Abstract
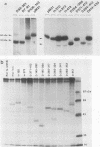
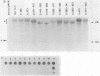
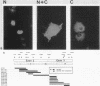
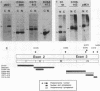
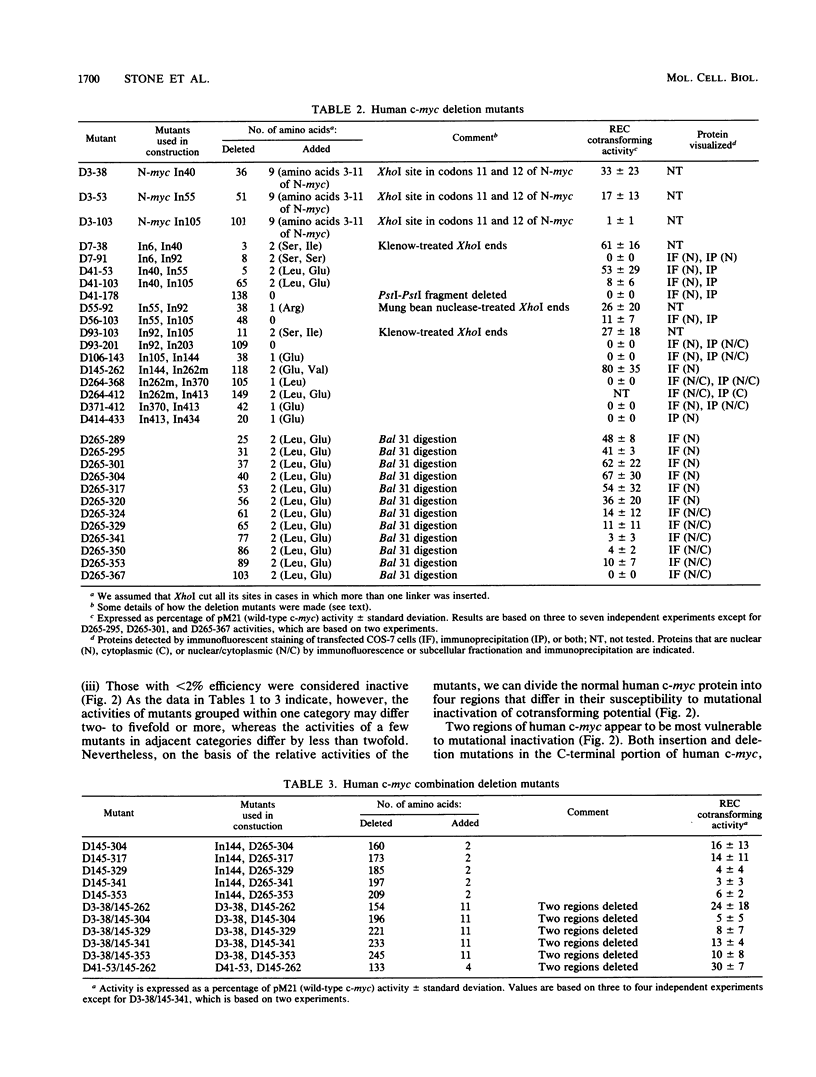
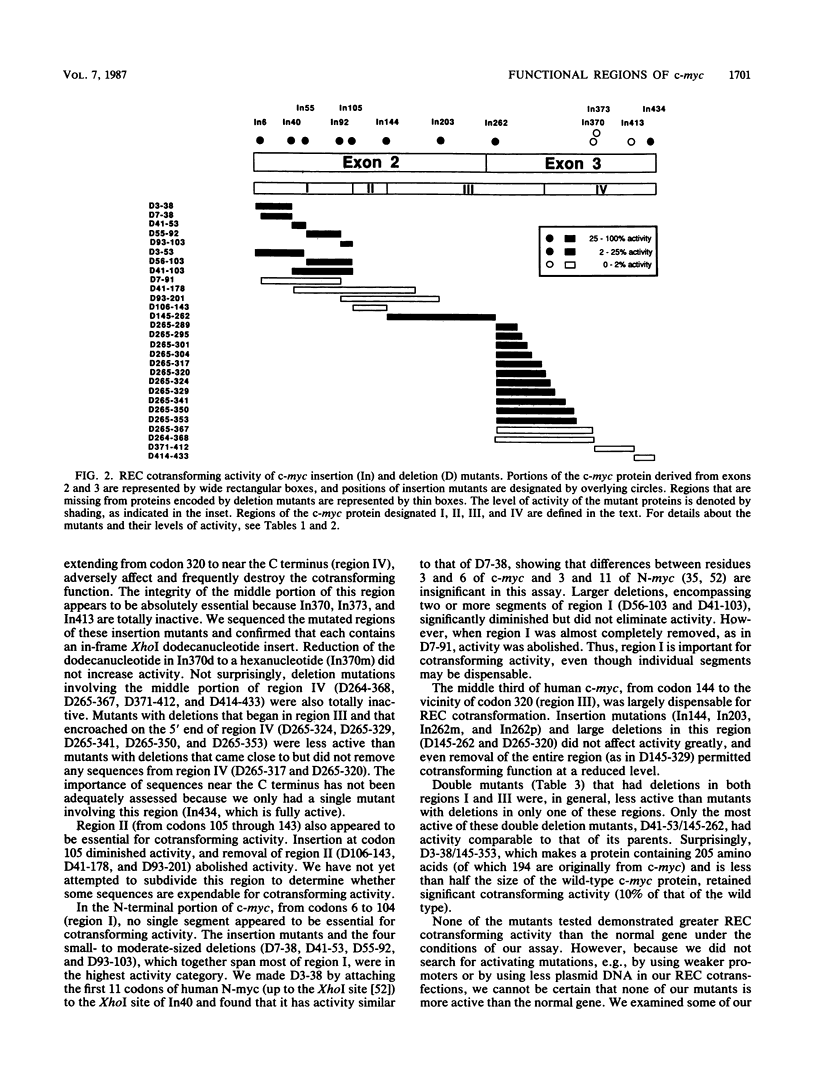
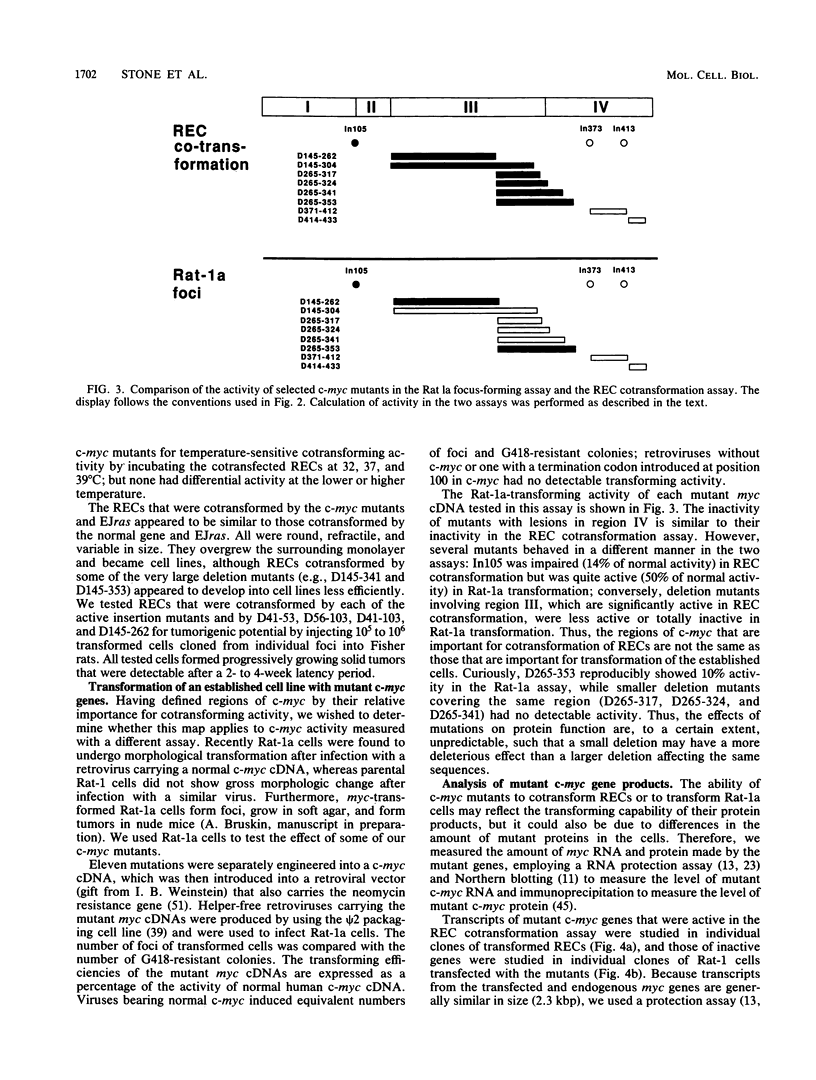
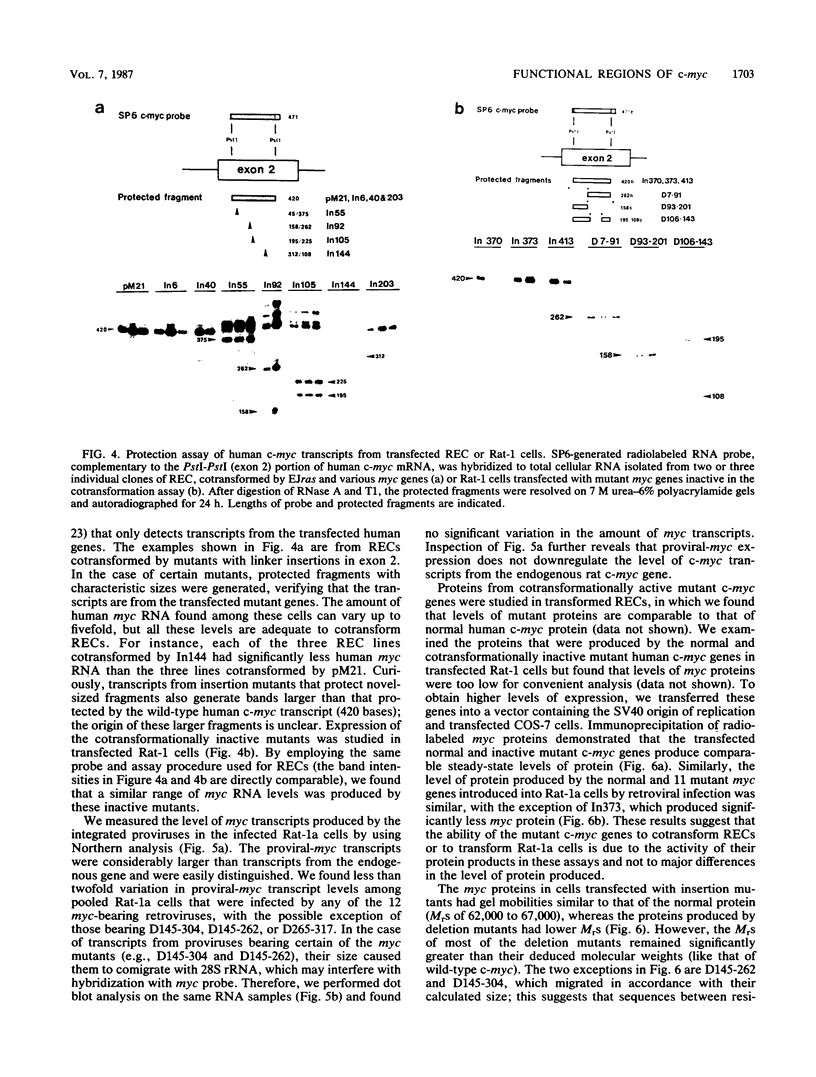
To study the relationship between the primary structure of the c-myc protein and some of its functional properties, we made in-frame insertion and deletion mutants of the normal human c-myc coding domain that was expressed from a retroviral promoter-enhancer. We assessed the effects of these mutations on the ability of c-myc protein to cotransform normal rat embryo cells with a mutant ras gene, induce foci in a Rat-1-derived cell line (Rat-1a), and localize in nuclei. Using the cotransformation assay, we found two regions of the protein (amino acids 105 to 143 and 321 to 439) where integrity was critical: one region (amino acids 1 to 104) that tolerated insertion and small deletion mutations, but not large deletions, and another region (amino acids 144) to 320) that was largely dispensable. Comparison with regions that were important for transformation of Rat-1a cells revealed that some are essential for both activities, but others are important for only one or the other, suggesting that the two assays require different properties of the c-myc protein. Deletion of each of three regions of the c-myc protein (amino acids 106 to 143, 320 to 368, and 370 to 412) resulted in partial cytoplasmic localization, as determined by immunofluorescence or immunoprecipitation following subcellular fractionation. Some abnormally located proteins retained transforming activity; most proteins lacking transforming activity appeared to be normally located.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Gerondakis S., Webb E., Corcoran L. M., Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Ramsay G., Bishop J. M., Pfeifer S. O., Colby W. W., Levinson A. D. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature. 1983 Nov 17;306(5940):274–277. doi: 10.1038/306274a0. [DOI] [PubMed] [Google Scholar]
- Armelin H. A., Armelin M. C., Kelly K., Stewart T., Leder P., Cochran B. H., Stiles C. D. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984 Aug 23;310(5979):655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Beimling P., Benter T., Sander T., Moelling K. Isolation and characterization of the human cellular myc gene product. Biochemistry. 1985 Nov 5;24(23):6349–6355. doi: 10.1021/bi00344a005. [DOI] [PubMed] [Google Scholar]
- Bister K., Ramsay G. M., Hayman M. J. Deletions within the transformation-specific RNA sequences of acute leukemia virus MC29 give rise to partially transformation-defective mutants. J Virol. 1982 Mar;41(3):754–766. doi: 10.1128/jvi.41.3.754-766.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M. J., Deininger P. L., Casey J. W. Nucleotide sequence of a transduced myc gene from a defective feline leukemia provirus. J Virol. 1985 Jul;55(1):177–183. doi: 10.1128/jvi.55.1.177-183.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Coughlin S. R., Lee W. M., Williams P. W., Giels G. M., Williams L. T. c-myc gene expression is stimulated by agents that activate protein kinase C and does not account for the mitogenic effect of PDGF. Cell. 1985 Nov;43(1):243–251. doi: 10.1016/0092-8674(85)90029-7. [DOI] [PubMed] [Google Scholar]
- Crews S., Barth R., Hood L., Prehn J., Calame K. Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science. 1982 Dec 24;218(4579):1319–1321. doi: 10.1126/science.7146913. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Bregni M., Erikson J., Patterson D., Gallo R. C., Croce C. M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Campisi J. c-myc regulation during retinoic acid-induced differentiation of F9 cells is posttranscriptional and associated with growth arrest. Mol Cell Biol. 1986 Feb;6(2):518–524. doi: 10.1128/mcb.6.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einat M., Resnitzky D., Kimchi A. Close link between reduction of c-myc expression by interferon and, G0/G1 arrest. Nature. 1985 Feb 14;313(6003):597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- Eisenman R. N., Tachibana C. Y., Abrams H. D., Hann S. R. V-myc- and c-myc-encoded proteins are associated with the nuclear matrix. Mol Cell Biol. 1985 Jan;5(1):114–126. doi: 10.1128/mcb.5.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enrietto P. J., Hayman M. J. Restriction enzyme analysis of partially transformation-defective mutants of acute leukemia virus MC29. J Virol. 1982 Nov;44(2):711–715. doi: 10.1128/jvi.44.2.711-715.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Hancock D. C. Studies on the interaction of the human c-myc protein with cell nuclei: p62c-myc as a member of a discrete subset of nuclear proteins. Cell. 1985 Nov;43(1):253–261. doi: 10.1016/0092-8674(85)90030-3. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Hamlyn P. H., Rabbitts T. H. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature. 1983 Jul 14;304(5922):135–139. doi: 10.1038/304135a0. [DOI] [PubMed] [Google Scholar]
- Hancock R., Boulikas T. Functional organization in the nucleus. Int Rev Cytol. 1982;79:165–214. doi: 10.1016/s0074-7696(08)61674-5. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R., Thompson C. B., Eisenman R. N. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. 1985 Mar 28-Apr 3Nature. 314(6009):366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Heaney M. L., Pierce J., Parsons J. T. Site-directed mutagenesis of the gag-myc gene of avian myelocytomatosis virus 29: biological activity and intracellular localization of structurally altered proteins. J Virol. 1986 Oct;60(1):167–176. doi: 10.1128/jvi.60.1.167-176.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek L., Hyland J. K., Watt R., Rosenberg M., Baserga R. Microinjected c-myc as a competence factor. Science. 1985 Jun 14;228(4705):1313–1315. doi: 10.1126/science.4001943. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Keath E. J., Caimi P. G., Cole M. D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984 Dec;39(2 Pt 1):339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Symonds G., Evan G. I., Bishop J. M. Subcellular localization of proteins encoded by oncogenes of avian myeloblastosis virus and avian leukemia virus E26 and by chicken c-myb gene. Cell. 1984 Jun;37(2):537–547. doi: 10.1016/0092-8674(84)90384-2. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Legouy E., DePinho R. A., Nisen P. D., Smith R. K., Gee C. E., Alt F. W. Human N-myc is closely related in organization and nucleotide sequence to c-myc. Nature. 1986 Jan 2;319(6048):73–77. doi: 10.1038/319073a0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lee W. M., Schwab M., Westaway D., Varmus H. E. Augmented expression of normal c-myc is sufficient for cotransformation of rat embryo cells with a mutant ras gene. Mol Cell Biol. 1985 Dec;5(12):3345–3356. doi: 10.1128/mcb.5.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little C. D., Nau M. M., Carney D. N., Gazdar A. F., Minna J. D. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983 Nov 10;306(5939):194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Murphy W., Sarid J., Taub R., Vasicek T., Battey J., Lenoir G., Leder P. A translocated human c-myc oncogene is altered in a conserved coding sequence. Proc Natl Acad Sci U S A. 1986 May;83(9):2939–2943. doi: 10.1073/pnas.83.9.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L. F., Tabin C. J., Shih C., Weinberg R. A. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982 Jun 10;297(5866):474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Persson H., Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984 Aug 17;225(4663):718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- Rabbitts T. H., Forster A., Hamlyn P., Baer R. Effect of somatic mutation within translocated c-myc genes in Burkitt's lymphoma. Nature. 1984 Jun 14;309(5969):592–597. doi: 10.1038/309592a0. [DOI] [PubMed] [Google Scholar]
- Ramsay G. M., Hayman M. J. Isolation and biochemical characterization of partially transformation-defective mutants of avian myelocytomatosis virus strain MC29: localization of the mutation to the myc domain of the 110,000-dalton gag-myc polyprotein. J Virol. 1982 Mar;41(3):745–753. doi: 10.1128/jvi.41.3.745-753.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G., Evan G. I., Bishop J. M. The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7742–7746. doi: 10.1073/pnas.81.24.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. D., Roberts B. L., Smith A. E. Nuclear location signals in polyoma virus large-T. Cell. 1986 Jan 17;44(1):77–85. doi: 10.1016/0092-8674(86)90486-1. [DOI] [PubMed] [Google Scholar]
- Sarid J., Halazonetis T. D., Murphy W., Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Ong G. L., Keath E. J., Piccoli S. P., Cole M. D. Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell. 1982 Dec;31(2 Pt 1):443–452. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- Shih C. K., Linial M., Goodenow M. M., Hayward W. S. Nucleotide sequence 5' of the chicken c-myc coding region: localization of a noncoding exon that is absent from myc transcripts in most avian leukosis virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4697–4701. doi: 10.1073/pnas.81.15.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stanton L. W., Schwab M., Bishop J. M. Nucleotide sequence of the human N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1772–1776. doi: 10.1073/pnas.83.6.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. C., Atkinson T., Smith M., Pawson T. Identification of functional regions in the transforming protein of Fujinami sarcoma virus by in-phase insertion mutagenesis. Cell. 1984 Jun;37(2):549–558. doi: 10.1016/0092-8674(84)90385-4. [DOI] [PubMed] [Google Scholar]
- Taub R., Kirsch I., Morton C., Lenoir G., Swan D., Tronick S., Aaronson S., Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D. K., Psallidopoulos M. C., Samuel K. P., Dalla-Favera R., Papas T. S. Nucleotide sequence analysis of human c-myc locus, chicken homologue, and myelocytomatosis virus MC29 transforming gene reveals a highly conserved gene product. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3642–3645. doi: 10.1073/pnas.80.12.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D. K., Reddy E. P., Duesberg P. H., Papas T. S. Nucleotide sequence analysis of the chicken c-myc gene reveals homologous and unique coding regions by comparison with the transforming gene of avian myelocytomatosis virus MC29, delta gag-myc. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2146–2150. doi: 10.1073/pnas.80.8.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Nishikura K., Sorrentino J., ar-Rushdi A., Croce C. M., Rovera G. The structure and nucleotide sequence of the 5' end of the human c-myc oncogene. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6307–6311. doi: 10.1073/pnas.80.20.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- Welsh J. D., Swimmer C., Cocke T., Shenk T. A second domain of simian virus 40 T antigen in which mutations can alter the cellular localization of the antigen. Mol Cell Biol. 1986 Jun;6(6):2207–2212. doi: 10.1128/mcb.6.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway D., Payne G., Varmus H. E. Proviral deletions and oncogene base-substitutions in insertionally mutagenized c-myc alleles may contribute to the progression of avian bursal tumors. Proc Natl Acad Sci U S A. 1984 Feb;81(3):843–847. doi: 10.1073/pnas.81.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]