Abstract
Advances in our understanding of the human genome and next-generation technologies have facilitated the use of genome-wide sequencing to decipher the genetic basis of Mendelian disease and hereditary cancer syndromes. The application of genome-wide sequencing in hereditary cancer syndromes has had mixed success, in part, due to complex nature of the underlying genetic architecture. In this review we discuss the use of genome-wide sequencing in both Mendelian diseases and hereditary cancer syndromes, highlighting the potential and challenges of this approach using familial pancreatic cancer as an example.
1. Introduction
The publication of the draft human reference genome and advent of highthroughput sequencing technology have ushered in the genomics era. These new sequencing technologies have driven down the cost of genome sequencing to within a hair’s breadth of the $1000 dollar genome. Over a decade, per genome cost of sequencing has dropped precipitously, from approximately $70 million dollars in 2002, to nearly $7000 dollars in 2012 [1]. Similarly, the time to generate genomic sequence has also decreased, with over 1010 bases per instrument per day with the latest next generation sequencing technology. These advances resulted in the application of genome-wide sequencing to elucidate the underlying genetic basis of hereditary disease. Numerous examples of high-throughput, genome-wide approaches for disease gene identification are available for Mendelian diseases and these provide a starting framework for the analysis of complex diseases. Such gene discovery approaches share common elements, such as case selection and sample acquisition, genome-wide sequencing, data analysis to identify candidate genes with subsequent functional or population-based validation (Fig. 1). However, the application genomewide sequencing in the background of complex disease faces unique challenges due to underlying genetic heterogeneity of these diseases, in particular the identification of candidate genes using either filter-based or statistical based tests in the face of underlying genetic heterogeneity. In this review, we: 1) discuss the current use of genome-wide sequencing to identify the genetic basis of Mendelian diseases and hereditary cancer syndromes, 2) outline current knowledge about the genetic basis of familial pancreatic cancer, and 3) highlight the challenges of using genome-wide sequencing to identify pancreatic cancer susceptibility genes. We focus on familial pancreatic cancer because while genomic sequencing approaches have successfully identified familial pancreatic cancer genes, the challenges of using these approaches, specifically underlying genetic heterogeneity and its impact in study size requirements and candidate variant selection are faced by researchers using these approaches to study other cancer syndromes or other complex diseases
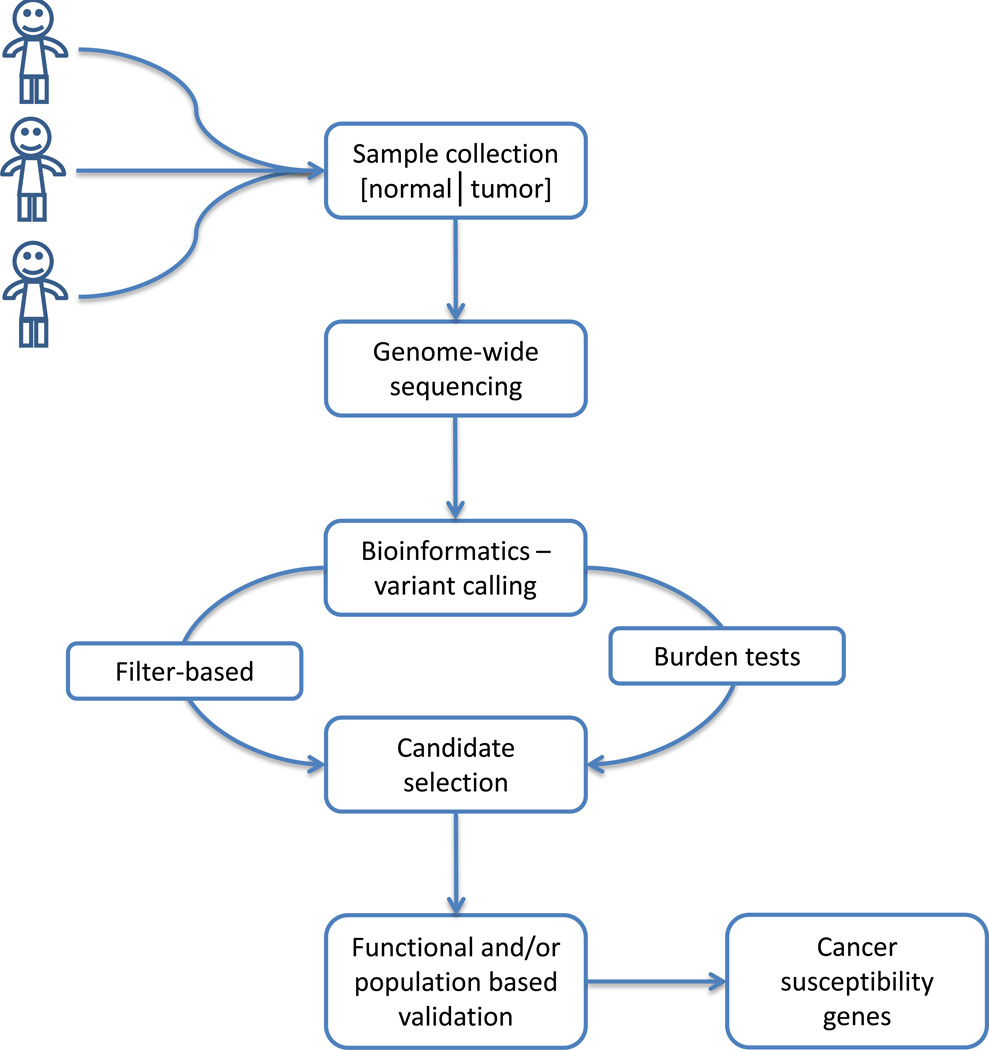
Figure 1. Schematic representation of cancer susceptibility gene discovery with genome-wide sequencing.
Discovery relies on careful case identification and collection of matched tumor and normal samples. Genome-wide sequencing is conducted on a cohort of samples and bioinformatic analyses performed to identify individual variation. Candidate selection carried out with either filterbased approaches or burden tests before functional or population-based validation. Validated cancer susceptibility genes may be used for screening, risk assessment, prognostics and therapeutic targets.
2. Genome-wide sequencing and hereditary disease
The first use of genome-wide sequencing to identify the genetic cause of a hereditary disease used Sanger sequencing to analyze the coding genes in the germline and tumor of a patient with familial pancreatic cancer [2]. Jones and colleagues employed a filter-based approach using the hypothesis that any susceptibility gene will be inactivating and heterozygous in the germline of the patient and contain a second mutation in the gene of the tumor, that is, it obeys the classical two-hit model of a tumor suppressor gene. Using this filter-based approach, 15,461 germline variants not seen in the human reference genome were narrowed to 3 in genes SERPINB12, RAGE and PALB2. SERPINB12 and RAGE were eliminated from further analysis as nonsense variants were common in healthy individuals. This left PALB2 as the putative susceptibility gene, highlighting the power of genome-wide sequencing to identify the genetic cause of a hereditary disease and the need of a good detective when approaching such a study.
Shortly after the discovery of PALB2 as a pancreatic cancer susceptibility gene, genome-wide sequencing was successfully employed to identify the genetic basis of a Mendelian disorder. Ng and colleagues used next-generation technology to sequence the exome of 4 unrelated individuals with Freeman-Sheldon Syndrome, an autosomal dominant disorder of congenital arthrogryposis [3]. This study provided a powerful demonstration of the ability of next-generation technology to identify the genetic cause of a Mendelian disorder. Central to this proof-of-principle study were computational methods to find the single causative variant in the 20,000 rare variants per exome, the proverbial needle in the haystack. To do this, Ng and colleagues used a filter-based approach, eliminating common variants using control samples and the database of single nucleotide polymorphisms (dbSNP), and searching for genes where each affected individual harbored a rare variant. Their efforts identified one gene, MYH3, previously identified as the cause of Freeman-Sheldon Syndrome [4]. Subsequently, numerous Mendelian disorders where traditional linkage studies were unsuitable have been subjected to genome-wide sequencing approaches and the genetic basis of the phenotype defined [5; 6; 7; 8]. Such studies predominantly use whole-exome sequencing which limit search to exons and small segments of adjacent sequence, that is, coding and splice site variants. Fewer studies use whole-genome sequencing due to added cost and computational difficulties associated with a 20-fold increase in data per individual. However, studies utilizing whole-genome sequencing are not limited to coding variants and as such can find associations between copy number variation or non-coding variation and disease. A study by Jaeger and colleagues [9] used wholegenome sequencing in individuals with hereditary mixed polyposis syndrome (HMPS) to identify a duplication, 5’ to GREM1, that results in ectopic expression of GREM1 in colonic epithelium. As the authors note, further evidence for the causal nature of GREM1 duplications in HMPS patients is given by juvenile polyposis syndrome (JPS). The majority of JPS patients harbor germline mutations in SMAD4 or BMPR1A [10], members of the bone morphogenetic protein (BMP) signaling pathway [11]. Interestingly, GREM1 is thought to negatively regulate BMP pathway signaling, providing a contemporaneous link between HMPS and JPS.
Hot on the heels of these successes were further large scale genome-wide sequencing studies to identify novel susceptibility genes in hereditary cancer syndromes. In 2012, Roberts and colleagues [12] reported on the whole-genome and whole-exome sequencing of 38 familial pancreatic cancer cases. Here they describe the aggregation of deleterious mutations in ATM in the germline of 6 pancreatic cancer cases representing 2 different kindreds. Similar to studies of Mendelian disorders described previously, Roberts and colleagues employ a filter-based approach using assumptions about the genetic architecture of susceptibility genes in familial pancreatic cancer to narrow down candidate genes for further study. In this study, germline variants present in the Human Gene Mutation Database (HGMD) [13] and associated with disease in people, were classified as functionally deleterious. This led to the identification of ATM as a candidate susceptibility gene and its validation in a larger sample set where 4 additional deleterious variants in 166 familial pancreatic cancer probands. Of note was the absence of deleterious ATM variants in a set of 190 controls which resulted in a statistically significant association of deleterious ATM variants in familial pancreatic cancer (Fisher’s exact test p-value = 0.046).
This was not the only successful application of genome-wide sequencing approaches to hereditary cancer syndromes. Park and colleagues [14] describe the use of whole-exome sequencing to identify XRCC2 as a breast cancer susceptibility gene. Again, a set of hereditary cases from 13 families, presumably enriched for susceptibility genes, were subjected to high-throughput sequencing, leading to the identification of a truncating and missense variant in the XRCC2 gene of 2 families. Validation in a case-control study of early onset breast cancer cases resulted in a statistically significant association of pathogenic coding variants in cases (6/1,308) vs. controls (0/1,120) (Fisher’s exact test p-value = 0.02). Interestingly, Park and colleagues used the computational algorithms: SIFT, Polyphen 2.1 and Align-GVGD, to assign functional significance to missense mutations. The contrast between the methods of assigning functional significance to missense variants by Roberts and colleagues and Park and colleagues are stark and occur due to the difficuly in assigning functional significance to the many rare and private germline variants discovered through genome-wide sequencing.
A study by Snape and colleagues [15] highlights perfectly the complexity of genetic risk determinants in hereditary cancer syndromes and the difficultly in identifying predisposition genes in hereditary breast cancers; even when an estimated 60 percent of familial risk for breast cancer is still to be explained [16]. Snape and colleagues conducted whole-exome sequencing in 50 individuals with familial breast cancer. Applying a filter-based approach and assumptions about the genetic basis of disease that were employed successfully in other genome-wide sequencing studies of hereditary cancer syndromes, Snape and colleagues failed to recognize susceptibility genes without a priori knowledge of breast cancer predisposition genetics. In their thorough study, an average of 26 truncating mutations per sample were identified and in 4 individuals the genetic basis of disease was assigned to truncating variants in three breast cancer susceptibility genes: BRCA2, CHEK2 and ATM. Validation of a subset of these truncating mutations with Sanger sequencing revealed a false-positive rate of 58.9 percent. However, once these false-positives were excluded from analysis, approximately 11 truncating variants per sample remained. Even in samples containing a truncating variant in a known susceptibility gene, other rare truncating variants occurred that may be linked to the disease phenotype e.g. regulators of apoptosis or transcriptional regulators, making interpretation difficult.
3. Lessons from hereditary pancreatic cancer
Genome-wide sequencing has proven useful with the identification of PALB2 and ATM as pancreatic cancer susceptibility genes. However, as described herein, the genetic basis of the overwhelming majority of familial cases remains undefined by convention linkage or candidate gene studies. Therefore, genome-wide sequencing provides an opportune approach to characterize elusive susceptibility genes essential for future genetic testing and targeted surveillance programs.
3.1. Evidence for a genetic basis to pancreatic cancer predisposition
Worldwide, more than 200,000 cases of pancreatic cancer are reported annually and the prognosis is particularly poor with a 5-year survival rate of less than 5 percent [17]. The disease is known to cluster in families, with 5–10 percent of cases in families with two first-degree relatives [18][19]. In 10–15 percent of these families, the genetic basis is attributed to known pancreatic cancer susceptibility, including: BRCA2, BRCA1, PALB2, STK11, CDKN2A, PRSS1, ATM, APC and mismatch repair genes (Table 1). However, the genetic basis in the remaining 85–90 percent of families with hereditary pancreatic cancer is unknown.
Table 1.
Known pancreatic cancer susceptibility genes and magnitude of their effect.
Co-occurrence of cases in a family may be due to underlying genetic basis, environmental risk factors or stochastic influences. Genetic contribution is thought to be the primary cause of aggregation [20] with evidence gleamed from family, twin, casecontrol and cohort studies. Initial case reports of familial clustering [21; 22; 23] were followed by observational epidemiological studies to assess the risk of pancreatic cancer in individuals with a family history of the disease. Fernandez and colleagues assessed 382 pancreatic cancers and 1408 controls and demonstrated excess risk of pancreatic cancer in individuals with a family history of the disease (relative risk 3.0) [24]. While the excess risk associated with a familial history of pancreatic cancer is beyond doubt, the magnitude of reported risk is variable between 2.3 and 32-fold increase [25]. Subsequently, the genetic contribution to pancreatic disease risk was highlighted and quantified by Lichtenstein and colleagues who surveyed a Swedish population-based twin registry, assessing disease concordance and discordance twins for a comprehensive list of cancers [26]. Estimates of the relative importance of heritable effects, the proportion of phenotypic liability attributable to genetic effects, was 0.36 (95 percent confidence interval: 0.00–0.53), suggesting a major role of genetic factors. However, the rarity of pancreatic cancer in the population prevented a statistically significant result. A population-based study of the Icelandic population, ascertained relative risk ratios for of different cancers for first, second, third, fourth and fifth-degree relatives of affected individuals [27]. For Pancreatic cancer, the risk ratio for first-degree relative of an affected was 2.3 (90 percent confidence interval: 1.8–3.0). This risk ratio was similar to other cancers assessed with a range of risk ratio for first degree relatives of 1.0 (90 percent confidence interval: 0.2–3.4) for myeloid leukemia and 5.0 (90 percent confidence interval: 2.8–9.5) for cancer of the lip. Interestingly, familial co-aggregation of pancreatic and ovarian cancer was significant (p-value < 0.05 percent) when considering first- to fifth-degree relatives. However, co-aggregation of pancreatic and breast cancers was not significant despite known susceptibility genes conferring risk to both breast and pancreatic cancer, for example, BRCA2. Similarly, a prospective study of pancreatic cancer families looked at the difference in observed and expected incidence of pancreatic cancers in families with at least one individual with pancreatic cancer. The ratio of observed to expected incidence of pancreatic cancers was 9.0 (95 percent confidence interval: 4.5–16.1) in familial pancreatic cancer kindreds versus sporadic pancreatic cancer kindreds with an observed-expected incidence ratio of 1.8 (95 percent confidence interval: 0.2–6.4) [18]. To quantify the magnitude of risk of pancreatic cancer in people with a family history of the disease, Wang and colleagues developed a Bayesian prediction model (PancPRO) has been developed to estimate the probability of carrying a pancreatic cancer susceptibility gene and the absolute risk of pancreatic cancer in people with a family history of the disease [18]. PancPRPO was validated using data from 6,134 individuals, across 961 families of the National Familial Pancreatic Tumor Registry (NFPTR). PancPRO allows for the selection of a high risk subset of individuals for more intense disease surveillance.
3.2 Pancreatic cancer susceptibility genes
3.2.1 BRCA2, PALB2 and BRCA1
BRCA2, PALB2 and BRCA1 are important components of DNA repair pathways [28; 29]. Germline mutations of BRCA1 and BRCA2 are the underlying cause for hereditary breast and ovarian syndrome, an autosomal dominant condition with a lifetime risk of up to 80 percent for breast cancer [30]. PALB2 mutations have been associated with hereditary breast cancer [31]. In addition to breast and ovarian cancers, families with a germline BRCA2 mutation have a 3.5 fold (95 percent confidence interval: 1.9–6.6) risk of pancreatic cancer [32]. In familial pancreatic cancer families, 6- 17 percent may harbor a germline BRCA2 mutation, making germline alterations in BRCA2 the most common identifiable cause of familial clustering of pancreatic cancer [33; 34]. As described previously, PALB2 was identified using genome-wide sequencing [2] and mutations have been reported in 1–3 percent of familial pancreatic cancer kindreds [35; 36] with carriers having an increased pancreatic cancer risk of 5.9- fold (95 percent confidence interval: 2.4–14.6) [37]. The risk of pancreatic cancer in carriers of BRCA1 germline mutations is less well-established, while some studies suggest a 2.2-fold increased risk of pancreatic cancer in BRCA1 families [38], other studies fail to report an association [39; 40; 41]. Still, molecular typing of BRCA1, BRCA2 and PALB2 status in pancreatic cancers may be provide therapeutic benefit. BRCA1, BRCA2 and PALB2 deficient cancers are more susceptible to chemotherapeutic agents that disrupt DNA [42] and Poly(ADP-ribose) polymerase inhibitors [43; 44]. Recent clinical trials of PARP inhibitors in breast and ovarian cancer patients indicate that treatment was well tolerated, with durable object responses seen in ~40 percent of cases [45; 46; 47]. Clinical trials are underway to examine the effects of treatment with PARP inhibitors in BRCA1, BRCA2 or PALB2 deficient pancreatic cancers and may provide new treatment options for this subgroup of patients.
3.2.2 STK11/LKB1
Germline mutations in STK11/LKB1 are associated with the autosomal dominant condition Peutz-Jeghers Syndrome (PJS) [48]. PJS patients are at increased risk of a variety of neoplasms, including: lung, breast, pancreatic, colon, gastric and ovarian [49]. Lifetime risk for any neoplasm in PJS patients is greater than 85 percent, with the relative risk of pancreatic cancer estimated to be increased 132-fold (95 percent confidence interval: 44–261) [49; 50].
3.2.3 CDKN2A
The gene CDKN2A is a tumor suppressor gene encoding p16INK4a and p14ARF [51]. Germline mutations in CDKN2A that disrupt p16INK4a function result in aberrant cell cycle progression and are associated with familial atypical multiple mole melanoma (FAMMM) syndrome, an autosomal dominant condition accounting for approximately 40 percent of hereditary melanoma [52]. The link between FAMMM syndrome and pancreatic cancer was first identified by Lynch and colleagues [53]. The rate of pancreatic cancer in CDKN2A families is variable with up to 60 percent of kindreds being affected. Interestingly, this may be due to differential risk of pancreatic cancer associated with different CDKN2A mutations [51]. Overall, the relative risk of pancreatic cancer in carriers of CDKN2A mutations is thought to be 32-fold (95% confidence interval: 1.5–47.4) [54].
3.2.4 PRSS1 and SPINK1
Germline mutations of PRSS1 and SPINK1 are the cause of autosomal dominant and autosomal recessive hereditary pancreatitis respectively [55]. PRSS1 encodes cationic trypsinogen, a component of pancreatic juice and germline alterations, including: missense mutations, copy number alterations and microduplication, all perturb protein function through either increased trypsin stability or enhanced trypsinogen autoactivation [56]. Aberrant trypsin activity results in early-onset, severe, chronic pancreatitis, leading to permanent loss of endocrine and exocrine function and an increased risk of pancreatic cancer. Those affected by hereditary pancreatitis have up to an 80 fold increase risk of developing pancreatic cancer, with nearly 40 percent of individuals developing the disease by 70 years of age [57]. Similarly, SPINK1 encodes a trypsin inhibitor where biallelic germline mutations affecting expression or protein function removes a key checkpoint guarding against inappropriate trypsin activity, resulting in a chronic pancreatitis [56].
3.2.5 ATM
The ATM protein is a serine/threonine kinase integral to the repair of double strand breaks in DNA [58; 59]. Inheritance of biallelic deleterious variants in the ATM gene result in ataxia-telangietasia [59], a disease that is characterized by progressive cerebellar ataxia, oculomotor apraxia, telangiectasias of the conjunctiva and skin, immunodeficiency, sensitivity to ionizing radiation and an increased rate of malignancies, in particular lymphoma and leukemia [58; 60; 61]. Population and family based studies point to an increased risk of breast and pancreatic cancers in carriers [12; 60; 61; 62]. Although, the magnitude of this effect is controversial [63; 64; 65; 66] and deleterious ATM variants are relatively common in the population, occurring in 0.5–1 percent of people [58; 60]. Further study is required to delineate the effect of environmental factors and modifier genes on disease risk in carriers of deleterious ATM variants.
3.2.6 Familial adenomatous polyposis and Lynch syndrome
Familial adenomatous polyposis (FAP) and Lynch syndrome (hereditary non-polyposis colon cancer – HNPCC), are associated with a moderately increased risk of pancreatic cancer. FAP, the archetypal colorectal cancer syndrome, is found in people harboring a germline, inactivating mutation in APC. Similarly, Lynch syndrome is an autosomal dominant condition of colorectal cancer predisposition caused by germline mutations in mismatch repair genes (MLH1, MSH2, MSH6 and PMS2) [67]. Besides an increased risk of colorectal cancer, patients with Lynch syndrome and FAP are more likely to develop certain extra-colonic malignancies, such as tumors of the: endometrium, pancreas, urinary tract, liver, thyroid and central nervous system [68; 69]. Recent studies have suggested the risk of pancreatic cancer in Lynch syndrome mutation carriers to be 3.7 percent (95 percent confidence interval: 1.6–5.9) up to age 70 years, represent a 8.6-fold increased risk (95 percent confidence interval: 4.7–15.7) [70]. This risk of pancreatic cancer associated with FAP is less clear while some studies fail to observe and increased risk of pancreatic cancers in FAP kindreds, up to a 4-fold increased risk have been observed in others [71].
3.3 Unexplained pancreatic cancer families
The vast majority 85–90 percent of familial pancreatic cancer kindreds are not explained by mutations in any of the above-mentioned genes. Given the rapid fatality of pancreatic cancer, DNA is often not available on multiple affected members of the same kindred. The lack of key DNA samples along with the underlying genetic heterogeneity, limits the success of linkage approaches to map susceptibility genes. Therefore, genomic sequencing approaches (Fig. 1) provide the best opportunity to discover novel pancreatic cancer susceptibility genes as evidenced by the recent identification of PALB2 and ATM using these approaches. However, genomic sequencing approaches provide a unique set of challenges.
4. Challenges of genome-wide studies of Hereditary Cancers
4.1 Multiple variants – nonsense and high-quality missense variants
The major challenge of using genomic sequencing to identify the causes of hereditary diseases is recognition of the causal mutation amidst the 1,000s of genetic variants identified, the needle in the haystack. On average, 35,000 and 6,000,000 variants are identified per exome or genome sequenced [12]. These will represent true variation, PCR or sequencing related errors, and bioinformatics artifacts. While majority of the changes may be excluded from further analysis as either common variants present in known SNP databases or observational artifacts below physical coverage thresholds, the number of rare and private variants is still far too large to be examined using functional approaches. Numerous statistical approaches have been developed to help identify the causal variant [72; 73], however, most of these approaches have not been applied in gene discovery studies. Furthermore, the power of these approached will depend on the underlying biology, which is often unknown. The approaches proposed fall into one of two categories: filter-based approaches and statistical approaches commonly referred to as “burden tests”.
4.2 Filter-based approaches
To date, the sequencing studies that have successfully identified the causes of hereditary diseases, such as those outlined at the beginning of this review, primarily relied upon the filter-based approaches. As the name states, this approach uses a series of filters to eliminate benign variation resulting in a narrowed listed of potentially disease associated variants. Commonly used filters include variant type e.g. nonsense, missense and synonymous, allele frequency in publically available datasets e.g. HapMap, 1000 genomes [74; 75], and for missense variants, predicted function. Numerous functional prediction algorithms have been developed with the goal of discriminating between benign and functional genetic variation. These include SIFT, PolyPhen-2, GERP and others [76; 77; 78]. However, these algorithms have imperfect sensitivity and specificity [79]. Family data can also be used to filter-out variation by requiring variant-sharing among affected pedigree members and/or the absence of the variant in unaffected pedigree members.
4.3 Tumor sequencing
In addition to the above-mentioned approaches, knowledge of the underlying biology of the disease can also be used to help filter variants. When studying cancer syndromes, sequence analysis of the tumor itself can be especially powerful. Analysis of the tumor data not only can provide insight into the somatic mutations involved in tumor initiation and progression but also provide information on germ-line mutations. Data quality for tumor analysis provides its own set of challenges and is beyond the scope of this article, readers are directed to see Ding and Colleagues for a comprehensive review [80]. Given that tumors are very heterogeneous and represent a mixture of tumor and normal cells, identification of somatic mutations can be challenging. However, high-quality paired data from both the tumor and germline of a patient with hereditary cancer can be used to identify potential tumor suppressor genes by looking for genes in which there is a germline mutation coupled with either a somatic mutation of LOH in the cancer. While this approach may be ideal, at a minimum it doubles the cost and sequencing effort required. Furthermore, the amount of high quality tumor DNA needed for sequencing is often unavailable for most patients. If limited tumor DNA is available, tumor analysis can be limited to a small list of candidates. Tumors from patients carrying a variant in a given candidate can be analyzed for a second somatic mutation of LOH in the gene of interest. If paired data is not available, data from large-scale tumor sequencing such as those that have been conducted for breast, colon and pancreatic cancer [81; 82; 83; 84] can also be used as a filter. Variants that occur in genes that have been found to be somatically altered in other studies can be prioritized for follow-up either in functional studies or in large-scale candidate genes studies. The major limitation to all of these filtering approaches is that the true causative mutation can easily be filtered-out and/or a very large number of potentially deleterious variants will remain after all filters have been used.
4.4 Burden tests
In contrast to the filtering based approaches, burden tests are a group of statistical tests aimed at identification of disease-associated variation. Traditional tests of association are not well suited to the analysis of sequencing data, as they typically require modest to large sample sizes. Because the frequency of any given variant in a gene can be quite low, these traditional association tests have little power to detect disease-causing mutations. To overcome this limitation, collapsing approaches or “burden-tests” have been developed. In brief, these methods combine variants detected in a given gene, pathway or region and then compare the frequency of these variants in cases and controls. A variety of burden test statistics have been developed. The Cohort Allelic Sums Test (CAST) groups all rare variants together [73], and the Combined Multivariate and Collapsing Method (CMC) groups rare variants and jointly examines them along with common variants [72]. These methods equally weight rare variants and thus assume all rare variants have a similar effect on the disease phenotype. Alternatively, variants can be weighted. While the best weighting strategy remains unclear and will likely depend on the underlying biology of the disease, common weighting strategies include the inverse of the allele frequency, functional information from prediction algorithms such as SIFT etc…, or estimates of genetic fitness.
4.5. Locus heterogeneity and incomplete penetrance of variants
The etiology of hereditary cancer syndromes can be quite complex with the majority of hereditary cancer syndromes exhibit locus heterogeneity, incomplete penetrance and phenocopies [85]. Locus heterogeneity occurs when mutations in more than one gene can lead to the same hereditary cancer phenotype. As locus heterogeneity increases the proportion of individuals sequenced who are likely to carry a disease causing mutation decreases. Because genome sequencing studies look for evidence of a mutation in a given gene across families, the presence of locus heterogeneity greatly limits the ability to identify disease associated genes. Similarly, not all individuals who inherit a mutation in hereditary cancer genes will go on to develop disease. While the probability of disease often increases with increasing age, even by age 80 a significant proportion of individuals may be disease free. For example, it is estimated that 29 and 54 percent of BRCA2 carriers develop breast cancer by age 50 and 70 respectively [86]. Because of incomplete penetrance, the interpretation of data from “unaffected” members of hereditary cancer families in genome sequencing studies must be carefully considered. Furthermore, many sequencing studies rely on publically available databases of “control” individuals. These controls represent either individual’s without disease, or in the case of 1000 genomes data [75] individuals with unknown cancer status. Therefore, it would not be unexpected to observe disease-causing variants in these control populations. Finally, sporadic cancers or phenocopies are not uncommon in hereditary pancreatic cancer families. These are cancers that develop in family members who did not inherit the disease predisposing mutations. Therefore, not all affected relatives will carry the disease causing mutations. Because the phenotype of the sporadic cancer mimics that of the hereditary cancer these phenocopies cannot be excluded a priori. The probability of a phenocopy depends on the frequency of the cancer in the general population.
5. Conclusion
Recent technological advancements allow for the broad application of genomic sequencing to discovery of the causes of hereditary diseases, including inherited cancer syndromes. While these methods are extremely powerful, as demonstrated by the identification of PALB2 and ATM as hereditary pancreatic cancer genes, identification of the single genetic change that leads to a hereditary cancer amidst the 1,000 of genetic changes identified though this approach can be elusive. For complex phenotypes, such as many hereditary cancers, locus heterogeneity, decreased penetrance and a high photocopy rate further complicate the interpretation of the sequence data. Despite these challenges, genome sequencing approaches will lead to the discovery of many cancer predisposition genes in near future.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors do not have any conflicts of interest.
References
- 1.Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Program. Available from: www.genome.gov/sequencingcosts.
- 2.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, Iacobuzio-Donahue CA, Maitra A, Parmigiani G, Kern SE, Velculescu VE, Kinzler KW, Vogelstein B, Eshleman JR, Goggins M, Klein AP. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toydemir RM, Rutherford A, Whitby FG, Jorde LB, Carey JC, Bamshad MJ. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nat Genet. 2006;38:561–565. doi: 10.1038/ng1775. [DOI] [PubMed] [Google Scholar]
- 5.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2011;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, Gumbs CE, Pevsner J, Thomas G, Valle D, Hoover-Fong JE, Goldstein DB. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaeger E, Leedham S, Lewis A, Segditsas S, Becker M, Cuadrado PR, Davis H, Kaur K, Heinimann K, Howarth K, East J, Taylor J, Thomas H, Tomlinson I. Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet. 2012;44:699–703. doi: 10.1038/ng.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosens LA, Langeveld D, van Hattem WA, Giardiello FM, Offerhaus GJ. Juvenile polyposis syndrome. World J Gastroenterol. 2011;17:4839–4844. doi: 10.3748/wjg.v17.i44.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick JC, Kodach LL, Offerhaus GJ, van den Brink GR. Bone morphogenetic protein signalling in colorectal cancer. Nat Rev Cancer. 2008;8:806–812. doi: 10.1038/nrc2467. [DOI] [PubMed] [Google Scholar]
- 12.Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, Gallinger S, Schwartz AG, Syngal S, Cote ML, Axilbund J, Schulick R, Ali SZ, Eshleman JR, Velculescu VE, Goggins M, Vogelstein B, Papadopoulos N, Hruban RH, Kinzler KW, Klein AP. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, Neuhausen SL, John EM, Andrulis IL, Terry MB, Daly M, Buys S, Le Calvez-Kelm F, Lonie A, Pope BJ, Tsimiklis H, Voegele C, Hilbers FM, Hoogerbrugge N, Barroso A, Osorio A, Giles GG, Devilee P, Benitez J, Hopper JL, Tavtigian SV, Goldgar DE, Southey MC. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snape K, Ruark E, Tarpey P, Renwick A, Turnbull C, Seal S, Murray A, Hanks S, Douglas J, Stratton MR, Rahman N. Predisposition gene identification in common cancers by exome sequencing: insights from familial breast cancer. Breast Cancer Res Treat. 2012;134:429–433. doi: 10.1007/s10549-012-2057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shuen AY, Foulkes WD. Inherited mutations in breast cancer genes--risk and response. J Mammary Gland Biol Neoplasia. 2011;16:3–15. doi: 10.1007/s10911-011-9213-5. [DOI] [PubMed] [Google Scholar]
- 17.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 18.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, Griffin C, Cameron JL, Yeo CJ, Kern S, Hruban RH. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 19.Shi C, Hruban RH, Klein AP. Familial pancreatic cancer. Arch Pathol Lab Med. 2009;133:365–374. doi: 10.5858/133.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein AP, Beaty TH, Bailey-Wilson JE, Brune KA, Hruban RH, Petersen GM. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol. 2002;23:133–149. doi: 10.1002/gepi.1102. [DOI] [PubMed] [Google Scholar]
- 21.MacDermott RP, Kramer P. Adenocarcinoma of the pancreas in four siblings. Gastroenterology. 1973;65:137–139. [PubMed] [Google Scholar]
- 22.Reimer RR, Fraumeni JF, Jr, Ozols RF, Bender R. Pancreatic cancer in father and son. Lancet. 1977;1:911. doi: 10.1016/s0140-6736(77)91244-2. [DOI] [PubMed] [Google Scholar]
- 23.Friedman JM, Fialkow PJ. Familial carcinoma of the pancreas. Clin Genet. 1976;9:463–469. doi: 10.1111/j.1399-0004.1976.tb01598.x. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez E, La Vecchia C, D'Avanzo B, Negri E, Franceschi S. Family history and the risk of liver, gallbladder, and pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 1994;3:209–212. [PubMed] [Google Scholar]
- 25.Hruban RH, Canto MI, Goggins M, Schulick R, Klein AP. Update on familial pancreatic cancer. Adv Surg. 2010;44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 27.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, Stefansson K. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domchek SM, Weber BL. Clinical management of BRCA1 and BRCA2 mutation carriers. Oncogene. 2006;25:5825–5831. doi: 10.1038/sj.onc.1209881. [DOI] [PubMed] [Google Scholar]
- 31.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Breast Cancer Linkage Consortium, Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 33.Murphy KM, Brune KA, Griffin C, Sollenberger JE, Petersen GM, Bansal R, Hruban RH, Kern SE. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 34.Couch FJ, Johnson MR, Rabe KG, Brune K, de Andrade M, Goggins M, Rothenmund H, Gallinger S, Klein A, Petersen GM, Hruban RH. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:342–346. doi: 10.1158/1055-9965.EPI-06-0783. [DOI] [PubMed] [Google Scholar]
- 35.Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, Taherian N, Srivastava A, Holter S, Rothenmund H, Ghadirian P, Foulkes WD, Gallinger S. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slater EP, Langer P, Niemczyk E, Strauch K, Butler J, Habbe N, Neoptolemos JP, Greenhalf W, Bartsch DK. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 37.Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71:2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 39.Moran A, O'Hara C, Khan S, Shack L, Woodward E, Maher ER, Lalloo F, Evans DG. Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam Cancer. 2011;11:235–242. doi: 10.1007/s10689-011-9506-2. [DOI] [PubMed] [Google Scholar]
- 40.Axilbund JE, Argani P, Kamiyama M, Palmisano E, Raben M, Borges M, Brune KA, Goggins M, Hruban RH, Klein AP. Absence of germline BRCA1 mutations in familial pancreatic cancer patients. Cancer Biol Ther. 2009;8:131–135. doi: 10.4161/cbt.8.2.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrone CR, Levine DA, Tang LH, Allen PJ, Jarnagin W, Brennan MF, Offit K, Robson ME. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol. 2009;27:433–438. doi: 10.1200/JCO.2008.18.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stecklein SR, Jensen RA. Identifying and exploiting defects in the Fanconi anemia/BRCA pathway in oncology. Transl Res. 2012;160:178–197. doi: 10.1016/j.trsl.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 43.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 44.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 45.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M, Carmichael J. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 46.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S, Messiou C, A'Hern R, Tutt A, Ashworth A, Stone J, Carmichael J, Schellens JH, de Bono JS, Kaye SB. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 47.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 48.Beggs AD, Latchford AR, Vasen HF, Moslein G, Alonso A, Aretz S, Bertario L, Blanco I, Bulow S, Burn J, Capella G, Colas C, Friedl W, Moller P, Hes FJ, Jarvinen H, Mecklin JP, Nagengast FM, Parc Y, Phillips RK, Hyer W, Ponz de Leon M, Renkonen-Sinisalo L, Sampson JR, Stormorken A, Tejpar S, Thomas HJ, Wijnen JT, Clark SK, Hodgson SV. Peutz-Jeghers syndrome: a systematic review and recommendations for management. Gut. 2012;59:975–986. doi: 10.1136/gut.2009.198499. [DOI] [PubMed] [Google Scholar]
- 49.Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJ, Keller JJ, Westerman AM, Scott RJ, Lim W, Trimbath JD, Giardiello FM, Gruber SB, Offerhaus GJ, de Rooij FW, Wilson JH, Hansmann A, Moslein G, Royer-Pokora B, Vogel T, Phillips RK, Spigelman AD, Houlston RS. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res. 2006;12:3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- 50.Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, Booker SV, Cruz-Correa M, Offerhaus JA. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 51.Eckerle Mize D, Bishop M, Resse E, Sluzevich J. Familial Atypical Multiple Mole Melanoma Syndrome. 2009 [PubMed] [Google Scholar]
- 52.Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, Azizi E, Bianchi-Scarra G, Bishop DT, Bressac-de Paillerets B, Bruno W, Calista D, Cannon Albright LA, Demenais F, Elder DE, Ghiorzo P, Gruis NA, Hansson J, Hogg D, Holland EA, Kanetsky PA, Kefford RF, Landi MT, Lang J, Leachman SA, Mackie RM, Magnusson V, Mann GJ, Niendorf K, Newton Bishop J, Palmer JM, Puig S, Puig-Butille JA, de Snoo FA, Stark M, Tsao H, Tucker MA, Whitaker L, Yakobson E. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66:9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 53.Lynch HT, Krush AJ. Heredity and malignant melanoma: implications for early cancer detection. Can Med Assoc J. 1968;99:17–21. [PMC free article] [PubMed] [Google Scholar]
- 54.Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH, Jr, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 55.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2011;51:14–24. doi: 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen JM, Ferec C. Genetics and pathogenesis of chronic pancreatitis: The 2012 update. Clin Res Hepatol Gastroenterol. 2012 doi: 10.1016/j.clinre.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Lowenfels AB, Maisonneuve P, DiMagno EP, Elitsur Y, Gates LK, Jr, Perrault J, Whitcomb DC. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 58.Taylor AM, Byrd PJ. Molecular pathology of ataxia telangiectasia. J Clin Pathol. 2005;58:1009–1015. doi: 10.1136/jcp.2005.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 60.Swift M, Sholman L, Perry M, Chase C. Malignant neoplasms in the families of patients with ataxia-telangiectasia. Cancer Res. 1976;36:209–215. [PubMed] [Google Scholar]
- 61.Swift M, Reitnauer PJ, Morrell D, Chase CL. Breast and other cancers in families with ataxia-telangiectasia. N Engl J Med. 1987;316:1289–1294. doi: 10.1056/NEJM198705213162101. [DOI] [PubMed] [Google Scholar]
- 62.Geoffroy-Perez B, Janin N, Ossian K, Lauge A, Croquette MF, Griscelli C, Debre M, Bressac-de-Paillerets B, Aurias A, Stoppa-Lyonnet D, Andrieu N. Cancer risk in heterozygotes for ataxia-telangiectasia. Int J Cancer. 2001;93:288–293. doi: 10.1002/ijc.1329. [DOI] [PubMed] [Google Scholar]
- 63.Athma P, Rappaport R, Swift M. Molecular genotyping shows that ataxiatelangiectasia heterozygotes are predisposed to breast cancer. Cancer Genet Cytogenet. 1996;92:130–134. doi: 10.1016/s0165-4608(96)00328-7. [DOI] [PubMed] [Google Scholar]
- 64.FitzGerald MG, Bean JM, Hegde SR, Unsal H, MacDonald DJ, Harkin DP, Finkelstein DM, Isselbacher KJ, Haber DA. Heterozygous ATM mutations do not contribute to early onset of breast cancer. Nat Genet. 1997;15:307–310. doi: 10.1038/ng0397-307. [DOI] [PubMed] [Google Scholar]
- 65.Swift M, Su Y. Link between breast cancer and ATM gene is strong. BMJ. 1999;318:400. doi: 10.1136/bmj.318.7180.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prokopcova J, Kleibl Z, Banwell CM, Pohlreich P. The role of ATM in breast cancer development. Breast Cancer Res Treat. 2007;104:121–128. doi: 10.1007/s10549-006-9406-6. [DOI] [PubMed] [Google Scholar]
- 67.Kastrinos F, Syngal S. Inherited colorectal cancer syndromes. Cancer J. 2011;17:405–415. doi: 10.1097/PPO.0b013e318237e408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galiatsatos P, Foulkes WD. Familial adenomatous polyposis. Am J Gastroenterol. 2006;101:385–398. doi: 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 69.Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kastrinos F, Mukherjee B, Tayob N, Wang F, Sparr J, Raymond VM, Bandipalliam P, Stoffel EM, Gruber SB, Syngal S. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302:1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giardiello FM, Offerhaus GJ, Lee DH, Krush AJ, Tersmette AC, Booker SV, Kelley NC, Hamilton SR. Increased risk of thyroid and pancreatic carcinoma in familial adenomatous polyposis. Gut. 1993;34:1394–1396. doi: 10.1136/gut.34.10.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morgenthaler S, Thilly WG. A strategy to discover genes that carry multiallelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.1000 Genomes Project Consortium, A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding nonsynonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 78.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaffe A, Wojcik G, Chu A, Golozar A, Maroo A, Duggal P, Klein AP. Identification of functional genetic variation in exome sequence analysis. BMC Proc. 2012;5(Suppl 9):S13. doi: 10.1186/1753-6561-5-S9-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet. 2010;19:R188–R196. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 82.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 83.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hindorff LA, Gillanders EM, Manolio TA. Genetic architecture of cancer and other complex diseases: lessons learned and future directions. Carcinogenesis. 2011;32:945–954. doi: 10.1093/carcin/bgr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]