Abstract
Canine distemper virus (CDV; Paramyxoviridae, Morbillivirus) is the etiologic agent of a multisystemic infectious disease affecting all terrestrial carnivore families with high incidence and mortality in domestic dogs. Sequence analysis of the hemagglutinin (H) gene has been widely employed to characterize field strains, permitting the identification of nine CDV lineages worldwide. Recently, it has been established that the sequences of the fusion protein signal-peptide (Fsp) coding region are extremely variable, suggesting that analysis of its sequence might be useful for strain characterization studies. However, the divergence of Fsp sequences among worldwide strains and its phylogenetic resolution has not yet been evaluated. We constructed datasets containing the Fsp-coding region and H gene sequences of the same strains belonging to eight CDV lineages. Both datasets were used to evaluate their phylogenetic resolution. The phylogenetic analysis revealed that both datasets clustered the same strains into eight different branches, corresponding to CDV lineages. The inter-lineage amino acid divergence was fourfold greater for the Fsp peptide than for the H protein. The likelihood mapping revealed that both datasets display strong phylogenetic signals in the region of well-resolved topologies. These features indicate that Fsp-coding region sequence analysis is suitable for evolutionary studies as it allows for straightforward identification of CDV lineages.
Introduction
Canine distemper virus (CDV) is a member of the Morbillivirus genus within the Paramyxoviridae family. The viral genome consists of a nonsegmented single-stranded negative RNA of 15.7 kb and encodes six structural proteins. The nucleocapsid protein, viral polymerase, and phosphoprotein are associated with the genomic RNA forming the ribonucleoprotein complex, which performs viral transcription and replication. The hemagglutinin (H) and fusion (F) proteins are the antigenic determinants of the virus and are located at the viral envelope, whereas the matrix protein is membrane associated [1].
CDV is the etiologic agent of canine distemper (CD), a severe multisystemic infectious disease, which is distributed worldwide and affects all terrestrial carnivores [2]–[4]. CD has been controlled by attenuated vaccines; however, in recent decades, several outbreaks in properly vaccinated dogs and an expansion in the host range have been reported [5]–[11]. These outbreaks might be a consequence of the emergence of new field strains able to avoid the immune response generated by the “old strains” currently used in the vaccines and/or because of the capacity of new field strains to infect other carnivore hosts [12]–[20].
The H gene shows the greatest genetic variation within the Morbillivirus genus, and it has been widely employed to characterize field strains [21]. In CDV, the amino acid divergence of the H protein reaches 10% among field strains [12], [22]. This variability in conjunction with a phylogenetic analysis provide the necessary information for the classification of CDV strains into genetic lineages: two strains belong to the same lineage if they cluster together and share an amino acid divergence of <3.5%; the strains belong to different lineages if they appear in separate clades and show values of divergence >4% [12], [22]. Currently, nine lineages have been characterized worldwide according mainly to their geographic origin [9], [22]–[25]. In South America there are two co-circulating lineages, the South America 2 (SA2) lineage exclusive to South America and the Europe 1/South America 1 (EU1/SA1) lineage, which is spread in Europe and South America [25].
The H gene can be more difficult to amplify directly from field samples because of its larger size (1824 bp) [17], [23]–[25], and its transcription level is proportionately lower in comparison to the genes located in the 3′ terminal region of the genome [1].
It has been recently established that a short region of the F gene that encodes the signal peptide of the F protein (Fsp; residues 1–135) is extremely variable. Comparative analysis of the Fsp peptide from Asian strains shows that it has substantial genetic variability, suggesting that this region could be a marker for classifying CDV strains [26], [27].
The aim of this work was to evaluate the phylogenetic resolution of the Fsp-coding region in comparison with the H gene. The analyses revealed that the Fsp-coding region is phylogenetically informative and quite useful for the identification of genetic CDV lineages, which will allow for rapid characterization of circulating strains.
Materials and Methods
Datasets
To analyze the phylogenetic resolution of the Fsp-coding region and compare it with that of the H gene, we constructed datasets for both genomic regions from the same 37 strains (Table 1). The H dataset included strains available at GenBank for eight of the nine CDV lineages; sequences from the African lineage strains were not included because there are no records of the Fsp-coding region sequences in these strains. The Fsp dataset included 29 sequences available at GenBank and eight sequences obtained here from the EU1/SA1 and SA2 lineages (Table 1).
Table 1. Strains employed for the construction of the Fsp and H datasets.
Denomination, GenBank accession number for the Fsp-coding region and H gene, and lineage for each strain are detailed. AS1, Asia 1; AS2, Asia 2; EU1/SA1, Europe 1/South America 1; EU2, Europe 2; EU3, Europe 3; NA1, North America 1; NA2, North America 2; SA2, South America 2.
Amplification of the Fsp-coding Region
Urine, ocular discharge, and clotted blood samples from dogs were subjected to Trizol RNA isolation (Invitrogen) to isolate CDV RNA (Table 1). The RT-PCR products of 681 bp encompassing the Fsp-coding region (405 bp) were amplified using the SuperScript One-Step RT-PCR kit reagents (Invitrogen) (the cycling conditions are available from the authors on request) through a set of newly designed primers, F4854∶5′-TCCAGGACATAGCAAGCCAACA-3′and R5535∶5′-GGTTGATTGGTTCGAGGACTGAA-3′ (strain N° AF378705 position numbering). The PCR products were sequenced bidirectionally on an ABI3130 automated sequencer (Applied Biosystems), and nucleotide sequences were submitted to the GenBank database (http://www.ncbi.nlm.nih.gov).
Comparative Analyses
To evaluate the phylogenetic resolution of the Fsp dataset and compare it with the H dataset, two bioinformatic approaches were employed: phylogenetic and likelihood mapping (LM) analyses.
Phylogenetic Analysis
The phylogenetic relationships among the strains were established for both datasets by maximum-likelihood trees using MEGA 5 software [28]. The substitution model used was Hasegawa-Kishino-Yano with gamma distribution (G) for both datasets.
Internal-node uncertainties were assessed using 500 bootstrap replications. Amino acid p-distance analysis was implemented for both datasets with MEGA 5 software [28].
LM Analysis
The LM method was implemented for both datasets using TREE-PUZZLE software [29]. LM assesses if a dataset is suitable for phylogenetic reconstruction by the analysis of groups of four randomly chosen sequences (quartets). For each quartet, three unrooted tree topologies are possible. The phylogenetic signals are computed as probabilities that are represented in a triangle surface [30].
Results
Amplification of the Fsp-coding Region
The 681-bp amplicon that encompasses the 405-bp Fsp-coding region was obtained directly from samples of different origin. The sequences of the eight strains from South American lineages (EU1/SA1 and SA2) were added to the Fsp dataset. The Fsp and H datasets each comprised 37 sequences of strains from eight of the nine CDV lineages characterized (Table 1) and were used to perform the subsequent analyses.
Comparative Analyses: Phylogenetic and LM Analysis
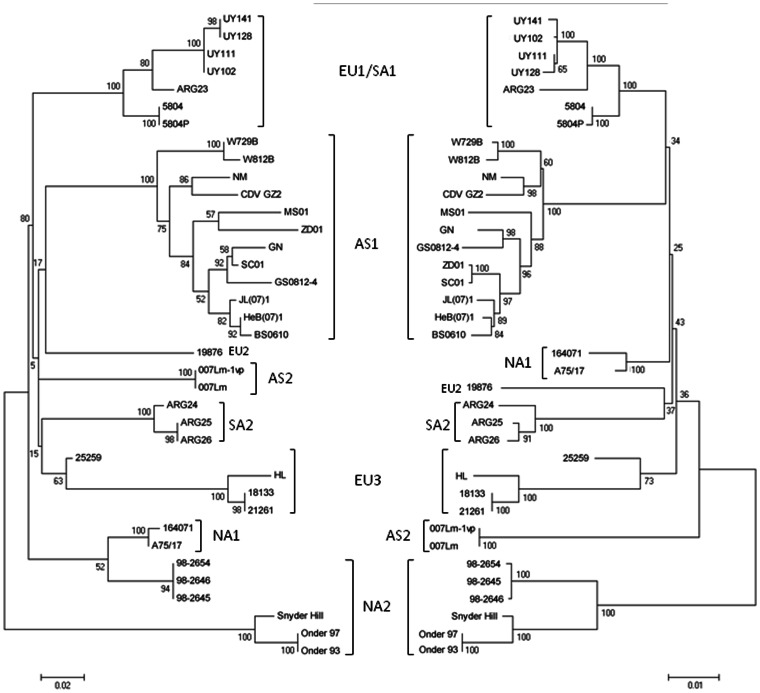
Phylogenetic analysis of the Fsp and H datasets was performed to compare the relationships among the CDV strains. The Fsp and H trees clustered the same strains corresponding to eight CDV lineages with similar bootstrap values. The branch lengths were measured according to the number of substitutions per site for the substitution model (0.01 for both datasets) (Figure 1). In both trees, similar relationships were observed among most lineages. The positions of the North American lineages (NA1 and NA2) were quite different between the trees: the NA1 and NA2 lineages clustered in the same clade in the Fsp tree, whereas these lineages were not related in the H tree (Figure 1). Inter-lineage amino acid divergence (p-distance) for the Fsp dataset ranged from 19.6% to 36.9%, whereas for the H dataset it ranged from 3.4% to 9.9% (Table 2). The relationships among strains within each CDV lineage were slightly different for the two trees. Intra-lineage amino acid divergence was from 1.8% to 20.7% for the Fsp dataset and from 1.3% to 6.3% for the H dataset (Table 2). The EU2 lineage was not considered because it was represented by a single strain.
Figure 1. Phylogenetic analysis of CDV isolates.
Thirty-seven nucleotide sequences of the Fsp (left) and H (right) datasets were used. Maximum likelihood trees were constructed using the Hasegawa-Kishino-Yano (G) substitution model for both datasets and were inferred through 500 replicates. Branch lengths are measured in the number of substitutions per site, as shown by the scale bars. Unrooted trees were depicted facing each other for comparison. AS1, Asia 1; AS2, Asia 2; EU1/SA1, Europe 1/South America 1; EU2, Europe 2; EU3, Europe 3; NA1, North America 1; NA2, North America 2; SA2, South America 2; Onder, Onderstepoort strain; Snyder, Snyder-Hill strain.
Table 2. Inter- and intra-lineage amino acid divergences (p-distances).
| EU1/SA1 | 11.9 | ||||||||
| 3.3 | |||||||||
| SA2 | 23.5 | 1.8 | |||||||
| 4.7 | 2.2 | ||||||||
| EU2 | 25 | 25.8 | – | ||||||
| 4.8 | 5.7 | – | |||||||
| EU3 | 29.5 | 31.1 | 30.5 | 18.5 | |||||
| 5.9 | 6.8 | 6.2 | 4.6 | ||||||
| NA1 | 19.6 | 22.6 | 23.2 | 27.3 | – | ||||
| 3.4 | 4.6 | 4.2 | 5 | 1.3 | |||||
| NA2 | 33.6 | 33.8 | 31.1 | 31.9 | 23.6 | 20.7 | |||
| 8.6 | 9.8 | 9.2 | 9.6 | 8.4 | 6.3 | ||||
| AS1 | 26.6 | 27.3 | 32.5 | 36.9 | 25.6 | 27.8 | 18.5 | ||
| 5.5 | 6.9 | 5.9 | 6.8 | 4.7 | 8.7 | 3.1 | |||
| AS2 | 24.6 | 28.4 | 31 | 26.5 | 23.2 | 22.2 | 33.6 | – | |
| 6.9 | 8.2 | 6.9 | 7.3 | 6.1 | 9.9 | 6.9 | – | ||
| EU1/SA1 | SA2 | EU2 | EU3 | NA1 | NA2 | AS1 | AS2 | ||
The upper values represent the divergence for Fsp; the lower values represent the divergence for the H protein. AS1, Asia 1; AS2, Asia 2; EU1/SA1, Europe 1/South America 1; EU2, Europe 2; EU3, Europe 3; NA1, North America 1; NA2, North America 2; SA2, South America 2.
The LM analysis showed that the Fsp and H datasets presented probabilities of 92.5% and 97.1%, respectively, in the three corners of the triangle, which represented the tree-like topologies (well-resolved phylogenies). The probabilities in the center region, representing the star-like topologies (unresolved phylogenies), were 4.9% for the Fsp dataset and 0.7% for the H dataset. The probabilities at the sides, representing the net-like region (partially unresolved phylogenies), were 1.8% and 1.5% for the Fsp and H datasets, respectively (Figure 2).
Figure 2. Likelihood mapping of the Fsp (A) and H (B) datasets.

The probabilities close to the triangle corners represent tree-like topologies (well-resolved). Those in the center and on the sides represent star-like (unresolved) and network-like signals (partially unresolved), respectively.
Discussion
The worldwide spread of canine distemper in all terrestrial carnivore families and some marine carnivores has lead to several studies of this disease in recent decades [4], [5], [6], [11], [18]. Most of the characterization studies of CDV strains have been based on H gene analysis. The direct amplification of the H gene from field samples is difficult without prior propagation of the virus in cell culture [9], [12], [13], [15], [31], [32]. Moreover, its extension (1824 bp) requires the use of several primers sets for amplification and sequencing of the full-length gene, which limits the rapid characterization of CDV strains. The genetic diversity of the Fsp-coding region in Asian strains [26], [27] encouraged us to evaluate the robustness of this region for phylogenetic analysis of the CDV lineages distributed worldwide.
For this work, we constructed Fsp and H datasets and analyzed their phylogenetic resolution. To include the South American strains, we designed a new set of primers, obtaining for the first time, the Fsp-coding region sequences of eight samples belonging to the EU1/SA1 and the SA2 lineages (Table 1). The Fsp-coding region consists of only 405 bp, making it easy to amplify directly from field samples of different origin using conventional methods for RNA isolation.
Phylogenetic analysis performed for the Fsp dataset clustered the same strains into eight well-defined clades according to the CDV lineages based on the H gene (Figure 1). The Fsp amino acid inter-lineage divergence was about fourfold greater with respect to the H protein. This result confirms previous reports regarding Asian strains [26], [27] and also allows us to extend the analysis to the rest of the CDV lineages worldwide, except for the African lineage.
The relationships among the lineages revealed slight differences in the Fsp and the H trees. The positions of the North American lineages (NA1 and NA2) were notably different between the trees: the Fsp tree showed that the NA1 and NA2 lineages were related, whereas the H tree did not show any relationship between them. The Fsp tree might reflect the actual relationships among the North American strains according to the geographical pattern of CDV lineage distribution [9], [12], [22]–[25] (Figure 1). Moreover, the NA1 lineage includes only field strains, whereas the NA2 lineage clusters field and vaccine strains into a main clade in both trees. The field and vaccine strains of the NA2 lineage presented amino acid divergence of up to 6.3% for the H protein, being this value higher than the 4% currently accepted to define lineages [22]. The NA2 main clade would then correspond to two different lineages: the vaccine cluster formed by “old” CDV strains isolated in 1930–1950 and the current field strain clade. For Fsp, the NA2 intra-lineage divergence is 20.7%, which represents the greatest value among all lineages analyzed (Table 2). This might be attributed to the clustering of the field and vaccine strains into this clade, which includes two different lineages (Table 2).
The intra-lineage divergence of Fsp was greater than that of the H protein for most of the lineages, except for the SA2 and NA1 lineages (Table 2). The SA2 lineage is composed of three strains isolated from the same geographical area (Buenos Aires, Argentina), which may have influenced their genomic homogeneity (Table 2). The NA1 lineage includes only two strains A75/17 and 164071, which showed two nucleotide synonymous changes for the Fsp-coding region, and 15 synonymous and two non-synonymous changes for the H gene. Thus, it is necessary to analyze more strains to establish if the variability of the Fsp-coding region is greater than that detected for the H gene as was observed for most of the lineages.
Despite the widespread use of the H gene for phylogenetic analysis of CDV and other Paramyxovirus [33]–[35], its phylogenetic signal has not been weighted before. Herein, we applied LM analysis, which revealed a strong phylogenetic signal for the H dataset in the region of tree-like topologies (Figure 2). Thus, this genomic region is phylogenetically informative. The LM analysis for the Fsp dataset showed values >90% in the region of tree-like topologies, indicating that this genomic region is also phylogenetically informative (Figure 2). We propose that the Fsp-coding region could be used as an alternative genetic marker for evolutionary studies and the rapid identification of CDV lineages because of its easy amplification, higher variability, and phylogenetic robustness. The current criteria take into account the H protein amino acid divergence and the clustering of the strains [12], [22]. Based on the Fsp analysis, we established that two strains belong to the same lineage if they cluster in the same clade with values of amino acid divergence <19%, whereas they belong to different lineages if they cluster in different clades with divergence >19%.
In conclusion, we have shown the suitability of the Fsp-coding region for phylogenetic analysis and CDV molecular characterization, which leads us to suggest this genomic region as an alternative marker to identify CDV lineages. The wide employment of the Fsp-coding region will improve the rapid and unequivocal classification of the circulating CDV strains and provide complementary information to understand CDV epidemiology.
Acknowledgments
We thank the clinical practitioners from Uruguayan and Argentinean veterinaries for generously providing samples for testing and Gregorio Iraola for the bioinformatic analyses assistance.
Funding Statement
This study was supported in part by “Programa de Desarrollo de las Ciencias Básicas” (PEDECIBA), Agencia Nacional de Investigación e Innovación (ANII) and Programa Amsud-Pasteur from Uruguay. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lamb RA, Parks GD (2007) Paramyxoviridae: the viruses and their replication. In: Fields BN, Knipe DV, Howley, PM, editors. Fields Virology. Lippincott Williams & Wilkins, 5th Ed. 1449–1496.
- 2. Martella V, Elia G, Buonavoglia C (2008) Canine distemper virus. Vet Clin North Am Small Anim Pract. 38: 787–797. [DOI] [PubMed] [Google Scholar]
- 3. Deem SL, Spelman LH, Yates RA, Montali RJ (2000) Canine distemper in terrestrial carnivores: a review. J Zoo Wildl Med 31: 441–451. [DOI] [PubMed] [Google Scholar]
- 4. Frolich K, Czupalla O, Haas L, Hentschke J, Dedek J, et al. (2000) Epizootiological investigations of canine distemper virus in free-ranging carnivores from Germany. Vet Microbiol 74: 283–292. [DOI] [PubMed] [Google Scholar]
- 5. Blixenkrone-Møller M, Svansson V, Have P, Orvell C, Appel M, et al. (1993) Studies on manifestations of canine distemper virus infection in an urban dog population. Vet Microbiol 37: 163–173. [DOI] [PubMed] [Google Scholar]
- 6. Gemma T, Watari T, Akiyama K, Miyashita N, Shin YS, et al. (1996) Epidemiological observations on recent outbreaks of canine distemper in Tokyo area. J Vet Med Sci 58: 547–550. [DOI] [PubMed] [Google Scholar]
- 7. Gebara CM, Wosiacki SR, Negrao FJ, de Oliveira DB, Beloni SN, et al. (2004) Detection of canine distemper virus nucleoprotein gene by RT-PCR in urine of dogs with distemper clinical signs. Arq Bras Med Vet Zootec 56: 480–487. [Google Scholar]
- 8. Calderón MG, Remorini P, Periolo O, Iglesias M, Mattion N, et al. (2007) Detection by RT-PCR and genetic characterization of canine distemper virus from vaccinated and non-vaccinated dogs in Argentina. Vet Microbiol 125: 341–349. [DOI] [PubMed] [Google Scholar]
- 9. Woma TY, van Vuuren M, Bosman AM, Quan M, Oosthuizen M (2009) Phylogenetic analysis of the haemagglutinin gene of current wild-type canine distemper viruses from South Africa: lineage Africa. Vet Microbiol 143: 126–132. [DOI] [PubMed] [Google Scholar]
- 10. Sarute N, Pérez R, Francia L, Hernández M, Bedó G, et al. (2011) Primer diagnóstico molecular y caracterización parcial del gen de la nucleoproteína del Virus Distemper Canino en Uruguay. Veterinaria 47: 7–13. [Google Scholar]
- 11. Keller SM, Gabriel M, Terio KA, Dubovi EJ, Vanwormer E, et al. (2012) Canine Distemper in an Isolated Population of Fishers (Martes pennanti) from California. J Wildl Dis 48: 1035–1041. [DOI] [PubMed] [Google Scholar]
- 12. Bolt G, Jensen TD, Gottschalck E, Arctander P, Appel M, et al. (1997) Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virology 78: 367–372. [DOI] [PubMed] [Google Scholar]
- 13. Iwatsuki K, Miyashita N, Yoshida E, Gemma T, Shin Y, et al. (1997) Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J Gen Virology 78: 373–380. [DOI] [PubMed] [Google Scholar]
- 14. Iwatsuki K, Tokiyoshi S, Hirayama N, Nakamura K, Ohashi K, et al. (2004) Antigenic differences in the H proteins of canine distemper viruses. Vet Microbiol 71: 281–286. [DOI] [PubMed] [Google Scholar]
- 15. Hirama K, Goto Y, Uema M, Endo Y, Miura R, et al. (2004) Phylogenetic analysis of the hemagglutinin (H) gene of canine distemper viruses isolated from wild masked palm civets (Paguma larvata). J Vet Med Sci 66: 1575–1578. [DOI] [PubMed] [Google Scholar]
- 17. Müller A, Silva E, Santos N, Thompson G (2011) Domestic dog origin of canine distemper virus in free-ranging wolves in Portugal as revealed by hemagglutinin gene characterization. J Wildl Dis 47: 725–729. [DOI] [PubMed] [Google Scholar]
- 18. Origgi FC, Plattet P, Sattler U, Robert N, Casaubon E, et al. (2012) Emergence of canine distemper virus strains with modified molecular signature and enhanced neuronal tropism leading to high mortality in wild carnivores. Vet Pathol 49: 913–929. [DOI] [PubMed] [Google Scholar]
- 19. Martella V, Bianchi A, Bertoletti I, Pedrotti L, Gugiatti A, et al. (2010) Canine distemper epizootic among red foxes, Italy, 2009. Emerg Infect Dis 16: 2007–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao JJ, Yan XJ, Chai XL, Martella V, Luo GL, et al. (2010) Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet Microbiol 140: 34–42. [DOI] [PubMed] [Google Scholar]
- 21. Rota JS, Hummel KB, Rota PA, Bellini WJ (1992) Genetic variability of the glycoprotein genes of current wild-type measles isolates. Virology 188: 135–142. [DOI] [PubMed] [Google Scholar]
- 22. Martella V, Elia G, Lucente M, Decaro N, Lorusso E, et al. (2006) Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet Microbiol 116: 301–309. [DOI] [PubMed] [Google Scholar]
- 23. Martella V, Elia G, Lucente M, Decaro N, Lorusso E, et al. (2007) Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet Microbiol 122: 32–42. [DOI] [PubMed] [Google Scholar]
- 24. An DJ, Yoon SH, Park JY, No IS, Park B (2008) Phylogenetic characterization of Canine Distemper Virus isolates from naturally infected dogs and a marten in Korea. Vet Microbiol 132: 389–395. [DOI] [PubMed] [Google Scholar]
- 25. Panzera Y, Calderón MG, Sarute N, Guasco S, Cardeillac A, et al. (2011) Evidence of two co-circulating genetic lineages of canine distemper virus in South America. Virus Res 163: 401–404. [DOI] [PubMed] [Google Scholar]
- 26. Lee MS, Tsai KJ, Chen LH, Chen CY, Liu YP, et al. (2008) The identification of frequent variations in the fusion protein of canine distemper virus. Vet Journal 183: 184–190. [DOI] [PubMed] [Google Scholar]
- 27. Sultan S, Charoenvisal N, Lan NT, Yamaguchi R, Maeda K, et al. (2009) The Asia 2 specific signal peptide region and other domains in fusion protein genes characterized Asia 1 and Asia 2 canine distemper viruses. Virol J 6: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA 5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol and Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt HA, von Haeseler A (2007) Maximum-likelihood analysis using TREE-PUZZLE. Curr Protoc Bioinformatics. doi:10.1002/0471250953.bi0606s17. [DOI] [PubMed]
- 30. Strimmer K, von Haeseler A (1997) Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA 94: 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lednicky JA, Dubach J, Kinsel MJ, Meehan TP, Bocchetta M, et al. (2004) Genetically distant American Canine distemper virus lineages have recently caused epizootics with somewhat different characteristics in raccoons living around a large suburban zoo in the USA. Virol J doi:10.1186/1743-422X-1-2. [DOI] [PMC free article] [PubMed]
- 32. Lan NT, Yamaguchi R, Inomata A, Furuya Y, Uchida K, et al. (2006) Comparative analyses of canine distemper viral isolates from clinical cases of canine distemper in vaccinated dogs. Vet Microbiol 115: 32–42. [DOI] [PubMed] [Google Scholar]
- 33. Kindermann J, Kübber-Heiss A, Kerschbaumer P, Nowotny N (2001) Phylogenetic analysis of the L and HN gene of ophidian paramyxoviruses. Brief report. Arch Virol 146: 1021–1035. [DOI] [PubMed] [Google Scholar]
- 34. Mahmood S, Alexander DJ, Slomka MJ, Manvell RJ, Hanna A, et al. (2010) Phylogenetic analysis of the nucleotide sequences for the HN gene of 22 avian paramyxovirus type 2 viruses reveals marked heterogeneity. Avian Pathol 39: 453–458. [DOI] [PubMed] [Google Scholar]
- 35. Shi J, Zheng J, Huang H, Hu Y, Bian H, et al. (2011) Measles incidence rate and a phylogenetic study of contemporary genotype H1 measles strains in China: is an improved measles vaccine needed? Virus Genes 43: 319–326. [DOI] [PubMed] [Google Scholar]