Summary
The insulin/IGF1 signaling pathways affect lifespan in several model organisms, including worms, flies and mice. To investigate whether common genetic variation in this pathway influences lifespan in humans, we genotyped 291 common variants in 30 genes encoding proteins in the insulin/IGF1 signaling pathway in a cohort of elderly Caucasian women selected from the Study of Osteoporotic Fractures (SOF), including 293 long-lived cases (lifespan ≥ 92 years (y), mean ± standard deviation (SD) = 95.3 ± 2.2y) and 603 average-lifespan controls (lifespan ≤ 79y, mean=75.7 ± 2.6y). Variants were selected for genotyping using a haplotype tagging approach. We found a modest excess of variants nominally associated with longevity. We then replicated nominally significant variants in two additional Caucasian cohorts containing both males and females: the Cardiovascular Health Study (CHS) and Ashkenazi Jewish Centenarians (AJC). An intronic single nucleotide polymorphism (SNP) in AKT1, rs3803304, was significantly associated with lifespan in a meta-analysis across the three cohorts (odds ratio (OR)=0.78 (95% confidence interval (CI)=0.68-0.89), adjusted p=0.043); two intronic SNPs in FOXO3A demonstrated a significant lifespan association among women only (rs1935949, OR=1.35, 95% CI=1.15-1.57, adjusted p=0.0093). Conclusion: common variants in several insulin/IGF1 pathway genes are associated with human lifespan.
Keywords: IGF1, longevity, gene, SNP, AKT1, FOXO3A
Introduction
Genes in the insulin/IGF1 signaling pathway affect lifespan in yeast, nematodes, fruit flies and mice (Kuningas et al., 2008). Mutation of genes in this signaling pathway confers greater resistance to oxidative stress and phenotypic characteristics consistent with delayed or slowed aging (Tatar et al., 2003).
The insulin/IGF1 pathway was first implicated in aging in C.elegans (Kenyon et al., 1993; Larsen et al., 1995; Morris et al., 1996). Mutation of daf-2, the nematode ortholog of the IGF1 and insulin receptors, results in extended lifespan and increased stress resistance (Kenyon et al., 1993; Kimura et al., 1997); these effects are mediated by the nuclear transcription factor, DAF-16, which is a homolog of the mammalian forkhead (FKHRL or FOXO) transcription factors (Ogg et al., 1997). Mutations in age-1, the homologue of mammalian PI3Kinase, which is involved in signal transduction from the insulin and IGF1 receptors (Dorman et al., 1995), also increase lifespan in C. elegans. Similarly, in Drosophila, mutations of the insulin receptor homolog, its substrate, chico (Clancy et al., 2001), and of the daf-16/foxo homolog (Hwangbo et al., 2004) reduce insulin signaling and extend lifespan.
In vertebrates, growth and glucose metabolism are regulated by two parallel pathways, with separate receptors for insulin and IGF1. In mice, mutations that result in growth hormone deficiency or mutations in growth hormone receptors cause reduced size, lower insulin levels, increased stress resistance and longer lifespan (Bartke, 2005). Heterozygous deletion of the IGF1R gene in mice causes a modest reduction in size, and improves stress resistance and extends lifespan in females. Disruption of the INSR gene globally or in most tissues leads to insulin resistance and shortened lifespan (Joshi et al., 1996; Okamoto & Accili, 2003); however, mice homozygous for INSR deletion specific to fat cells have extended lifespan in both sexes (Bluher et al., 2002), indicating the presence of tissue-specific effects.
Results from model organisms suggest the hypothesis that genes in the insulin/IGF1 pathway may be involved in the control of human lifespan and variants that affect their activity may be associated with human longevity. Although a few associations of insulin/IGF1 pathway genes with longevity have been reported (Bonafe et al., 2003; van Heemst et al., 2005; Kuningas et al., 2007; Suh et al., 2008; Willcox et al., 2008), most of these findings have not been replicated and the studies did not analyze genetic variation in the insulin/IGF1 pathway systematically.
We have comprehensively evaluated common variation in genes in the insulin/IGF1 signaling pathway for association with human lifespan. We performed a nested case-control association study of longevity in a cohort of elderly Caucasian women from the Study of Osteoporotic Fractures (SOF). We then replicated significant associations in two independent Caucasian cohorts of men and women.
Results
All subjects were Caucasian. The SOF cohort was all women, the CHS cohort was 47% female, and the AJC cohort was 66% female (Table 1). More than half (55.6%) of the longevity cases were still alive in SOF, about a third (33.7%) were still alive in CHS and 24% of cases were still alive in AJC.
Table 1.
Characteristics of the three study cohorts.
| Characteristic | SOF | CHS | AJC | |||
|---|---|---|---|---|---|---|
| cases | controls | cases | controls | cases | controls | |
| N | 293 | 603 | 279 | 797 | 383 | 363 |
| Age (years) | 95.3 ± 2.2 | 75.7 ± 2.6 | 94.5 ± 2.1 | 76.1 ± 3.1 | 99.8 (95-108) | 85.5 (43-94) |
| Sex (% male) | 0 | 0 | 47.0 | 52.6 | 25.2 | 42.7 |
| % Alive | 55.6 | 0 | 33.7 | 0 | 24 | 96 |
| % Atherosclerosis death | 17.7 | 30.2 | 16.8 | 26.1 | NA | NA |
| % Cancer death | 4.1 | 40.8 | 12.9 | 37.5 | NA | NA |
| % Other death | 22.5 | 29.0 | 36.6 | 39.1 | NA | NA |
SOF, Study of Osteoporotic Fractures, CHS, Cardiovascular Health Study, AJC, Ashkenazi Jewish Centenarians. Age is the age at death for deceased subjects, and age at last followup for subjects still alive. NA, not available
Association of genotype with longevity in SOF
We evaluated 291 SNPs in 30 genes in the insulin/IGF1 signaling pathway (Supplemental Table A). Twenty one (7%) SNPs in 9 (30%) genes (AKT1, FOXO1A, FOXO3A, GHR, GHRHR, IGF1R, IGFBP3, IGFBP4, PTEN) were associated with longevity at a nominal p<0.05, a modest excess over the expected ~15 (Table 2). Eight SNPs had a p value < 0.01 (an excess of ~5 over the expected number) and 3 SNPs had p<0.005 (~1.5 more than expected). FOXO3A (7/16) and GHR (7/26) had the most SNPs associated with longevity.
Table 2.
Association of variants in insulin/IGF1 pathway genes with longevity.
| Gene | # SNPs | # p<0.05 | # p<0.01 | # p<0.005 |
|---|---|---|---|---|
| AFX | 7 | 0 | 0 | 0 |
| AKT1 | 8 | 1 | 0 | 0 |
| AKT2 | 5 | 0 | 0 | 0 |
| AKT3 | 13 | 0 | 0 | 0 |
| FOXO1A | 7 | 1 | 0 | 0 |
| FOXO3A | 16 | 7 | 4 | 1 |
| GHR | 26 | 7 | 3 | 1 |
| GHRH | 1 | 0 | 0 | 0 |
| GHRHR | 13 | 1 | 0 | 0 |
| IGF1 | 6 | 0 | 0 | 0 |
| IGF1R | 34 | 1 | 1 | 1 |
| IGFBP1 | 11 | 0 | 0 | 0 |
| IGFBP2 | 8 | 0 | 0 | 0 |
| IGFBP3 | 5 | 1 | 0 | 0 |
| IGFBP4 | 7 | 1 | 0 | 0 |
| IGFBP5 | 11 | 0 | 0 | 0 |
| IGFBP6 | 4 | 0 | 0 | 0 |
| INS | 2 | 0 | 0 | 0 |
| INSR | 32 | 0 | 0 | 0 |
| INSRR | 4 | 0 | 0 | 0 |
| IRS1 | 6 | 0 | 0 | 0 |
| IRS2 | 13 | 0 | 0 | 0 |
| IRS4 | 1 | 0 | 0 | 0 |
| PDPK1 | 3 | 0 | 0 | 0 |
| PIK3CA | 5 | 0 | 0 | 0 |
| PIK3CB | 3 | 0 | 0 | 0 |
| PIK3R1 | 28 | 0 | 0 | 0 |
| PIK3R2 | 2 | 0 | 0 | 0 |
| PTEN | 9 | 1 | 0 | 0 |
| SHC1 | 1 | 0 | 0 | 0 |
| All genes, observed | 291 | 21 | 8 | 3 |
| All genes, expected | - | 15 | 2.9 | 1.5 |
Number of SNPs in each gene associated with longevity at a nominally significant level is shown. Results shown are for an additive model corrected for self-reported ethnicity. Total number of SNPs observed at 3 different significance thresholds is compared with the number expected.
For all genes except GHR, haplotypes containing longevity-associated SNPs exhibited a similar effect size and significance level for association with longevity, suggesting that in each case, the putative causative variant was in linkage disequilibrium (LD) with a single haplotype-tagging SNP (data not shown). For GHR, we observed the most significant association for a three-SNP haplotype comprising rs9292854, rs13182117 and rs4866941 (p=0.0005, compared to the best single-SNP association of p=0.007 for rs4866941; analysis not corrected for race/ethnicity), suggesting that the putative causative allele resides on this haplotype.
Replication of association of AKT1 and FOXO3A SNPs with longevity
Since the case-control associations we observed in SOF were not significant after correction for multiple hypothesis testing, we replicated the nominally associated SNPs in two independent Caucasian cohorts, the Cardiovascular Health Study (CHS) and Ashkenazi Jewish Centenarians (AJC). We selected all 21 SNPs individually associated with longevity in SOF (p<0.05) as well as 4 SNPs significantly associated with longevity (p<0.001) in 2-SNP interaction analyses for replication genotyping (Table 3). We observed the most consistent results across all 3 cohorts for 1 SNP in AKT1 (rs3803304), which was nominally associated with longevity in each of the 3 cohorts, with a consistent effect size (OR) of 0.77-0.79. In a meta-analysis of results from all three cohorts, this association was statistically significant (p=0.0004; permutation-based correction for multiple testing p = 0.043).
Table 3.
Meta-analysis of association with longevity for SNPs genotyped in SOF, CHS, and AJC.
| All (males + females) | SOF (n=896) | CHS (n=1076) | AJC (n=746) | Meta-analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Reason | Gene | p | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI |
| rs3803304 | longevity | AKT1 | 0.049 | 0.79 | 0.62-0.99 | 0.033 | 0.77 | 0.61-0.98 | 0.041 | 0.77 | 0.59-0.98 | 0.00040 | 0.78 | 0.68-0.89 |
| rs1334241 | longevity | FOXO1A | 0.032 | 1.30 | 1.02-1.65 | 0.17 | 0.84 | 0.65-1.08 | 0.48 | 0.90 | 0.67-1.20 | 0.87 | 1.01 | 0.87-1.17 |
| rs1044891 | longevity | FOXO3A | 0.0083 | 1.48 | 1.10-1.97 | 0.53 | 0.91 | 0.66-1.22 | NA | NA | NA | 0.14 | 1.17 | 0.95-1.45 |
| rs1935949 | longevity | FOXO3A | 0.043 | 1.25 | 1.00-1.56 | 0.23 | 1.14 | 0.92-1.41 | 0.016 | 1.36 | 1.05-1.74 | 0.0014 | 1.24 | 1.09-1.41 |
| rs2153960 | longevity | FOXO3A | 0.015 | 1.31 | 1.05-1.63 | 0.19 | 1.15 | 0.93-1.41 | 0.81 | 1.03 | 0.81-1.30 | 0.019 | 1.16 | 1.02-1.32 |
| rs3778588 | longevity | FOXO3A | 0.0047 | 1.51 | 1.13-1.99 | 0.84 | 0.97 | 0.73-1.29 | 0.19 | 1.24 | 0.89-1.72 | 0.023 | 1.22 | 1.03-1.45 |
| rs4946935 | longevity | FOXO3A | 0.041 | 1.26 | 1.00-1.57 | 0.11 | 1.19 | 0.96-1.47 | 0.029 | 1.33 | 1.03-1.72 | 0.00087 | 1.25 | 1.10-1.43 |
| rs9486902 | longevity | FOXO3A | 0.0050 | 1.47 | 1.12-1.92 | 0.93 | 0.99 | 0.74-1.31 | 0.99 | 1.00 | 0.66-1.50 | 0.073 | 1.18 | 0.98-1.40 |
| rs9486913 | longevity | FOXO3A | 0.0051 | 1.49 | 1.12-1.96 | 0.60 | 0.93 | 0.69-1.23 | 0.32 | 1.19 | 0.84-1.66 | 0.053 | 1.18 | 0.99-1.40 |
| rs12153009 | longevity | GHR | 0.017 | 0.76 | 0.60-0.95 | 0.55 | 0.94 | 0.75-1.16 | 0.47 | 0.91 | 0.71-1.16 | 0.033 | 0.87 | 0.76-0.99 |
| rs2940923 | longevity | GHR | 0.0094 | 1.31 | 1.06-1.61 | 0.44 | 1.08 | 0.88-1.31 | 0.023 | 1.28 | 1.03-1.59 | 0.0013 | 1.21 | 1.09-1.37 |
| rs2940935 | interaction | GHR | 0.61 | 1.07 | 0.82-1.38 | 0.011 | 1.37 | 1.07-1.73 | 0.046 | 1.30 | 1.00-1.67 | 0.0031 | 1.24 | 1.08-1.44 |
| rs3764451 | longevity | GHR | 0.0093 | 0.68 | 0.50-0.90 | 0.86 | 1.03 | 0.77-1.35 | 0.61 | 0.92 | 0.66-1.26 | 0.094 | 0.86 | 0.73-1.03 |
| rs4292454 | longevity | GHR | 0.043 | 1.25 | 1.00-1.54 | 0.49 | 0.93 | 0.76-1.13 | NA | NA | NA | 0.39 | 1.07 | 0.92-1.23 |
| rs4866941 | longevity | GHR | 0.0027 | 1.42 | 1.12-1.78 | 0.27 | 0.88 | 0.70-1.10 | NA | NA | NA | 0.19 | 1.11 | 0.95-1.31 |
| rs6883523 | longevity | GHR | 0.042 | 1.30 | 1.00-1.67 | 0.48 | 0.91 | 0.70-1.17 | 0.64 | 0.92 | 0.63-1.32 | 0.52 | 1.05 | 0.90-1.24 |
| rs6887528 | longevity | GHR | 0.012 | 1.72 | 1.12-2.62 | 0.38 | 0.83 | 0.54-1.26 | NA | NA | NA | 0.25 | 1.19 | 0.88-1.61 |
| rs2228078 | longevity | GHRHR | 0.048 | 2.68 | 1.00-7.14 | 0.96 | 1.02 | 0.46-2.21 | NA | NA | NA | 0.21 | 1.48 | 0.81-2.72 |
| rs2272037 | longevity | IGF1R | 0.0048 | 1.39 | 1.10-1.74 | 0.55 | 1.06 | 0.86-1.29 | NA | NA | NA | 0.021 | 1.19 | 1.03-1.39 |
| rs6670 | longevity | IGFBP3 | 0.047 | 1.28 | 1.00-1.63 | 0.36 | 0.89 | 0.70-1.13 | 0.63 | 0.94 | 0.74-1.19 | 0.75 | 1.02 | 0.89-1.18 |
| rs1009728 | interaction | IGFBP4 | 0.43 | 1.09 | 0.87-1.36 | 0.78 | 1.03 | 0.82-1.28 | 0.35 | 1.10 | 0.89-1.35 | 0.24 | 1.08 | 0.95-1.22 |
| rs7214466 | longevity | IGFBP4 | 0.012 | 0.75 | 0.59-0.93 | 0.68 | 1.04 | 0.84-1.28 | 0.37 | 0.91 | 0.73-1.12 | 0.094 | 0.90 | 0.79-1.02 |
| rs11575134 | interaction | IGFBP5 | 0.71 | 1.05 | 0.80-1.37 | 0.40 | 0.90 | 0.70-1.15 | 0.043 | 1.28 | 1.00-1.63 | 0.36 | 1.07 | 0.93-1.24 |
| rs3770472 | interaction | IGFBP5 | 0.91 | 1.01 | 0.78-1.31 | 0.23 | 0.87 | 0.68-1.09 | 0.042 | 1.29 | 1.00-1.63 | 0.60 | 1.04 | 0.90-1.20 |
| rs1022427 | longevity | PTEN | 0.033 | 1.65 | 1.04-2.61 | 0.99 | 1.00 | 0.63-1.58 | NA | NA | NA | 0.13 | 1.28 | 0.93-1.78 |
We show OR, 95% CI and p values for an additive model for each study and for the meta-analysis. SOF analyses are adjusted for self-reported ethnicity, CHS and AJC analyses are adjusted for sex. Nominal p-values are shown for all analyses. For replication, only AKT1 rs3803304 meets the permutation-based threshold for statistical significance in the entire cohort; FOXO3A rs1935949 and rs4946935 meet the threshold for women only (data not shown). Reason = reason for replication. NA = not available.
Two SNPs in FOXO3A were nominally associated with longevity in SOF and in AJC, but not in CHS, although the magnitude and direction of the effect were consistent in CHS. The p values in the meta-analysis were 0.0014 for rs1935949 and 0.0009 for rs4946935; these approached statistical significance in the meta-analysis after correction for multiple testing.
We observed a borderline effect of sex on the FOXO3A rs1935949 SNP association with longevity, suggesting a stronger effect of female sex (OR=0.72, 95% CI=0.52-0.99, p=0.049), as also seen when comparing the odds ratio for association with longevity between the mixed-sex (OR=1.24, 95%CI=1.09-1.41) and female-only subsets (OR=1.35, 95% CI=1.15-1.57). When restricted to female subjects, the association of FOXO3A rs1935949 and rs4946935 with longevity in the meta-analysis achieved statistical significance (adjusted P values were 0.0093 and 0.019 respectively).
Next, we evaluated the association with longevity for the replicated SNPs in a genotypic model (Table 4). For AKT1 rs3803304, the association with longevity appeared driven by the minor allele homozygous genotype (CC). Subjects homozygous for the minor allele were underrepresented among long-lived cases (OR=0.41-0.5, p=0.00016 in the meta-analysis). A single allele did not confer a significant effect (p=0.12), suggesting a recessive effect, with homozygosity for the minor allele decreasing the probability of longevity. For the two FOXO3A SNPs, the effect appeared to be consistent with a multiplicative model; the probability of surviving to exceptional longevity increased with each additional copy of the minor allele. Results were similar when evaluated in women only (rs1935949: CT, OR=1.33 (1.08-1.63), p=0.067; TT, OR=1.82 (1.25-2.65), p=0.0017).
Table 4.
Meta-analysis of association with longevity for top SNPs genotyped in SOF, CHS, and AJC.
| SNP | Geno type | SOF (n=896) | CHS (n=1076) | AJC (n=746) | Meta-analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | P | OR | 95% CI | N | P | OR | 95% CI | N | P | OR | 95% CI | OR | 95% CI | P | |||
| AKT1 rs3803304 G/C | GG | 465 | ref | - | - | 607 | - | - | - | 455 | - | - | - | - | - | ||
| GC | 309 | 0.53 | 0.90 | 0.66-1.23 | 345 | 0.25 | 0.84 | 0.62-1.13 | 249 | 0.36 | 0.86 | 0.63-1.18 | 0.87 | 0.73-1.04 | 0.12 | ||
| CC | 70 | 0.025 | 0.50 | 0.27-0.91 | 67 | 0.040 | 0.50 | 0.25-0.97 | 34 | 0.023 | 0.43 | 0.20-0.89 | 0.48 | 0.32-0.70 | 0.00016 | ||
| FOXO3A rs1935949 C/T | CC | 420 | ref | - | - | 531 | - | - | - | 429 | - | - | - | - | - | ||
| CT | 366 | 0.10 | 1.29 | 0.95-1.75 | 394 | 0.15 | 1.24 | 0.92-1.67 | 274 | 0.10 | 1.30 | 0.95-1.77 | 1.27 | 1.07-1.52 | 0.0064 | ||
| TT | 81 | 0.10 | 1.52 | 0.92-2.52 | 87 | 0.56 | 1.16 | 0.70-1.94 | 39 | 0.039 | 2.08 | 1.04-4.16 | 1.46 | 1.07-2.01 | 0.019 | ||
| FOXO3A rs4946935 G/A | GG | 415 | ref | - | - | 521 | - | - | - | 437 | - | - | - | - | - | ||
| GA | 358 | 0.16 | 1.25 | 0.92-1.69 | 403 | 0.050 | 1.34 | 1.00-1.80 | 211 | 0.090 | 1.34 | 0.96-1.87 | 1.31 | 1.09-1.56 | 0.0035 | ||
| AA | 78 | 0.066 | 1.61 | 0.97-2.67 | 84 | 0.47 | 1.21 | 0.72-2.04 | 39 | 0.10 | 1.78 | 0.90-3.52 | 1.48 | 1.07-2.03 | 0.018 | ||
OR and 95% CI from a genotypic model are shown in men and women combined, adjusted for sex.
Follow-up of significant findings in AKT1 and FOXO3A
Complete coverage (at r2>0.8) of common variation in both AKT1 and FOXO3A based on HapMap Caucasians (CEU) has been achieved.
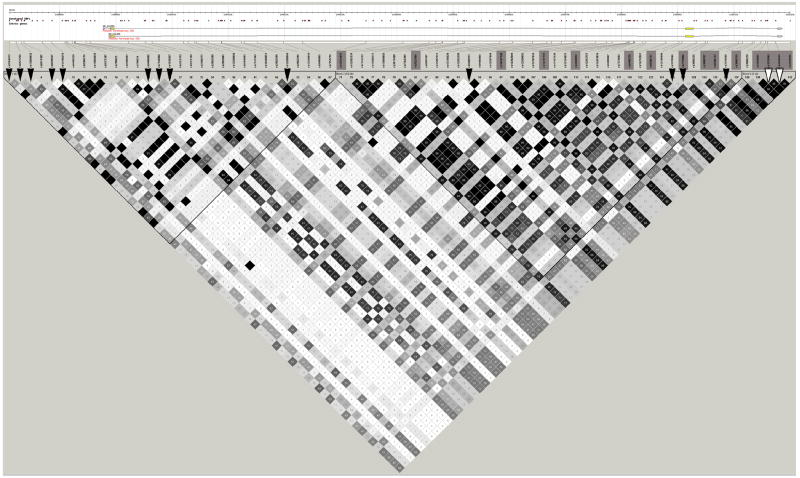
The longevity-associated AKT1 rs3803304 SNP is not in LD with any other HapMap SNPs. Therefore to follow-up we selected other variants in the surrounding area from dbSNP. We genotyped 8 additional AKT1 SNPs in SOF, including 5 coding SNPs, (1 rare, 4 unvalidated), 2 synonymous SNPs, and 1 intronic SNP located 22bp from an intron-exon junction. Three of these 8 SNPs were polymorphic in SOF and none were associated with longevity (Supplementary Table A). Since no coding SNPs in AKT1 were associated with longevity, we searched for potential regulatory sites in the region. AKT1 rs3803304 lies in an intron, 70bp away from a conserved exon-intron boundary and in an area of high predicted regulatory potential (0.3 score on the evolutionary and sequence pattern extraction through reduced representations (ESPERR) prediction metric (King et al., 2005). Data from the Affymetrix Transcriptome Phase 3 gene tiling arrays (Kapranov et al., 2007) show a long (>200bp) RNA signal at this intron/exon border encompassing rs3803304 (Figure 2A); a CpG island is also found here. It is possible that rs3803304 disrupts an RNA regulatory mechanism.
Figure 2. Genomic context and linkage disequilibrium for (A) AKT1 and (B, C) FOXO3A.
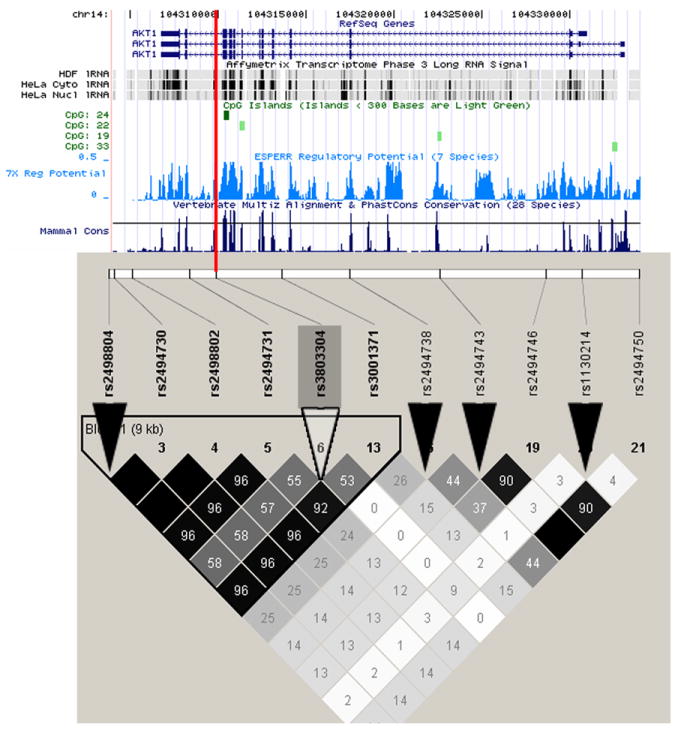
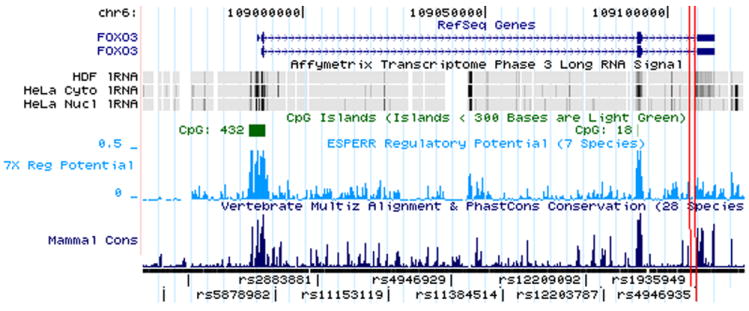
UCSC genome browser tracks show mRNA isoforms for both genes, CpG islands, ESPERR regulatory potential and conservation across 28 mammalian species (indicator line at 70%). Linkage disequilibrium plots below show r2 in HapMap Caucasians. All HapMap variants are tagged at r2>0.8. SNPs genotyped in SOF are marked by black triangles. Longevity-associated SNPs are marked by white triangles and red lines and other SNPs in LD with them are boxed.
Initial SNP discovery in FOXO3A was by sequencing – no other common exonic SNPs were found. The two SNPs most strongly associated with longevity, FOXO3A rs1935949 and rs4946935, lie in intron 3, 1.7 kilo-base-pairs (kb) from the intron-exon boundary of the 3’UTR, but not in an area of evolutionary conservation (Figure 2B and C).
Interaction analyses
We included 4 SNPs in the replication genotyping that were not individually associated with longevity in SOF, but had a significant association with longevity in interaction analyses with another SNP. Two IGFBP5 SNPs (rs3770472 and rs11575134) had significant interactions with GHR SNP rs2940935 in SOF (both P < 0.00004); these interactions appeared to replicate in the AJC cohort (p=0.012 for rs3770472 and 0.026 for rs11575134), but not in the CHS cohort. SNP rs1009728 was included in the replication genotyping because of a significant interaction with a SNP in PIK3R1, but this latter SNP could not be genotyped in the replication cohorts.
Association of lifespan-associated SNPs with cause-specific mortality
We evaluated associations of the longevity-associated SNPs with reported cause of death in SOF and CHS. AKT1 rs3803304 was associated with deaths due to cardiovascular disease and deaths from non-cardiovascular, non-cancer causes. The FOXO3A SNPs rs4946935 was associated with cardiovascular causes of death in SOF and with non-cardiovascular, non-cancer other causes of death in CHS. The FOXO3A SNP rs1935949 was associated with cardiovascular and cancer causes of death in SOF, and with other causes of death in CHS.
Discussion
We have comprehensively evaluated common variants in 30 genes in the insulin/IGF1 signaling pathway for their associations with human lifespan in a cohort of elderly Caucasian women. Replication of nominally significant findings in two independent Caucasian cohorts revealed a SNP in AKT1 which was significantly associated with lifespan, and 2 SNPs in FOXO3A which were significantly associated with lifespan in women. Overall, the longevity-associated SNPs were associated with the three major categories (cardiovascular, cancer, other) of cause of death, suggesting that their association with lifespan is due to a general effect rather than to a specific disease process.
Several previous studies have demonstrated associations between variants in other genes in the insulin/IGF1 signaling pathway and human aging phenotypes. Most of these studies investigated only a small number of SNPs or genes, and their results have not been replicated. Our current study analyzed 291 SNPs in 30 genes providing the first comprehensive screen of the insulin/IGF1 signaling pathway in humans.
Association of genetic variation in AKT1 with human lifespan has not been previously reported; however several studies have reported associations with FOXO3A SNPs or haplotypes. A recent case-control association study in elderly Japanese men found an association with 3 SNPs in FOXO3A and longevity (Willcox et al., 2008); this study did not report replication of findings in independent cohorts. While the tag SNPs genotyped were different in our study, 2 of the 3 associated SNPs share strong LD (r2>0.8 in HapMap Caucasians) with the FOXO3A SNPs associated with longevity in our study. The effect size is larger in the Japanese cohort, but, as in our study, homozygosity for the minor allele is associated with longevity. The most strongly associated SNP from the Japanese cohort lies in the first intron, and has lower LD in Caucasians (r2=0.48) than in Japanese (r2=0.71) with the best SNPs in our study, which are located in the last intron; this difference in LD patterns may account for the different localization of the top signal in our two studies.
The population-based Leiden 85-plus study (van Heemst et al., 2005) also found a FOXO3A haplotype, but no single SNPs, associated with an increase in mortality (Kuningas et al., 2007) in Caucasians. The FOXO3A haplotype identified contains some of the SNPs we found nominally associated with longevity in SOF, but resides in a different part of the gene from the two FOXO3A SNPs we found associated with longevity in our 3-cohort meta-analysis. Taken together, these results suggest that 5 independent cohorts may be detecting an association between FOXO3A variants and longevity. A combined analysis in which all cohorts genotype the same SNPs will be useful to confirm whether these findings are true replications of the same effect, and to identify the region of the gene with the strongest signal in order to prioritize fine mapping efforts.
Other insulin/IGF1-pathway genes implicated in human longevity include FOXO1A and IGF1R. A genome-wide association scan in the Framingham Study showed association between age at death with two FOXO1A SNPs (Lunetta et al., 2007), which are in complete LD with FOXO1A rs1334241, which we found to be nominally associated with longevity in SOF. However, this finding did not replicate in CHS or AJC. In the AJC cohort, rare nonsynonymous mutations of IGF1R were significantly more common in female centenarians (Suh et al., 2008). Female centenarians carrying those mutations had lower IGF1R levels and decreased IGF signaling compared with other female centenarians (Suh et al., 2008). In a study of long lived Italians, those carrying a common variant allele at the IGF1R locus had lower plasma IGF1 levels than those without the allele; this allele was over-represented in long-lived Caucasians (Bonafe et al., 2003). We genotyped this variant but did not replicate the association with lifespan in the SOF cohort.
The association of lifespan with FOXO3A variants in several studies and the novel association we identified with a SNP in AKT1 strongly suggest that the same components of the insulin/IGF1 signaling pathway that affect lifespan in C. elegans, Drosophila and mice are also involved in determining lifespan in humans. FOXO3A is one of the three mammalian homologues of the C. elegans transcription factor DAF-16, which is required for the lifespan effects of the insulin/IGF1 signaling pathway. AKT1 is one of the tyrosine kinases in the signaling cascade that links IGF1 binding to its receptor to transcriptional activation and downstream effects on cell metabolism and oxidative stress response; AKT1 phosphorylates FOXO3A and prevents it from entering the nucleus thus inhibiting transcriptional activation (Brunet et al., 1999; Zheng et al., 2000). Thus the two genes most strongly associated with longevity in our results are also functionally related.
How the variants we identified affect lifespan remains uncertain. All three longevity-associated SNPs that replicated across 3 independent cohorts are intronic SNPs of no known function and they do not reside in motifs conserved through evolution. In the case of FOXO3A, the two longevity-associated SNPs are in linkage disequilibrium with other variants across an area of approximately 67kb. Since the linkage disequilibrium is so strong in this area, it may not be possible to discern which is causative from association studies in Caucasians alone; association studies in other populations, locus re-sequencing and functional studies are needed.
The AKT1 SNP associated with lifespan is not in high LD with any other HapMap variants; evaluation of 8 unlinked variants in the nearby area, including 5 amino-acid coding polymorphisms, did not reveal any further associations with lifespan. Since AKT1 rs3803304 resides 70 base-pairs (bp) away from an intron-exon boundary, and in an area of high predicted regulatory potential (King et al., 2005), which potentially encodes a non-coding regulatory RNA (Kapranov et al., 2007), it may be a candidate SNP for influencing RNA splicing or other regulatory mechanisms. In vitro functional studies will be needed to explore such potential mechanisms.
Our study has several potential limitations. First, although we adjusted for self-reported ethnicity in the SOF study, population stratification may have affected our results. In addition, we did not have self-reported ethnicity information in CHS. Second, subtle effects on longevity might have been missed because of our relatively small sample size (896 participants in SOF); we had adequate (80%) power to detect an odds ratio of ~1.7. As with other complex traits (Easton et al., 2007; Frayling, 2007; Harley et al., 2008), much larger sample sizes will be required to identify modest risk modifiers. Third, like most studies of longevity in humans, our study may be influenced by birth cohort effects, since controls were generally born later (e.g., in the 1910’s-20’s) than the cases (in the 1890’s-1900’s). Finally, we only tested common SNPs and small insertion-deletions. We did not genotype copy number variants or large insertion-deletions and, with the exception of a few amino-acid coding variants, we only evaluated the effects of common variants (minor allele frequency (MAF)>0.05). The relative dearth of common SNPs associated with longevity in the genes we analyzed does not exclude the existence of other genetic effects in the pathway. To obtain a complete picture of the association of genetic variation in the insulin/IGF1 pathway with phenotypes, discovery of rare variants by resequencing of the same set of candidate genes will be necessary.
Despite these limitations, replication among three independent Caucasian samples, including a founder population (AJC), strengthens our findings. It is unlikely that population stratification could lead to false-positive results that would be consistent across a European American/Caucasian American population and the Ashkenazi population, since the patterns of stratification would be different. Any birth cohort effects should be relatively modest, since the differences in birth-year were small, and since both groups were born before there were substantial changes in early-life mortality rates due to advances in medical care.
In summary, we screened the insulin/IGF1 signaling pathway for association with human longevity and identified common variants in 2 genes, AKT1 and FOXO3A, that are consistently associated with longer lifespan in 3 independent Caucasian cohorts. Our results extend the remarkably consistent effect of this pathway on lifespan in model organisms to human populations. Although the effects we observed are subtle, they implicate the insulin/IGF1 signaling pathway in human aging, confirming that studies in model organisms can help us to gain insights into the biology of human aging.
Experimental Procedures
Samples
Data from 3 studies were analyzed. Two are large cohort studies of older adults: SOF and CHS. The third study is a case-control study (AJC).
Study of Osteoporotic Fractures (SOF)
A total of 9704 ambulatory community-dwelling Caucasian women age 65 and older were recruited from September 1986 through October 1988 from 4 areas in the United States: Baltimore, MD: Monongahela Valley, PA; Minneapolis, MN; and Portland, OR (Cummings et al., 1990). The study protocol was approved by the appropriate institutional review committees and written informed consent was obtained from all participants. Self-reported ethnicity was recorded as Northern, Southern, or Central European, or mixed/other. The cohort has been followed for over 20 years with age and cause of death recorded for participants who have died. 896 SOF participants were selected for the present study. These included 293 long-lived cases (lifespan ≥ 92y, mean ± SD = 95.3 ± 2.2y) of whom approximately 56% were still living at the last study visit. We selected 603 controls who were the youngest to die in this cohort (lifespan ≤ 79y, mean=75.7 ± 2.6y); however, the age of death in the controls is not substantially different compared to the general population, since all women had to be relatively healthy to enter the cohort (Cummings et al., 1990). Cause of death was adjudicated by an experienced internist based on review of death certificates and hospital records.
Cardiovascular Health Study (CHS)
The original CHS cohort includes 5201 Caucasian and African-American participants recruited from 1989 to 1990 in 4 communities in the United States (Fried et al., 1991; Tell et al., 1993) and 684 additional African-American participants recruited from 1992 to 1993. All participants were 65 years of age or older. At enrollment, participants were not institutionalized, wheelchair-bound at home, or receiving treatment for cancer. Informed consent was obtained from all participants. Deaths were ascertained through surveillance and at semiannual contacts. For all deaths, the CHS collected information from medical records, physician interviews, autopsy reports, coroner reports, and next of kin interviews. Data were reviewed by the CHS Events Committee and cause of death classified according to pre-specified criteria (Ives et al., 1995). From the 4383 Caucasian CHS participants, we defined a case-control replication sample of 1076 for the longevity analysis using the same age cutoffs as in the SOF study. This subset included 279 long-lived cases (lifespan ≥ 92y, mean = 94.5 ± 2.1 years (y)) and 797 average-lifespan controls (lifespan ≤ 79y, mean=76.1 ± 3.1 y).
Ashkenazi Jewish Centenarians (AJC)
Subjects were of self-reported Ashkenazi Jewish ancestry (Barzilai et al., 2003; Atzmon et al., 2006; Suh et al., 2008). Informed written consent was obtained in accordance with the policy of the Committee on Clinical Investigations of the Albert Einstein College of Medicine. The 383 cases (74.8% female) were required to be living independently at 95 years of age as a reflection of good health (mean age 97.7y, range 95–108y). The 363 controls (mean age 79.5 years, age range 43-94y, 57% female), all living at time of recruitment, were of self-reported Ashkenazi Jewish descent without a family history of unusual longevity (parents’ age <85y). Participants’ ages were defined by birth certificates or passports. 4% of controls and 76% of centenarians have died since recruitment.
Gene and Variant Selection
Variants were selected for genotyping in 30 genes encoding the key proteins in the human insulin/IGF1 signaling pathway, including ligands (IGF1, insulin, GH1, GHRH), binding proteins (IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP6), receptors (IGF1R, INSR, INSRR, GHR, GHRHR), signal transduction proteins (AKT1, AKT2, AKT3, IRS1, IRS2, IRS4, PDPK1, PIK3CA, PIK3CB, PIK3R1, PIK3R2, PTEN, SHC1) and transcription factors (AFX, FOXO1A, FOXO3A). GH1 was included initially, but all 3 SNPs available in dbSNP failed genotyping attempts. Initially, variants in FOXO1A and FOXO3A were identified by sequencing exons, intron-exon boundaries and the promoter in 80 Caucasians and selecting haplotype-tagging variants. Subsequently, we utilized the Human Haplotype Consortium data from CEU Caucasians (2005; Frazer et al., 2007) (www.hapmap.org) to select haplotype-tagging SNPs. Briefly, tag SNPs were picked for the gene ± approximately 10 kb genomic flanking sequence using the Tagger algorithm (de Bakker et al., 2005) implemented in the Haploview program (Barrett et al., 2005) (www.broad.mit.edu/mpg/haploview/download.php). The entire genomic sequence was tagged for all genes except IGF1R and INSR, where most of the large intron 1 was excluded. We set the r2 cutoff at 0.8 and minor allele frequency cutoff at 0.05 (except for amino acid coding variants, which were all included). Pair-wise tag selection was used for all genes except IGFBP2, IGFBP5, IRS2 and PIK3R1, where 2-marker haplotypes were used to reduce the number of tag SNPs. Amino-acid coding variants were prioritized and previously published candidate variants were included (Bonafe et al., 2003). Eight additional SNPs in AKT1 were selected for follow up. We selected 339 SNPs overall; 293 SNPs were successfully genotyped.
To analyze the genomic regions surrounding FOXO3A and AKT1, we used the UCSC Genome Browser (www.genome.ucsc.edu) including data from several analysis tracks (ESPRR regulatory potential (King et al., 2005), Phastcons conservation, CpG islands, Affymetrix Transcriptome Phase 3 (Kapranov et al., 2007)) on the March 2006 assembly of the human genome.
Genotyping
SNPs were genotyped in the SOF and CHS cohorts using two genotyping platforms: template directed primer extension with fluorescence polarization detection (FP-TDI, AcycloPrime II detection kit, Perkin Elmer, Boston, MA) (Hsu & Kwok, 2003) and SNPstream 48plex (Beckman Coulter, Fullerton, CA). Oligonucleotide primers were designed using www.autoprimer.com, primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and as previously described (Hsu & Kwok, 2003). Assay conditions are available upon request. Genotyping in AJC was performed at the Albert Einstein College of Medicine using the Sequenom platform and pyrosequencing.
Samples were genotyped by investigators blinded to phenotype. All data was scored by at least 2 investigators. DNA samples were genotyped on 384-well plates with negative and positive controls (duplicates) included in each plate. There were 19 duplicates among the SOF and 267 among the CHS samples. Plates with fewer than 90% successful genotypes or with duplicate discrepancies were repeated. Plates where duplicate discrepancies persisted after re-genotyping were excluded. Overall genotyping accuracy was >99.5%.
SNP genotype distributions in SOF controls were tested for consistency with Hardy Weinberg equilibrium using a γ2 test (Bonferroni-corrected significance threshold p=1.7E-04). 2/293 SNPs: IRS4 rs1801162 and IGFBP2 rs9341156, were not in Hardy-Weinberg equilibrium and were removed from further analysis.
Statistical Analyses
Association of SNP genotype with longevity
For all 291 SNPs analyzed, minor allele homozygotes were coded 2, heterozygotes were coded 1, and major allele homozygotes were coded 0. Logistic regression was used to evaluate the association between genotype and longevity case/control status. We assumed an additive model as our baseline model; significant associations were also tested with a genotypic model. In SOF, the logistic regression model was adjusted for self-reported ethnicity. In CHS and AJC, models were adjusted for sex; we also analyzed data separately for women in these two cohorts. Association of significant genotypes with the three main categories of cause of death (cardiovascular disease, cancer, or other) was evaluated in the 2 cohorts (SOF and CHS) that had this information. OR and 95% CI are reported. We also performed haplotype case-control association analyses using 2- and 3-SNP sliding window and haplotype analysis across entire LD-blocks using the Haploview program (Barrett et al., 2005).
Replication and meta-analysis
The logistic regression results from all 3 cohorts (or from 2 cohorts only in some cases) were combined using inverse variance weighting. To evaluate statistical significance in the replication we corrected for all of the SNPs we replicated and used only the data from the 2 replication cohorts (AJC+CHS) to assess significance. We determined statistical significance by permutation. Case/control status was permuted randomly for all participants for the 25 SNPs genotyped in the replication stage. In each permutation, we recorded the maximum absolute value of the Z scores. We performed 2000 permutations. We then compared the Z scores from the observed data with the distribution of the maximum Z scores from the permutations. The percentile of the observed absolute z score among the permuted z scores was calculated as the adjusted P value for determining statistical significance corrected for multiple testing. Statistical significance in the replication was defined as an adjusted p<0.05 in the AJC+CHS replication cohorts.
Supplementary Material
Figure 1. Graphic of insulin/IGF1 signaling pathway showing proteins encoded by scanned genes.
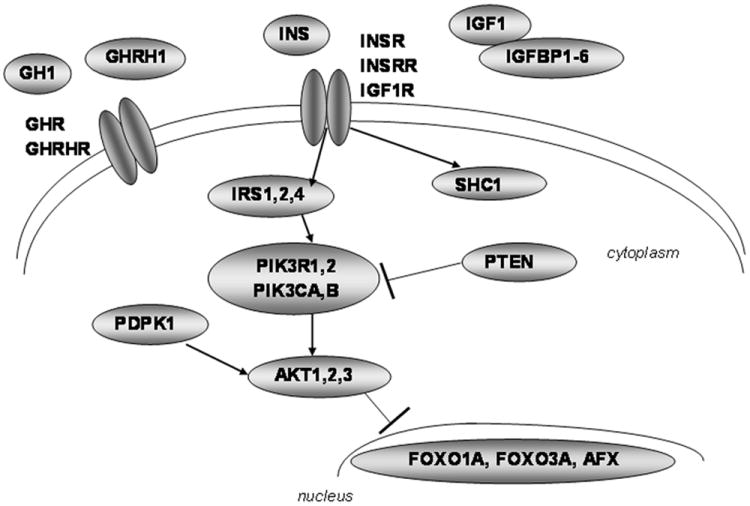
Table 5.
Association of genotype with cause of death in SOF and CHS.
| Cause of death | Cohort | Significance | AKT1 rs3803304 | FOXO3A rs1935949 | FOXO3A rs4946935 |
|---|---|---|---|---|---|
| Atherosclerosis | SOF | P | 0.069 | 0.013 | 0.017 |
| OR (95% CI) | 1.33 (0.98-1.81) | 0.68 (0.51-0.92) | 0.69 (0.51-0.94) | ||
| CHS | P | 0.040 | 0.49 | 0.73 | |
| OR (95% CI) | 1.38 (1.02-1.89) | 1.10 (0.84-1.45) | 1.05 (0.79-1.40) | ||
| Meta-analysis | P | 0.0062 | 0.25 | 0.17 | |
| OR (95% CI) | 1.36 (1.09-1.69) | 0.89 (0.73-1.09) | 0.87 (0.70-1.07) | ||
| Cancer | SOF | P | 0.087 | 0.045 | 0.12 |
| OR (95% CI) | 1.28 (0.97-1.67) | 0.76 (0.58-0.99) | 0.81 (0.62-1.06) | ||
| CHS | P | 0.37 | 0.38 | 0.16 | |
| OR (95% CI) | 1.14 (0.85-1.53) | 0.89 (0.70-1.15) | 0.83 (0.64-1.08) | ||
| Meta-analysis | P | 0.064 | 0.044 | 0.037 | |
| OR (95% CI) | 1.21 (0.99-1.48) | 0.83 (0.69-0.99) | 0.82 (0.68-0.99) | ||
| Other | SOF | P | 0.10 | 0.32 | 0.48 |
| OR (95% CI) | 1.30 (0.95-1.78) | 0.86 (0.64-1.16) | 0.90 (0.66-1.21) | ||
| CHS | P | 0.036 | 0.035 | 0.048 | |
| OR (95% CI) | 1.34 (1.02-1.75) | 0.76 (0.58-0.98) | 0.76 (0.58-1.00) | ||
| Meta-analysis | P | 0.0079 | 0.025 | 0.051 | |
| OR (95% CI) | 1.32 (1.08-1.62) | 0.80 (0.66-0.97) | 0.82 (0.67-1.00) |
For this analysis, the case group comprises participants who died of specific causes and the control group comprises long-lived participants.
Results for an additive model are shown. Analyses in SOF are adjusted for self-reported ethnicity; analyses in CHS are adjusted for sex.
Acknowledgments
We acknowledge D. Lind for coordination of pilot studies, J. Atilano, S.-W. Chan, E. Lovins and T.J. Nguyen for technical assistance and the staff of SOF and CHS Coordinating Centers for study support. This work was supported by the Longevity Consortium, www.longevityconsortium.org (National Institute on Aging (NIA) 5U19AG023122 to S.R.C. and W.S.B.). The Study of Osteoporotic Fractures (SOF) is supported by the NIA (AG05407, AR35582, AG05394, AR35584, AR35583, R01 AG005407, R01 AG027576-22, 2 R01 AG005394-22A1, and 2 R01 AG027574-22A1). The Cardiovascular Health Study is supported by the National Heart, Lung, and Blood Institute (N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, U01 HL080295) with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
Footnotes
Author Contributions
Ludmila Pawlikowska: Study design, acquisition of data, interpretation of data, preparation of manuscript
Donglei Hu: Analysis and interpretation of data, preparation of manuscript
Scott Huntsman: Analysis and interpretation of data
Andrew Sung: Acquisition of data, preparation of manuscript
Catherine Chu: Acquisition of data, preparation of manuscript
Justin Chen: Acquisition of data, preparation of manuscript
Alex Joyner: Interpretation of data, preparation of manuscript
Nicholas J. Schork: Interpretation of data, preparation of manuscript
Wen-Chi Hsueh: Interpretation of data, preparation of manuscript
Alexander P. Reiner: Interpretation of data, preparation of manuscript
Bruce M. Psaty: Interpretation of data, preparation of manuscript
Gil Atzmon: Interpretation of data, preparation of manuscript
Nir Barzilai: Interpretation of data, preparation of manuscript
Steven R. Cummings: Study design, interpretation of data, preparation of manuscript
Warren S. Browner: Study design, interpretation of data, preparation of manuscript
Pui-Yan Kwok: Interpretation of data, preparation of manuscript
Elad Ziv: Study design, analysis and interpretation of data, preparation of manuscript
Contributor Information
Ludmila Pawlikowska, Email: pawlikowskal@anesthesia.ucsf.edu.
Donglei Hu, Email: dhu@medicine.ucsf.edu.
Scott Huntsman, Email: shuntsman@medicine.ucsf.edu.
Andrew Sung, Email: ajsung@gmail.com.
Catherine Chu, Email: ChuCatherine@derm.ucsf.edu.
Justin Chen, Email: Justin.Chen@ucsf.edu.
Alex Joyner, Email: ajoyner@ucsd.edu.
Nicholas J. Schork, Email: nschork@scripps.edu.
Wen-Chi Hsueh, Email: hsueh@medicine.ucsf.edu.
Alexander P. Reiner, Email: apreiner@u.washington.edu.
Bruce M. Psaty, Email: psaty@u.washington.edu.
Gil Atzmon, Email: gatzmon@aecom.yu.edu.
Nir Barzilai, Email: barzilai@aecom.yu.edu.
Steven R. Cummings, Email: scummings@sfcc-cpmc.net.
Warren S. Browner, Email: warren@cpmcri.org.
Pui-Yan Kwok, Email: Pui.Kwok@ucsf.edu.
References
- Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 2006;4:e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bartke A. Minireview: role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology. 2005;146:3718–3723. doi: 10.1210/en.2005-0411. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. Unique lipoprotein phenotype and genotype associated with exceptional longevity. Jama. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- Bonafe M, Barbieri M, Marchegiani F, Olivieri F, Ragno E, Giampieri C, Mugianesi E, Centurelli M, Franceschi C, Paolisso G. Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J Clin Endocrinol Metab. 2003;88:3299–3304. doi: 10.1210/jc.2002-021810. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Cummings SR, Black DM, Nevitt MC, Browner WS, Cauley JA, Genant HK, Mascioli SR, Scott JC, Seeley DG, Steiger P, et al. Appendicular bone density and age predict hip fracture in women The Study of Osteoporotic Fractures Research Group. Jama. 1990;263:665–668. [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schurmann P, Dork T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TM, Kwok PY. Homogeneous primer extension assay with fluorescence polarization detection. Methods Mol Biol. 2003;212:177–187. doi: 10.1385/1-59259-327-5:177. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- IHMC. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives DG, Fitzpatrick AL, Bild DE, Psaty BM, Kuller LH, Crowley PM, Cruise RG, Theroux S. Surveillance and ascertainment of cardiovascular events. The Cardiovascular Health Study. Ann Epidemiol. 1995;5:278–285. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- Joshi RL, Lamothe B, Cordonnier N, Mesbah K, Monthioux E, Jami J, Bucchini D. Targeted disruption of the insulin receptor gene in the mouse results in neonatal lethality. Embo J. 1996;15:1542–1547. [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuningas M, Magi R, Westendorp RG, Slagboom PE, Remm M, van Heemst D. Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur J Hum Genet. 2007;15:294–301. doi: 10.1038/sj.ejhg.5201766. [DOI] [PubMed] [Google Scholar]
- Kuningas M, Mooijaart SP, van Heemst D, Zwaan BJ, Slagboom PE, Westendorp RG. Genes encoding longevity: from model organisms to humans. Aging Cell. 2008;7:270–280. doi: 10.1111/j.1474-9726.2008.00366.x. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, D’Agostino RB, Sr, Karasik D, Benjamin EJ, Guo CY, Govindaraju R, Kiel DP, Kelly-Hayes M, Massaro JM, Pencina MJ, Seshadri S, Murabito JM. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S13. doi: 10.1186/1471-2350-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Accili D. In vivo mutagenesis of the insulin receptor. J Biol Chem. 2003;278:28359–28362. doi: 10.1074/jbc.R300009200. [DOI] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Tell GS, Fried LP, Hermanson B, Manolio TA, Newman AB, Borhani NO. Recruitment of adults 65 years and older as participants in the Cardiovascular Health Study. Ann Epidemiol. 1993;3:358–366. doi: 10.1016/1047-2797(93)90062-9. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng WH, Kar S, Quirion R. Insulin-like growth factor-1-induced phosphorylation of the forkhead family transcription factor FKHRL1 is mediated by Akt kinase in PC12 cells. J Biol Chem. 2000;275:39152–39158. doi: 10.1074/jbc.M002417200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.