Abstract
Background
A novel variant of the ST1-SCCmecIV methicillin-resistant Staphylococcus aureus (MRSA) lineage, mostly associated with nosocomial bloodstream infections (BSI), has emerged in Rio de Janeiro. Bacterial biofilm has been considered a major virulence factor in central venous catheter-associated BSI. The mechanisms involved in biofilm formation/accumulation are multifactorial and complex. Studies have suggested that biofilm production was affected in vitro and vivo for agr-null mutants of S. aureus.
Results
The impact of naturally occurring inhibition of agr signaling on virulence profiles and infections associated with the ST1 variant was investigated. agr dysfunction was detected in a significant percentage (13%) of the isolates with concomitant increase in biofilm accumulation in vitro and in vivo, and enhanced ability to adhere to and invade airway cells. The biofilm formed by these ST1 isolates was ica-independent and proteinaceous in nature. In fact, the improved colonization properties were paralleled by an increased expression of the biofilm-associated genes fnbA, spa and sasG. The transcription of sarA, a positive regulator of agr, was two-times reduced for the agr-dysfunctional MRSA. Remarkably, the agr inhibition was genetically stable. Indeed, agr-dysfunctional isolates succeed to colonize and cause both acute and chronic infections in hospitalized patients, and also to effectively accumulate biofilm in a mouse subcutaneous catheter implant model.
Conclusion
The ability of agr-dysfunctional isolates to cause infections in humans and to form biofilm in the animal model suggests that therapeutic approaches based on agr-inactivation strategies are unlikely to be effective in controlling human-device infections caused by ST1 isolates. The increased biofilm accumulation associated with the acquisition of multiple antimicrobial resistant traits might have influenced (at least in part) the expansion of this USA400 related clone in our hospitals.
Keywords: MRSA, ST1-SCCmecIV, USA400, agr, Biofilm, Virulence factors
Background
Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) lineage ST1- SCCmec IV was first reported in the 1980s among aborigines in Australia (WA-1 clone) and in the USA (MW2/USA400 clone) where cases of fatal infections were reported in Michigan, Minnesota and North Dakota [1-3]. Nowadays, CA-MRSA infections have been described in different countries involving a number of genetically distinct lineages [4,5].
Many CA-MRSA isolates (including USA300, USA400 and USA1100) carry lukSF encoding for Panton-Valentine leukocidin (PVL). Despite the controversy regarding the role of the PVL, this leukocidin has been linked to severe skin infections and necrotizing pneumonia [6-8]. In the USA, USA300 has replaced USA400 as the predominant clone in many communities [9]. However, USA400 isolates were the main cause of an outbreak of skin infections that occurred in rural southwestern Alaska, in 1996–2000 [10]. Indeed, USA400 was the far most common CA-MRSA clone recovered from three northern remote communities of Saskatchewan, Canada [11]. In 2005, a novel variant of the lineage ST1-SCCmecIV emerged in Rio de Janeiro city as an important cause of bloodstream infections (BSI) [12]. It is intriguing that despite the genetic relationship with Australian WA-1 and MW2/USA400, isolates of this novel clone were PVL-negative, multiresistant and mostly involved in hospital-associated BSI [12]. It is still poorly understood why isolates of CA-MRSA have become successful so quickly [13]. Nevertheless, for hospital-associated MRSA (HA-MRSA), the bacterial ability to produce biofilm has been recognized as an important virulence factor for the pathogenesis of intravenous catheter-related bacteremia and infections associated with the use of medical prosthesis. In addition, the bacterial ability to adhere to, colonize and invade host tissues is considered important factor associated with bacterial virulence, adaptation and spread [14,15]. Different surface proteins have been implicated in biofilm formation/accumulation and host colonization, including fibronectin-binding proteins A and B (FnBPAB), S. aureus surface protein G (SasG) and staphylococcal protein A (Spa) [16-19]. In addition, extracellular DNA (eDNA) has also been associated with bacterial biofilms [20].
It is also well known that virulence in S. aureus is modulated by an intricate regulatory network [21]. The accessory gene regulator (agr), the major S. aureus quorum sensing system, down-regulates a number of genes encoding for cell-surface proteins involved in colonization processes, and up-regulates (by an indirect mechanism involving RNAIII dependent down-regulation of Rot) different exoproteins associated with host-cell damages [22]. Previous works have suggested that inactivation of Agr could be very effective at inhibiting S. aureus infections [23], including those associated with implantable medical devices [24,25]. Studies have demonstrated that biofilm production, host cell adhesion and invasion as well as other mechanisms involved in the establishment and course of staphylococcal diseases were affected by knockout of the agr locus [26-28]. Despite the improvements achieved in staphylococcal virulence, most of the investigations have been carried out using relatively few laboratory constructions or clinical isolates [28]. In addition, those results have not been validated using current clinical isolates of MRSA. In this paper we characterized the biofilm formed by USA400-related (ST1-SCCmecIV) MRSA emergent in Rio de Janeiro, investigated the adhesive and invasive properties of naturally agr-dysfunctional isolates and analyzed the impact of the agr inhibition on S. aureus infections associated with the use of medical device. Our results suggest that strategies based on agr inactivation approaches may not be effective as an anti-biofilm strategy in the management of device-associated infections caused by these MRSA isolates.
Results
Biofilm
All sixty ST1 isolates tested were able to produce biofilm on inert surfaces. The majority (58.3% and 25%; respectively) exhibited a moderate (BU varying from 0.468 to 0.901) or strong (BU varying from 1.008 to 3.615) biofilm phenotypes (Figure 1, top). For 19 randomly selected isolates, the ability to accumulate biofilm on human Fn-coated surfaces increased significantly (p<0.01 to p<0.0001) when compared with that on inert surfaces (Figure 1, bottom).
Figure 1.
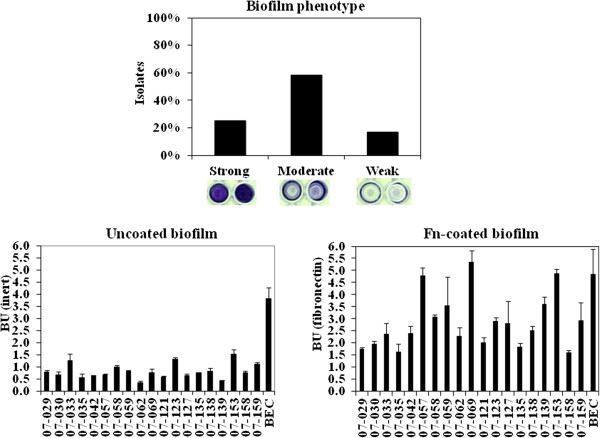
Biofilm formed by ST1 isolates. Top: Percentage of the total 60 ST1 isolates displaying strong, moderate and weak biofilm phenotypes. Wells show the different biofilm phenotypes formed on inert polystyrene surfaces by representative ST1 isolates. Bottom: Biofilm formed on inert or fibronectin-coated surfaces by 19 ST1 isolates.
Proteinaceous nature of the biofilm
Treatment with proteinase K virtually disrupted preformed biofilms for 12 ST1 isolates tested. However, the carbohydrate oxidant metaperiodate almost did not affect the biofilm accumulated by these isolates (Figure 2, top). CLSM studies revealed that the agr-dysfunctional 08–008 accumulated a denser and compact biofilm when compared to the heterogeneous film formed by the agr-functional isolate (96/05). Despite the stronger biofilm phenotype displayed by the isolate 08–008, proteinase K could significantly remove the biological film accumulated (Figure 2, bottom).
Figure 2.
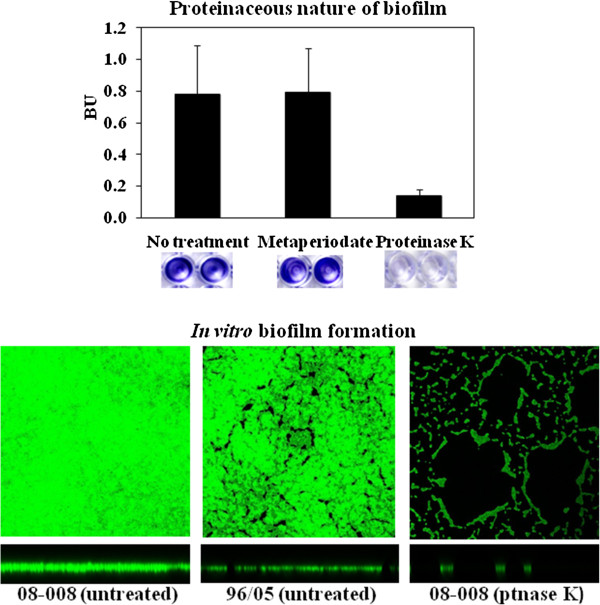
Proteinaceous nature of the biofilm. Top: Effect of 1mM/well sodium metaperiodate or 6U/well proteinase K on preformed biofilm. Wells show the effect of these compounds on biofilms preformed on inert polystyrene surfaces by representative ST1 isolates. Bottom: Confocal laser scanning microscopy (CLSM) images of proteinase K-treated and -untreated biofilms stained with SYTO 9. The square indicates the slice of the biofilm from which the XY image was taken. The horizontal bar indicates the location of the X plane from which the cross-section was taken. Isolate 08–008 (strong biofilm producer, agr-dysfunctional), 96/05 (moderate biofilm producer, agr-functional).
Role of eDNA in ST1 biofilm
No correlation was detected between the activity of bacterial DNase and the levels of biofilm accumulated by 17 USA400-related isolates displaying strong, moderate or weak biofilm phenotypes (Figure 3, top). The addition of 28U/well DNase I in the culture media did not significantly affect the biofilm formed by these ST1 isolates. However, when this concentration was increased to 56U/well, a significant (p=0.0078) reduction of 31% in biofilm accumulation was detected (BU untreated =0.91±0.1 and treated =0.63±0.078; Figure 3, left bottom). In addition, the concentration of eDNA recovered from the supernatant of the strong biofilm producer (BU=1.167 ±0.07) isolate 08–008 was 182 ng/mL, three-times higher than that determined for the weaker producer (BU=0.348±0.01) isolate 117/05 (Figure 3, right bottom). In agreement with these results, we have also detected a moderate correlation (r=0.59) between bacterial autolysis and biofilm accumulation, when 4 stronger biofilm producers were compared with the same number of weaker producers (Figure 4).
Figure 3.
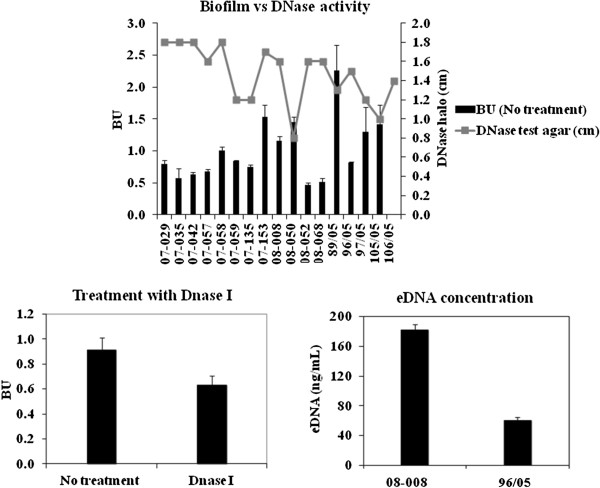
Bacterial DNase activity, treatment of the biofilm with DNase I and eDNA assay. Top: DNase activity was detected in culture supernatants of 16 ST1 isolates by measuring the halo size (cm) produced on Difco™ DNase Test Agar (BD). BU: Biofilm values for 16 ST1 isolates using inert polystyrene. Left bottom: For 16 ST1 isolates, 56U/well of DNase I were added to the culture media and the amount of biofilm accumulated determined. Right bottom: The concentration of eDNA determined in the biofilm supernatant. Isolate 08–008 (strong biofilm producer, agr-dysfunctional), 96/05 (moderate biofilm producer, agr-functional).
Figure 4.
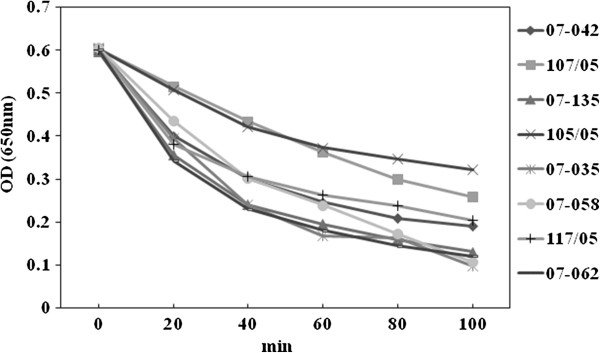
Autolysis assays for USA400-related isolates. 07–058, 105/05, 107/05 are strong biofilm producers; 07–035, 07–042, 07–135 moderate; and 07–062, 117/05 weak producers.
agrRNAIII inhibition
About 13% (8/60) of the USA400 related isolates exhibited no apparent hemolytic activity (Figure 5, top right). These 8 isolates had almost undetectable agr expression by RT-qPCR (Figure 5, top left). Of significance is the fact that 4 out of 8 agr-dysfunctional MRSA were recovered from BSI (50%). The RNAIII transcriptional levels for the 8 agr-functional isolates analyzed were significantly lower than that of strain RN6390B (Figure 5, top left). When we correlated the biofilm values (BU) with the levels of RNAIII transcription, we found that the population of clinical isolates with no hemolytic activity showed significant increase (p=0.01) in biofilm formation/accumulation (Figure 5, bottom). No significant difference could be detected in the values of oxacillin MIC when agr-functional (MIC90 = 128µg/mL) were compared with agr-dysfunctional isolates (MIC90 = 128µg/mL). Indeed, when we quantified mecA transcripts for 5 ST1 isolates, 08–008 (RQ=0.06±0.004), 89/05 (RQ=1.194±0.1), 08–068 (RQ=2.841±0.816), 07–135 (RQ=1.867±0.69), 07–058 (RQ=1±0.62), displaying different levels of agr expression (Figure 5, top left), we could not find a negative linear correlation between mecA and agr expressions (correlation coefficient, r = 0.823). Thus, an overexpression of mecA can not to be implicated in the inhibition of RNAIII transcription. Because agr is positively regulated by SarA, the expression of sarA gene was also analyzed by RT-qPCR. Our data showed a significant (p=0.0052) attenuation of sarA for the agr-dysfunctional isolate 08–008 when compared with the agr-functional 96/05 (Figure 6).
Figure 5.
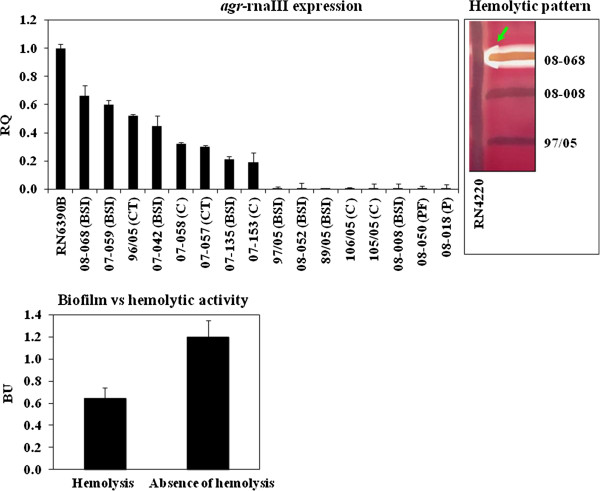
agr differential expression in USA400-related isolates. Top left:rnaIII expression was analyzed by RT-qPCR using ΔΔCT comparative method. RQ: Relative quantity, (BSI): bloodstream infection, (CT): catheter tip, (P): Pneumonia, (C): colonization and (PF): prosthesis fragment. Top right: The arrow indicates the arrow-tip-like zone of the δ-hemolysin activity on sheep blood agar. Bottom: Mean biofilm values (BU) for the populations formed by isolates showing hemolytic activity or absence of hemolysis.
Figure 6.
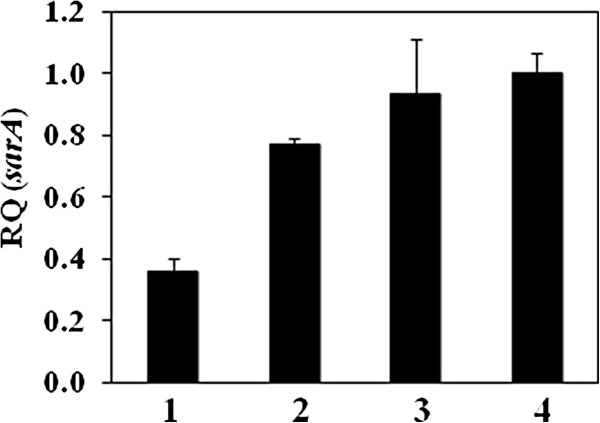
Transcriptional levels of sarA determined by using ΔΔCT comparative method. (1) USA400-related isolates 08–008 (agr-dysfunctional) and (2) 96/05 (agr-functional). (3) BMB9393 was used as a control and (4) RN6390B as calibrator. RQ: Relative quantity.
Animal model
The naturally agr-dysfunctional MRSA was able to colonize and grow on the surface of implanted catheter fragment, as well as to accumulate an increased amount of biofilm (2-log CFU/mL) when compared with the agr-functional isolate (Figure 7, top). The stability of the agr expression in the agr-dysfunctional MRSA was examined by observing the hemolytic activity of individual colonies. No hemolytic halo was detected before and after passages in mice (Figure 7, bottom).
Figure 7.
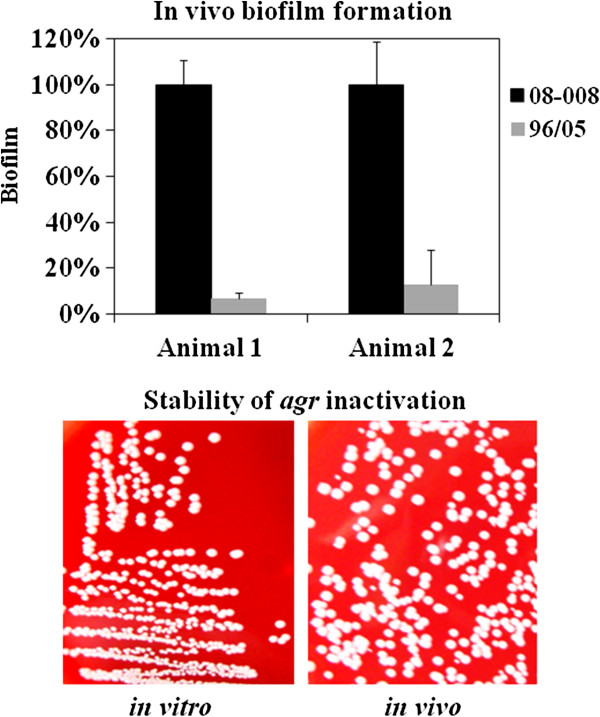
In vivo biofilm accumulation and stability of agr inhibition. Top: For the foreign body animal model, data were transformed in percentage considering the CFU/mL of the isolate 08–008 as the reference value (100%). Bottom: The stability of agr inhibition was tested by examining the hemolytic activity of individual colonies of the isolates 08–008 before (left) and after (right) passage in the animal.
Expression of agr-regulated genes
Total RNA obtained from isolates with significant differences (p<0.001) in the RNAIII transcription level (08–008; RQ=0.0001±0.16 and 96/05; RQ=0.53±0.13) was used to analyze the expression of genes that are well known to be regulated by agr. As expected, the agr-up-regulated hla was less expressed (p<0.01) in the isolate 08–008 (Figure 8) when compared with the isolate 96/05 (RQ=0.05±0.01 and RQ=0.33±0.05, respectively). Similar pattern of expression was found for another agr-up-regulated gene, psmα (RQ96/05=75.90±0.10 and RQ08-008=0.005±0.12; p<0.001), except that in this case we also observed a very high expression of psmα for 96/05 (Figure 8). To verify if this amplified expression was a characteristic of this MRSA clone, other agr-functional isolates were randomly selected for testing. High level of psmα transcripts was also detected for the isolates 07–035, 07–059 and 08–068 (RQ07 035=35.71±0.06; RQ07-059=48.90±0.07; RQ08-068=31.30±0.07). For all virulence genes tested, the expression of the agr-functional isolate BMB9393 was higher than that of USA400-related isolates, except for psmα gene (Figure 8). Accordingly, the RNAIII-down-regulated spa gene showed a very significant lower expression (p<0.001) in the agr-functional 96/05 (RQ=0.8±0.20) compared with the agr-dysfunctional isolate 08–008 (RQ= 52.8±0.17; Figure 8).
Figure 8.
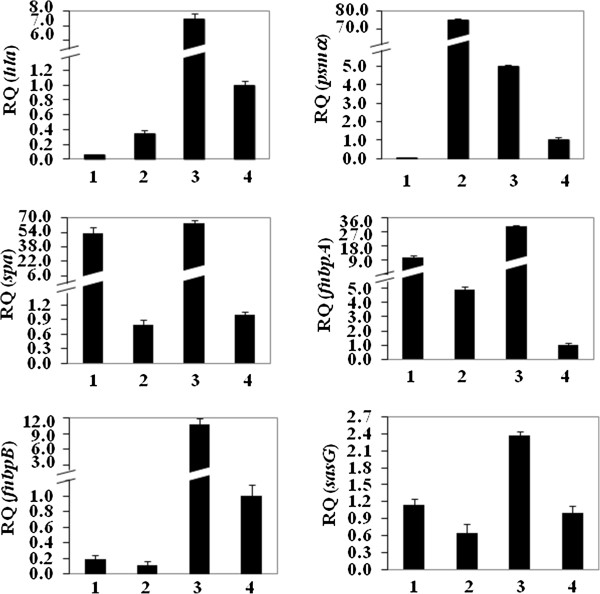
Transcriptional levels of virulence-associated genes determined by RT-qPCR, using ΔΔCT comparative method. (1) USA400-related isolates 08–008 (agr-dysfunctional) and (2) 96/05 (agr-functional). (3) BMB9393 was used as a control and (4) RN6390B as calibrator. RQ: Relative quantity.
Expression of biofilm-associated genes fnbAB, sasG and spa
The agr-dysfunctional isolate 08–008, which showed increased biofilm accumulation in vitro and in vivo, had a significant increase (p=0.02) in fnbA transcripts (RQfnbA=10.08±0.18) when compared with the isolate 96/05 RQfnbA=4.91±0.19; Figure 8). However, no significant difference was detected when fnbB expression were analyzed (RQ96/05 =0.11±0.04; RQ08-008 =0.18±0.05; Figure 8). Similarly to fnbA, the expression of sasG (Figure 8; p=0.03) and spa (Figure 8; p<0.001) was also increased in 08–008 (RQsasG=1.13±0.11; RQspa=52.8±0.17) compared with 96/05 isolate (RQsasG=0.65±0.14; RQspa=0.8±0.20).
Adherence and invasion
The naturally agr-dysfunctional isolate 08–008 showed significant increase (p<0.05) in the adherence to human airway cells, reaching 25.27%±0.4% at 3h30min of incubation. In contrast, at the same conditions, the adherence of the agr-functional (isolate 96/05) to airway cells occurred in much less extent (4.94%±0.2%). Similarly, invasion was also higher for the agr-dysfunctional isolate (6.37%±0.3%) when compared with the agr-functional (1.76%±0.2%) at 3h30min incubation (Figure 9, top). Likewise, an increased invasive ability in the stationary phase was observed for the agr-knockout MHC474 (10.6%±0.3%) when compared with the wild type (HC474; 2.8%±0.1%) and complemented construction CMHC474 (2.3%±0.1%; p=0.0033; Figure 9, bottom).
Figure 9.
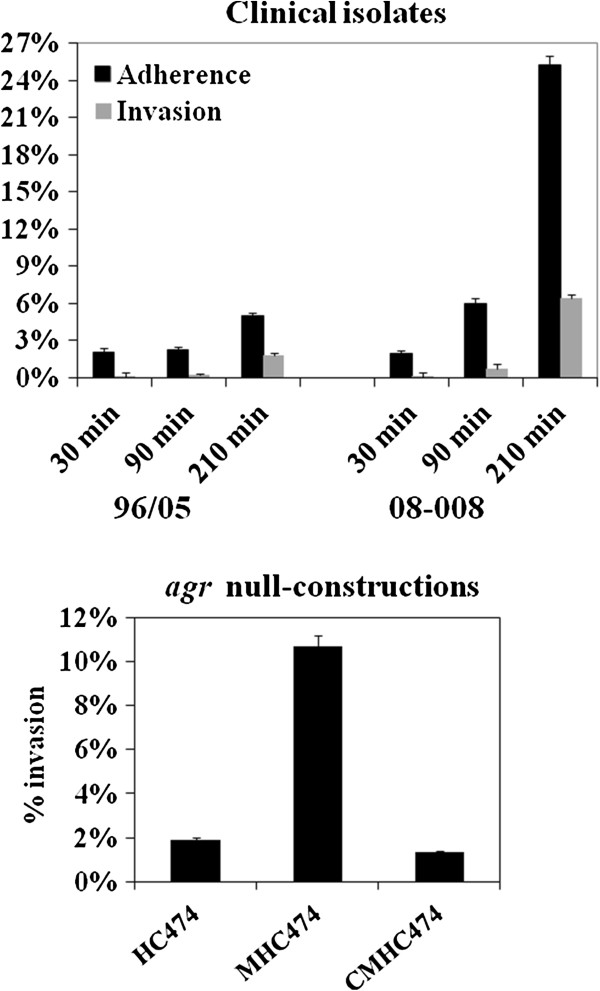
Adherence and invasion assays using human bronchial epithelial cell line (16HBe14o-). Top: 96/05 (agr-functional) and 08–008 (agr-dysfunctional). Bottom: Invasion assay was also determined after 3h30 min for the wild-type strain HC474, isogenic agr knockout MHC474 (Δagr::tetM) and the rnaIII-trans-complemented construction CMHC474 (Δagr::tetM, pbla-rnaIII).
Discussion
The great majority of the USA400-related isolates (50/60; 83.3%) were able to accumulate strong/moderate biofilms on polystyrene surfaces. The isolates remaining produced weak biofilms. The ability to accumulate biofilm increased when the surfaces were covered with human fibronectin, as also reported by others [19,29]. In opposition to our results, it was reported that MW2 MRSA had a weak biofilm phenotype [30,31]. Similarly, a slight biofilm accumulation (OD=0.25-0.3) was observed for another USA400 strain called BAA-1683 [32]. In addition, recent data from our laboratory (Ramundo MS & Figueiredo AMS, 2012; unpublished observations) showed that another SCCmecIV isolates (ST30 CA-MRSA) accumulated much lower amount of biofilm compared with ST1-SCCmecIV isolates.
Previous data from our group [12] have also demonstrated that the ST1 isolates from Rio de Janeiro do not carry lukSF genes and have acquired a number of antimicrobial resistance traits. Thus, it is possible that the enhanced ability to accumulate biofilm, associated with the biological cost of acquired resistance and the absence of PVL, might have been the results (at least in part) of the microevolutionary events that accounted for changes in a previously community pathogen, promoting enhanced bacterial fitness to spread in hospitals and cause health-care associated diseases. The ica-independent nature of the biofilm formed by USA 400-related isolates was revealed by the disruption of bacterial film by proteinase K. Similar results were also observed by others using different MRSA isolates [33,34]. Some researchers have suggested that the bacterial autolysis increases eDNA concentration and, consequently, enhances the level of biofilm accumulation [20]. In fact, in our study, we observed a moderate correlation between biofilm accumulation and autolysis. In addition, we detected threefold increase in eDNA for the ST1 MRSA displaying enhanced ability to accumulate biofilm. Indeed, the addition of DNase I (56U/Well) caused a significant reduction (about 30%) in biofilm accumulation, suggesting eDNA cooperatively contributes to the biofilm architecture of ST1 isolates.
The statistical analysis showed that the group of clinical isolates with no hemolytic activity (agr-dysfunctional) had significant increase in the level of biofilm accumulation when compared with agr-functional isolates. These data are in agreement with previous studies for agr-laboratory knockouts [27,35,36], which have indicated that some agr mutants can display increased levels of biofilm accumulation. In spite of that, using another S. aureus strain it was reported that inhibition of agr reduced biofilm accumulation significantly [24,25]. In fact, agrRNAIII is a negative regulator of different surface proteins [22,23], and consistent with this regulation, amplified expression of genes encoding for biofilm-associated proteins FnBPA, SasG and Spa was found for the agr-dysfunctional variant. Both FnBPA and B have been implicated as major proteins for biofilm formation/accumulation in S. aureus[19,33]. However, despite the detection of an enhanced expression of fnbA, we could not find a significant increase in the transcription of fnbB-mRNA for the agr-dysfunctional ST1-MRSA. Equally, a study from Wolz and collaborators suggested that fnbB was not significantly affected by agr[36].
Confirming the agr inhibition detected, the expression of two genes up-regulated by RNAIII, hla and psmα, was lower compared with the agr-functional MRSA. Both cytolysins (HLA and PSMα) seem to have remarkable roles in the pathogenesis of S. aureus. HLA has been associated with lethal pneumonia in USA400 and USA300 strains [37,38]. It was also previously found that psmα-deleted mutant of CA-MRSA exhibited attenuated virulence in animal models [39]. In this study, we detected a superior expression of pmsα by the agr-functional isolates of USA400-related clone detected in Rio de Janeiro. In fact, it was shown by others that the transcription of psmα-mRNA was increased in most prevalent CA-MRSA lineages, including MW2, compared with other S. aureus isolates [39]. However, the molecular mechanisms involved with the enhanced expression of PSMα were not clarified [39]. Despite the importance of these virulence factors for S. aureus pathogenicity, it is remarkable that among the agr-dysfunctional variants, 4 were recovered from cases of BSI, 2 from colonization, 1 from pneumonia and 1 from infected prosthesis, showing that these variants were able to colonize and cause both severe acute (pneumonia and BSI) and chronic (foreign-body infection) staphylococcal diseases in humans. These data demonstrated that regardless the reduced virulence of agr-laboratory knockouts in some animal models [40], the virulence of naturally dysfunctional agr variants was confirmed for hospitalized patients. In contrast to the assumption that agr-dysfunctional isolates may not be able to initiate infections [41], the isolate 08–008 was able to colonize polyurethane endovenous catheter in a foreign-body mouse model, forming a denser biofilm accumulation when compared with the agr-functional isolate. It is important to state that because the ST1 isolates studied were not isogenic, it is possible that factors other than the inhibition of agr might also have accounted for the increased biofilm accumulation observed. Nevertheless, supporting our data, similar increase of the biofilm formed on catheters implanted in mice was previously reported for an agr laboratory knockout [28]. In opposition to the results obtained by Traber et al. [41], all individual colonies formed by the agr-dysfunctional MRSA remained non-hemolytic before and after passages in mice, strongly suggesting the genetic stability of the phenotype. This stability was confirmed for all agr-dysfunctional isolates from our collection. Corroborating our findings, while we were finishing this manuscript, we noticed the work by Park et al. [42] that found agr dysfunction in S. aureus significantly associated with persistent bacteremia with eradicated foci, even though the predominant MRSA isolates showed SCCmecII, agrII (possible belonging to USA100-New York/Japan clone) while the isolates studied here displayed SCCmecIV, agrIII and clustered in USA400-MW2/WA-1 clone. In fact, the bacterial ability to adhere to and invade epithelial cells, and consequently evade host defense mechanisms, has already been associated with persistence in host cells and development of disseminated infections [43,44]. In the present study, the differential expression of agrRNAIII in MRSA clinical isolates had a significant impact on adherence and invasion at 3h30min incubation. The same impact was observed for the agr isogenic knockout, as previously showed by others using different cell lines and mostly laboratory mutants [26,45].
Recently, Pozzi et al. demonstrated that high level of PBP2a expression by the homogeneous methicillin-resistant derivative of the strain 8325–4 induced a proteinaceous biofilm and significant repression of the agr locus [46]. In addition, excision of the SCCmec element from the MRSA strain BH1CC, with consequent loss of oxacillin resistance, had the opposite effect on biofilm and lead to an increase of the agrRNAIII transcription. In addition, Rudkin et al. showed that methicillin resistance reduced the virulence of HA-MRSA by interfering with agr[47]. The great majority of ST1 isolates studied had MIC of 128 µg/mL (agr-functional or -dysfunctional), which is compatible with heterogeneous resistance to this drug. Indeed, mecA overexpression was not detected in the agr-dysfunctional isolates tested. SarA, a global transcriptional regulator of S. aureus, was previously found to be a positive regulator of agr and of biofilm formation/accumulation [21,48]. Thus, aiming to understand the mechanism involved in agr impairment in these clinical isolates, the level of sarA transcripts was also examined. It was observed that sarA expression was significantly diminished in the agr-dysfunctional compared with the agr-functional MRSA, suggesting the defect was upstream agr. Beeken et al. indicated that sarA repression inhibited biofilm accumulation due to SarA inhibition of both proteases and nucleases activity either in the presence or absence of agr mutations [49]. In contrast, the results obtained here demonstrated that agr-dysfunctional isolates showed increased biofilm accumulation, despite the fact that sarA-mRNA transcripts were reduced. In fact, other studies have showed that sarA or agr-sarA laboratory mutants had lower ability to bind to fibronectin due to sarA down-regulation of fnbA transcription [36]. Possible explanations for this apparent divergence could be the fact that the agr-dysfunctional ST1 studied showed only partial sarA inhibition, or may display strain-dependent variation in the genetic background affecting other genes apart to those studied.
Conclusion
Isolates of this novel hospital-associated USA400 clone were able to accumulate moderate/strong amount of biofilms, in vitro and in vivo, and could efficiently adhere to and invade human airway cells. Moreover, agr inhibition was an ordinary phenomenon among those isolates, which seems to have impacted the expression of some important virulence genes studied. Although it is difficult to interpret in vitro studies in the light of what occurs in an infected human host, it follows logical that the enhanced adhesive properties combined with the acquisition of multiple drug resistance traits by ST1 isolates could have provided fitness advantages for spreading in hospital environments. Indeed, agr-dysfunctional isolates were recovered from cases of hospital pneumonia, bacteremia and infected prosthesis. Finally, our results strongly suggest that strategies for controlling MRSA biofilm based on agr inhibition approaches are unlikely to be effective, at least for ST1 MRSA isolates.
Methods
Isolates
Sixty USA400-related isolates were obtained from patients located in different hospital wards in Rio de Janeiro as part of standard clinical care. Thirty isolates were recovered from BSI (50%) and 8 from catheter tips (CT; 13.3%). The remaining were from colonization (C; 13.3%), pneumonia (P; 6.7%), skin/soft tissue infections (SSTI; 5%), urinary tract infections (UTI; 3.3%) and prosthesis fragment (PF; 1.7%). The infection sites had not been reported for 4 isolates. The agr-knockout MNY474 (Δagr::tetM) and the rnaIII-trans-complemented mutant CMNY474 (Δagr::tetM, pbla-rnaIII) were previously constructed from the clinical S. aureus isolate NY474 [27]. BMB9393 (ST239-SCCmecIII) was used as positive control for biofilm and gene expression experiments [27]. The S. aureus RN4220 and RN6390B, a gift from Richard Novick (New York University), were used for hemolytic activity and gene expression analyses; respectively. This study was approved (#1055/09) by the Human Research Ethics Committee from Federal University of Rio de Janeiro, RJ, Brazil.
Minimal inhibitory concentration (MIC)
Oxacillin MIC was determined using Müller Hinton plates and performed in accordance with the Clinical Laboratories Standards Institutes (CLSI) guidelines [50].
In vitro biofilm assay
For all 60 isolates, biofilm was tested using 96-well inert polystyrene microtiter plates (Nunclon; Nunc A/S, Roskilde, Denmark) as previously described [28]. The biofilm unit (BU) was defined as indicated by Amaral et al. [14] and the isolates were classified as non-producers (BU≤0.230), weak (BU>0.230 and ≤0.460), moderate (BU>0.460 and ≤0.920) or strong producers (BU>0.920), as suggested [14]. For 19 isolates, biofilm assays were also carried out on surfaces covered with human fibronectin (Merck; Darmstadt, Germany) as previously described [28].
In some experiments, before treatment with crystal violet, the biofilm was treated with sodium metaperiodate (10mM/well; Sigma; St. Louis, MO, USA) or proteinase K (6U/well, Invitrogen; Carlsbad, California, EUA) [27]. Confocal laser scanning microscopy (CLSM) was employed to record and contrast structural images of the biofilm as described [28]. eDNA was quantified in biofilm supernatants using Qubit® 2.0 Fluorometer (Invitrogen; Eugene, Oregon, USA), after ethanol precipitation. For some experiments, biofilms were formed in the presence of DNase I (28U/well or 56U/well Invitrogen; Carlsbad, California, EUA).
Animal model
A pair of isolates showing differential agr expression (08–008, agr-dysfunctional, obtained from BSI and 96/05, agr-functional, from CT) was used. The mouse subcutaneous catheter implant model was described in detail by Ferreira et al. [28]. Briefly, two intravenous polyurethane catheter segments (C-UDLM-953J model; Cook Medical, Bloominaton, USA) were implanted in the back of each anesthetized young-adult BALB/c male mice. Infection was induced 24 h after the implantation procedure by injecting a mid-exponential growth phase culture (106 CFU/10 µL) into the lumen of the implanted catheter segment. The animal was euthanized after three days post-infection, and the catheter segments were surgically removed to assess the biofilm by counting catheter-adherent bacteria by CFU determination. Three independent experiments were performed. The animal study was approved (#IMPPG013) by The Ethics Committee for Animal Care and Use from Federal University of Rio de Janeiro, RJ, Brazil.
DNase activity
Difco™ DNase Test Agar (BD; Becton, Dickinson and Company, Sparks, USA) was used to screen 17 USA400-related MRSA, as recommended by the manufacturer.
Autolysis assay
Autolysin activity was measured in 8 selected isolates as previously described [51], except that cells were grown in TSB 1% Glc.
Hemolytic activity
The δ-hemolysin (Hld), encoded by the hld gene, is codified within the rnaIII region and, consequently, the detection of δ-hemolysin is an indicative of agr expression. Sixty USA400-related isolates were screened for hemolytic activity on sheep red blood (5%) agar plates (Plast Labor, RJ, Brazil) as previously described [52].
Gene expression
For RNA preparations, bacterial cells grown in TSB (18h/37°C; 250 rpm) were obtained in the exponential phase (OD600nm = 0.3) and in the stationary phase. Total RNA was prepared using the RNeasy Mini kit (Qiagen; Maryland, USA) and quantified by the Qubit 2.0 Fluorometer. The RNA quality was analyzed by running RNA-gel electrophoresis. The real-time quantitative PCR (RT-qPCR) was carried out using Power SYBR® Green RNA-to-CTTM 1-Step Kit (Applied Biosystems; Foster city, CA, USA) as recommended, using ΔΔCt comparative method. The primers and run conditions used for rnaIII, hla, psmα[53], sarA, mecA[54], spa, sasG, fnbA and fnbB genes and for the endogenous control rrna 16S are listed in Table 1. All primers designed for this study were validated as recommended (Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR; Applied Biosystems). The run was performed in the Step One™ Real Time PCR System (Applied Biosystems). Data were analyzed using the Step One Software 2.2 (Applied Biosystems).
Table 1.
Primers used in Real Time qPCR
| Target gene | Primer sequencea | Amplicon length (bp) | Reference |
|---|---|---|---|
|
rnaIII |
F: AATTTGTTCACTGTGTCGATAAT |
135 |
This study |
| R:TGGAAAATAGTTGATGAGTTGTT | |||
|
sarA |
F: TTCTTTCTCTTTGTTTTCGCTG |
115 |
This study |
| R: GTTATCAATGGTCACTTATGCT | |||
|
spa |
F: TGGTTTGCTGGTTGCTTCTTA |
116 |
This study |
| R: GCAAAAGCAAACGGCACTAC | |||
|
hla |
F: TTTGTCATTTCTTCTTTTTCCCA |
169 |
This study |
| R: AAGCATCCAAACAACAAACAAAT | |||
|
psmα |
F:TATCAAAAGCTTAATCGAACAATTC |
176 |
53 |
| R: CCCCTTCAAATAAGATGTTCATATC | |||
|
sasG |
F:GGTTTTCAGGTCCTTTTGGAT |
192 |
This study |
| R:CTGGTGAAGAGCGAGTGAAA | |||
|
fnbpA |
F: ACTTGATTTTGTGTAGCCTTTTT |
185 |
This study |
| R:GAAGAAGCACCAAAAGCAGTA | |||
|
fnbpB |
F:CGTTATTTGTAGTTGTTTGTGTT |
118 |
This study |
| R:TGGAATGGGACAAGAAAAAGAA | |||
|
rrna 16S |
F: AGAGATAGAGCCTTCCCCTT |
84 |
This study |
| R:TTAACCCAACATCTCACGACA | |||
|
mecA |
F:TCCAGATTACAACTTCACCAGG |
162 |
54 |
| R:CCACTTCATATCTTGTAACG |
aF and R: forward and reverse primers, respectively, in 5´→ 3´orientation. The cycling conditions for all primers were as follows: One cycle of 48°C/30min and 95°C/10 min, followed by 35 cycles of 95°C/30s, 55°C/45s and 72°C/45 s. Each run included a nontemplate and a gene-negative RNA controls.
Adherence and invasion kinetics
Bacterial adherence and invasion were investigated using human bronchial epithelial cells (16HBE14o- cell line) as described [14], except that monolayers were prepared using Dulbecco´s Modified Eagle Medium (DMEM, Low Glucose 1X; Gibco, Invitrogen, Grand Island, USA) and 10% Fetal Bovine Serum (Gibco, Invitrogen). For determining the colony forming units (CFU) of the total adhered and invasive bacteria (CFUAI), infected monolayers were washed twice in DMEM (to remove non-adherent bacteria), incubated (5 min/37°C) with 0.25% (wt/vol) trypsin (11,000 U/mg; Sigma; St. Louis, MO USA), lysed (5 min/37°C) with 0.025% (vol/vol) Triton X-100 (Sigma) and plated in TSA. For determining the CFU of invasive bacteria (CFUI), infected monolayers were washed twice in DMEM and incubated (20 min/37°C) with 100 µg/mL lysostaphin (500 U/mg; Sigma) to lyse adherent bacteria. Monolayers were washed twice and incubated (5 min/37°C) with 0.25% (wt/vol) trypsin. The epithelial cells were lysed (5 min/37°C) with 0.025% (vol/vol) triton X-100 and plated. For each aliquot, the total CFU in the supernatant was also determined (CFUS). The CFU of adherent bacteria (CFUA) was obtained by the formula: CFUA = CFUAI - CFUI. The percentages of invasive or adherent bacteria were calculated considering as 100% the total CFU obtained by the sum of CFUAI + CFUS for each aliquot. In addition to the USA400-related isolates, the wild-type HC474, and the isogenic Δagr::tetM and rnaIII-trans-complemented constructions were also used for investigating bacterial invasion.
Statistical calculations
Student’s t-test (unpaired data) was used to compare the means of the biofilm values and of the data from gene expression experiments. In addition, correlation coefficient (r) was used to test the relationship between the autolysis and the ability of ST1 isolates to accumulate strong or weaker biofilms. This last test was also used to determine the occurrence of linear correlation between mecA and agr expressions [55]. Data were expressed in terms of mean values obtained from at least three independent experiments and three repetitions of each set.
Abbreviations
Agr: Accessory gene regulator; BSI: Bloodstream infection; BU: Biofilm unit; C: Colonization; CA-MRSA: Community-acquired Methicillin-resistant Staphylococcus aureus; CFU: Colony-forming unit; CLSM: Confocal laser scanning microscopy; CT: Catheter tip; DNase: Desoxyribonuclease; DMEM: Dulbecco´s Modified Eagle Medium; eDNA: Extracellular DNA; Fn: Fibronectin; fnbAB: Genes encoding for fibronectin-binding protein A and B; FnBPA/FnBPB: Fibronectin-binding protein A and B; ica: Operon encoding for enzymes involved in the synthesis of the polysaccharide of intercellular adhesion (PIA); HA-MRSA: Hospital-associated methicillin-resistant Staphylococcus aureus; Hla: α-Hemolysin; Hld: δ-Hemolysin; lukSF: Genes involved in the synthesis of the subunits S and F of the Panton Valentine leukocidin; MIC: Minimal inhibitory concentration; mecA: Gene encoding for penicillin-binding protein 2A; MRSA: Methicillin-resistant Staphylococcus aureus; OD: Optical density; P: Pneumonia; PBP2A: Penicillin-binding protein 2A; PCR: Polymerase chain reaction; PF: Prosthesis fragment; Psm: Phenol-soluble modulin; PVL: Panton-Valentine leukocidin; r: Correlation coefficient; Rot: Repressor of toxins; RQ: Relative quantity; RT-qPCR: Real time quantitative polymerase chain reaction; rrna 16S: Gene encoding for ribosomal RNA 16S; SarA: Transcriptional regulator SarA; SasG: S. aureus surface protein G; SCCmec: Staphylococcal cassette chromosome mec; ST: Sequence-type; Spa: Protein A; SSTI: Skin and soft tissue infections; TSB: Tryptic soy broth; TSB 1% Glu: Tryptic soy broth supplemented with 1% glucose; UTI: Urinary tract infections.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FAF wrote the draft paper and carried out the experiments of biofilm formation/accumulation on inert polystyrene surfaces, DNase activity, autolysis assay, hemolytic activity, gene expression experiments, DNA Sequencing and statistical calculations. RRS, MAA and SELF carried out experiments of the animal model including animal surgery and observation, and biofilm determinations. RRS also carried out oxacillin MIC determinations. BSM carried out the experiments of biofilm formation/accumulation on inert polystyrene surfaces and also on implanted catheters. AMAF and JNS carried out studies of adherence and invasion kinetics. AMSF carried out the experiments on mecA gene expression and was responsible for the study design, methodology used, wrote and review the draft paper and gave final approval of the manuscript. All authors read and approved the final manuscript. All authors contributed significantly for the conduction of the studies and discussion of the results.
Contributor Information
Fabienne Antunes Ferreira, Email: fabienneaf@gmail.com.
Raquel Rodrigues Souza, Email: rosakel@globomail.com.
Bruno de Sousa Moraes, Email: bruno.sousa.moraes@gmail.com.
Ana Maria de Amorim Ferreira, Email: anamaria@micro.ufrj.br.
Marco Antônio Américo, Email: marcoamerico@hotmail.com.
Sérgio Eduardo Longo Fracalanzza, Email: fracalanzza@micro.ufrj.br.
José Nelson dos Santos Silva Couceiro, Email: jncouceiro@micro.ufrj.br.
Agnes Marie Sá Figueiredo, Email: agnes@micro.ufrj.br.
Acknowledgements
This work was supported in part by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and by European Commission's Seventh Framework Programme (FP7), through the Marie Curie International Research Staff Exchange Scheme NANO_GUARD (PIRSES-GA-2010-269138).
References
- Centers for Disease Control and Prevention. Community-acquired methicillin–resistant staphylococcus aureus infections-Michigan. MMWR Morb Mortal Wkly Rep. 1981;30:185–187. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant staphylococcus aureus: Minnesota and north Dakota, 1997–1999. MMWR Morb Mortal Wkly Rep. 1999;48:707–710. [PubMed] [Google Scholar]
- Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. Community-acquired methicillin-resistant staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- David MZ, Daum RS. Community-associated methicillin-resistant staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov R, Christiansen K, Dancer SJ, Daum RS, Dryden M, Huang YC, Lowy FD. Update on the prevention and control of community-acquired methicillin-resistant staphylococcus aureus (CA-MRSA) Int J Antimicrob Agents. 2012;39:193–200. doi: 10.1016/j.ijantimicag.2011.09.029. [DOI] [PubMed] [Google Scholar]
- Diep BA, Chan L, Tattevin P, Kajikawa O, Martin TR, Basuino L, Mai TT, Marbach H, Braughton KR, Whitney AR, Gardner DJ, Fan X, Tseng CW, Liu GY, Badiou C, Etienne J, Lina G, Matthay MA, DeLeo FR, Chambers HF. Polymorphonuclear leukocytes mediate staphylococcus aureus Panton-valentine leukocidin-induced lung inflammation and injury. Proc Natl Acad Sci USA. 2010;107:5587–5592. doi: 10.1073/pnas.0912403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler B, Hussain M, Grundmeier M, Brück M, Holzinger D, Varga G, Roth J, Kahl BC, Proctor RA, Peters G. Staphylococcus aureus Panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010;6:e1000715. doi: 10.1371/journal.ppat.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. A MRSA-terious enemy among us: end of the PVL controversy? Nat Med. 2011;17:169–170. doi: 10.1038/nm0211-169. [DOI] [PubMed] [Google Scholar]
- Hudson LO, Murphy CR, Spratt BG, Enright MC, Terpstra L, Gombosev A, Hannah P, Mikhail L, Alexander R, Moore DF, Huang SS. Differences in methicillin-resistant staphylococcus aureus (MRSA) strains isolated from pediatric and adult from hospitals in a large California county. J Clin Microbiol. 2011;50:573–579. doi: 10.1128/JCM.05336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS. Molecular epidemiology of methicillin-resistant staphylococcus aureus rural southwestern Alaska. Emerg Infect Dis. 2008;14:1693–1699. doi: 10.3201/eid1411.080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding GR, Levett PN, McDonald RR, Irvine J, Quinn B, Nsungu M, Woods S, Khan M, Ofner-Agostini M, Mulvey MR. Northern Antibiotic Resistance Partnership. High rates of staphylococcus aureus USA400 infection, northern Canada. Emerg Infect Dis. 2011;17:722–725. doi: 10.3201/eid1704.100482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Carvalho MC, Bonelli RR, Souza RR, Moreira S, dos Santos LC, de Souza Conceição M, de Mello Junior SJ, Carballido JM, Vieira VV, Teixeira LA, Sá Figueiredo AM. Emergence of multiresistant variants of the community-acquired methicillin-resistant staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, brazil. Diagn Microbiol Infect Dis. 2009;65:300–305. doi: 10.1016/j.diagmicrobio.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Shukla SK, Karow ME, Brady JM, Stemper ME, Kislow J, Moore N, Wroblewski K, Chyou PH, Warshauer DM, Reed KD, Lynfield R, Schwan WR. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 staphylococcus aureus isolates. J Clin Microbiol. 2010;48:3582–3592. doi: 10.1128/JCM.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral MM, Coelho LR, Flores RP, Souza RR, Silva-Carvalho MC, Teixeira LA, Ferreira-Carvalho BT, Figueiredo AM. The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J Infect Dis. 2005;192:801–810. doi: 10.1086/432515. [DOI] [PubMed] [Google Scholar]
- Datta R, Huang SS. Risk of infection and death due to methicillin-resistant staphylococcus aureus in long-term carriers. Clin Infec Dis. 2008;47:176–181. doi: 10.1086/589241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B, Herrmann M. Mechanism and consequences of invasion of endothelial cells by staphylococcus aureus. Thromb Haemost. 2005;94:266–277. doi: 10.1160/TH05-04-0235. [DOI] [PubMed] [Google Scholar]
- Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penadés JR, Lasa I. Protein a-mediated multicellular behavior in staphylococcus aureus. J Bacteriol. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O´Gara JP, Potts JR, Foster TJ. Role of surface protein SasG in biofilm formation in stapylococcus aureus. J Bacteriol. 2010;192:5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston P, Rowe SE, Pozzi C, Waters EM, O´Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. Nuclease modulates biofilm formation in community-associated methicillin-resistant staphylococcus aureus. PLoS One. 2011;6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner S, Monteil H, Prévost G. Regulation of virulence determinants in staphylococcus aureus: complexity and applications. FEMS Microbiol Rev. 2004;28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Figueiredo AMS, Abramochkin G, Muir T. Activation and inhibition of staphylococcal Agr system. Science. 2000;287:391a. doi: 10.1126/science.287.5452.391a. [DOI] [Google Scholar]
- Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N, Cirioni O, Giacometti A, Ghiselli R, Braunstein JB, Silvestri C, Mocchegiani F, Saba V, Scalise G. Treatment of staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob Agents Chemother. 2007;51:2226–2229. doi: 10.1128/AAC.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Leban F, Kiran MD, Wolcott R, Balaban N. Molecular mechanisms of RIP, an effective inhibitor of chronic infections. Int J Artif Organs. 2010;33:582–589. doi: 10.1177/039139881003300904. [DOI] [PubMed] [Google Scholar]
- Shenkman B, Varon D, Tamarin I, Dardik R, Peisachov M, Savion N, Rubinstein R. Role of agr (RNAIII) in staphylococcus aureus adherence to fibrinogen, fibronectin, platelets and endothelial cells under static and flow conditions. J Med Microbiol. 2002;51:747–754. doi: 10.1099/0022-1317-51-9-747. [DOI] [PubMed] [Google Scholar]
- Coelho LR, Souza RR, Ferreira FA, Guimarães MA, Ferreira-Carvalho BT, Figueiredo AMS. agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of staphylococcus aureus. Microbiology. 2008;154:3480–3490. doi: 10.1099/mic.0.2007/016014-0. [DOI] [PubMed] [Google Scholar]
- Ferreira FA, Souza RR, Bonelli RR, Américo MA, Fracalanzza SEL, Figueiredo AMS. Comparison of in vitro and in vivo systems to study ica-independent staphylococcus aureus biofilm. J Microbiol Methods. 2012;88:393–398. doi: 10.1016/j.mimet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Zautner AE, Krause M, Stropahl G, Holtfreter S, Frickmann H, Maletzki C, Kreikemeyer B, Pau HW, Podbielski A. Intracellular persisting staphylococcus aureus is the major pathogen in recurrent tonsillitis. PLoS One. 2010;5:e9452. doi: 10.1371/journal.pone.0009452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotonda MP, Tamber S, Memmi G, Cheung AL. MgrA represses biofilm formation in staphylococcus aureus. Infect Immun. 2008;76:5645–5654. doi: 10.1128/IAI.00735-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara-Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate staphylococcus aureus regulation. PLoS Pathog. 2011;7:e1001267. doi: 10.1371/journal.ppat.1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SG, Paff M, Friedman G, Fridman G, Fridman A, Brooks AD. Control of methicillin-resistant staphylococcus aureus planktonic form and biofilms: a biocidal efficacy study of nonthermal dielectric-barrier discharge plasma. Am J Infect Control. 2010;38:293–301. doi: 10.1016/j.ajic.2009.11.002. [DOI] [PubMed] [Google Scholar]
- O´Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. A novel staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara-Irigaray M, Valle J, Merino N, Latasa C, García B, Ruiz de Los Mozos I, Solano C, Toledo-Arana A, Penadés JR, Lasa I. Relevant role of fibronectin-binding proteins in staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. 2009;77:3978–3991. doi: 10.1128/IAI.00616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between sigma B, agr, and proteolytic activity in staphylococcus aureus biofilm maturation. Infect Immun. 2009;7:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz C, Pöhlmann-Dietze P, Steinhuber A, Chien YT, Manna A, van Wamel W, Cheung A. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in staphylococcus aureus. Mol Microbiol. 2000;36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]
- Bartlett AH, Foster TJ, Hayashida A, Park PW. Alpha-toxin facilitates the generation of CXC chemokine gradients and stimulates neutrophil homing in Staphylococcus aureus pneumonia. J Infect Dis. 2008;198:1529–1535. doi: 10.1086/592758. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Bae T, Otto M, DeLeo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-valentine leukocidin in staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- Wright JS 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. agr function in clinical staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Chong YP, Park HJ, Park KH, Moon SM, Jeong JY, Kim MN, Kim SH, Lee SO, Choi SH, Woo JH, Kim YS. agr dysfunction and persistent methicillin-resistant staphylococcus aureus bacteremia in patients with removed eradicable foci. Infection. 2013;41:111–119. doi: 10.1007/s15010-012-0348-0. [DOI] [PubMed] [Google Scholar]
- Falkow S. Bacterial entry into eukaryotic cells. Cell. 1991;65:1099–1102. doi: 10.1016/0092-8674(91)90003-H. [DOI] [PubMed] [Google Scholar]
- Garzoni C, François P, Huyghe A, Couzinet S, Tapparel C, Charbonnier Y, Renzoni A, Lucchini S, Lew DP, Vaudaux P, Kelley WL, Schrenzel J. A global view of staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics. 2007;8:171. doi: 10.1186/1471-2164-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesson CA, Liou LE, Todd KM, Bohach GA, Trumble WR, Bayles KW. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infect Immun. 1998;66:5238–5243. doi: 10.1128/iai.66.11.5238-5243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. Methicillin resistance alters the biofilm phenotype and attenuates virulence in staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, Chan WC, Williams P, O'Gara JP, Massey RC. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis. 2012;205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, Smeltzer MS. Epistatic relationships between sarA and agr in staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. 9. Wayne, PA: CLSI document M07-A9. Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- De Jonge BLM, De Lencastre H, Tomasz A. Supression of autolysis and cell wall turnover in heterogeneous Tn551 mutants of a methicillin-resistant staphylococcus aureus strain. J Bacteriology. 1991;173:1105–1110. doi: 10.1128/jb.173.3.1105-1110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari RP, Arvidson S, Novick RP. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of staphylococcus 8325–4 traP::kan. Infect Immun. 2007;75:4534–4540. doi: 10.1128/IAI.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cheung GYC, Hu J, Wang D, Joo H, De Leo FR, Otto M. Comparative analysis of virulence and toxin expression of global community-associated MRSA strains. J Infect Dis. 2010;202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn OJ. Basic statistics: a primer for the biomedical sciences. New York: John Wiley & Sons; 1964. [Google Scholar]


