolonged metabolic challenge in vitro, but caution must be exercised in the extrapolation of the present data to cerebral ischemia in vivo. No single cell culture system can model the complexity of the response of cerebral tissue to substrate deprivation. Astrocyte cultures and neuronal cells of neuroblastoma lineage (such as SH-SY5Y cells) can tolerate prolonged OGD, whereas primary neuronal and oligodendrocyte cultures are irreversibly damaged within a few hours.38, 39 The temporal susceptibility of these different cultures to metabolic challenge, their ability to generate energy from various sources (SH-SY5Y cells have considerable capacity to perform anaerobic glycolysis40) and their intrinsic energy demands will all impact on the evoked proteomic response to OGD.
Additional proteomic and biochemical analyses (not just LC-MS as in the present study) including protein–protein interaction assays, membrane potential recordings, and in vitro ATP levels across a range of different cell types would enhance our understanding of the true nature of the metabolic challenge posed by OGD. These studies would also provide more detail as to the subtleties of the proteomic changes, not only in terms of abundance, but also in cellular and subcellular localization, and the interacting partners of altered proteins.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
AGH is supported by the Medical Research Council. The research is supported by Age UK as part of the Disconnected Mind programme and performed under the aegis of the Centre for Cognitive Aging and Cognitive Epidemiology (part of the Lifelong Health and Wellbeing initiative of UK Research Councils). TLB and LEK are funded by SynthSys which is a Centre for Integrative Systems Biology (CISB) funded by BBSRC and EPSRC; reference BB/D019621/1.
Supplementary Material
References
- van der Worp HB, van Gijn J. Acute Ischemic Stroke. N Engl J Med. 2007;357:572–579. doi: 10.1056/NEJMcp072057. [DOI] [PubMed] [Google Scholar]
- Beal MF, Hyman BT, Koroshetz W. Do defecs in mitochondrial energy metabolism underlie the pathology of neurodegenerative diseases. Trends Neurosci. 1993;16:125–131. doi: 10.1016/0166-2236(93)90117-5. [DOI] [PubMed] [Google Scholar]
- de la Torre JC. The vascular hypothesis of Alzheimer's disease: bench to bedside and beyond. Neurodegener Dis. 2010;7:116–121. doi: 10.1159/000285520. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Sheng R, Qin ZH. The neuroprotective mechanism of brain ischemic preconditioning. Acta Pharmacol Sin. 2009;30:1071–1080. doi: 10.1038/aps.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soane L, Kahraman S, Kristian T, Fiskum G. Mechanisms of impaired mitochondrial energy metabolism in acute and chronic neurodegenerative disorders. J Neurosci Res. 2007;85:3407–3415. doi: 10.1002/jnr.21498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Kogure K, Kato H. Altered gene expression in cerebral ischemia. Stroke. 1993;24:2121–2127. doi: 10.1161/01.str.24.12.2121. [DOI] [PubMed] [Google Scholar]
- Scandalios J. Oxidative stress responses - what have genome-scale studies taught us. Genome Biol. 2002;3:1–6. doi: 10.1186/gb-2002-3-7-reviews1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer MM, McQueen J, Searcy L, Scullion G, Zonta B, Desmazieres A, et al. Rapid disruption of axon–glial integrity in response to mild cerebral hypoperfusion. J Neurosci. 2011;31:18185–18194. doi: 10.1523/JNEUROSCI.4936-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NJ, Green KN, Peers C, Vaughan PF. Altered processing of amyloid precursor protein in the human neuroblastoma SH-SY5Y by chronic hypoxia. J Neurochem. 2002;83:1262–1271. doi: 10.1046/j.1471-4159.2002.01236.x. [DOI] [PubMed] [Google Scholar]
- Almeida A, Delgado-Esteban M, Bolanos JP, Medina JM. Oxygen and glucose deprivation induces mitochondrial dysfunction and oxidative stress in neurones but not in astrocytes in primary culture. J Neurochem. 2002;81:207–217. doi: 10.1046/j.1471-4159.2002.00827.x. [DOI] [PubMed] [Google Scholar]
- Bayes A, Grant SG. Neuroproteomics: understanding the molecular organization and complexity of the brain. Nat Rev Neurosci. 2009;10:635–646. doi: 10.1038/nrn2701. [DOI] [PubMed] [Google Scholar]
- James R, Searcy JL, Le Bihan T, Martin SF, Gliddon CM, Povey J, et al. Proteomic analysis of mitochondria in APOE transgenic mice and in response to an ischemic challenge. J Cereb Blood Flow Metab. 2012;32:164–176. doi: 10.1038/jcbfm.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- Deighton RF, Kerr LE, Short DM, Allerhand M, Whittle IR, McCulloch J. Network generation enhances interpretation of proteomic data from induced apoptosis. Proteomics. 2010;10:1307–1315. doi: 10.1002/pmic.200900112. [DOI] [PubMed] [Google Scholar]
- Griffiths IR, Duncan ID, McCulloch M. Shaking pups: a disorder of central myelination in the spaniel dog. II. Ultrastructural observations on the white matter of the cervical spinal cord. J Neurocytol. 1981;10:847–858. doi: 10.1007/BF01262657. [DOI] [PubMed] [Google Scholar]
- Bell KF, Hardingham GE. CNS peroxiredoxins and their regulation in health and disease. Antioxid Redox Signal. 2011;14:1467–1477. doi: 10.1089/ars.2010.3567. [DOI] [PubMed] [Google Scholar]
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Kim SH, Vlkolinsky R, Cairns N, Lubec G. Decreased levels of complex III core protein 1 and complex V beta chain in brains from patients with Alzheimer's disease and Down syndrome. Cell Mol Life Sci. 2000;57:1810–1816. doi: 10.1007/PL00000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil V, Nicolas O, Mingorance A, Urena JM, Tang BL, Hirata T, et al. Nogo-A expression in the human hippocampus in normal aging and in Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:433–444. doi: 10.1097/01.jnen.0000222894.59293.98. [DOI] [PubMed] [Google Scholar]
- Murayama KS, Kametani F, Saito S, Kume H, Akiyama H, Araki W. Reticulons RTN3 and RTN4-B/C interact with BACE1 and inhibit its ability to produce amyloid beta-protein. Eur J Neurosci. 2006;24:1237–1244. doi: 10.1111/j.1460-9568.2006.05005.x. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA. Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, et al. Proteomic analysis of pharmacological preconditioning. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- Yao J, Chen S, Mao Z, Cadenas E, Brinton RD. 2-Deoxy-D-glucose treatment induces ketogenesis, sustains mitochondrial function, and reduces pathology in female mouse model of Alzheimer's disease. PLoS ONE. 2011;6:e21788. doi: 10.1371/journal.pone.0021788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- Du H, Yan SS. Mitochondrial permeability transition pore in Alzheimer's disease: Cyclophilin D and amyloid beta. Biochim Biophys Acta (BBA) 2010;1802:198–204. doi: 10.1016/j.bbadis.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W, Nijholt DA, Hoozemans JJ. The unfolded protein response and proteostasis in Alzheimer disease: preferential activation of autophagy by endoplasmic reticulum stress. Autophagy. 2011;7:910–911. doi: 10.4161/auto.7.8.15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiola N, Penas C, Minano-Molina A, Barneda-Zahonero B, Fado R, Sanchez-Opazo G, et al. Induction of ER stress in response to oxygen-glucose deprivation of cortical cultures involves the activation of the PERK and IRE-1 pathways and of caspase-12. Cell Death Dis. 2011;2:e149. doi: 10.1038/cddis.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci. 2008;105:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Valera AG, Diaz-Hernandez M, Hernandez F, Ortega Z, Lucas JJ. The ubiquitin-proteasome system in Huntington's disease. Neuroscientist. 2005;11:583–594. doi: 10.1177/1073858405280639. [DOI] [PubMed] [Google Scholar]
- Paschen W, Mengesdorf T, Aufenberg C. Suppression of protein synthesis after transient cerebral ischemia. Int Congr Ser. 2003;1252:179–191. [Google Scholar]
- Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66 (2 Suppl 1:S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- Wetzel M, Li L, Harms KM, Roitbak T, Ventura PB, Rosenberg GA, et al. Tissue inhibitor of metalloproteinases-3 facilitates Fas-mediated neuronal cell death following mild ischemia. Cell Death Differ. 2008;15:143–151. doi: 10.1038/sj.cdd.4402246. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li Y, Zheng X, Gao Q, Liu Z, Qu R, et al. Bone marrow stromal cells protect oligodendrocytes from oxygen-glucose deprivation injury. J Neurosci Res. 2008;86:1501–1510. doi: 10.1002/jnr.21617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzio EA, Soliman YI, Soliman KF. Variable toxicological response to the loss of OXPHOS through 1-methyl-4-phenylpyridinium-induced mitochondrial damage and anoxia in diverse neural immortal cell lines. Cell Biol Toxicol. 2010;26:527–539. doi: 10.1007/s10565-010-9161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.