Abstract
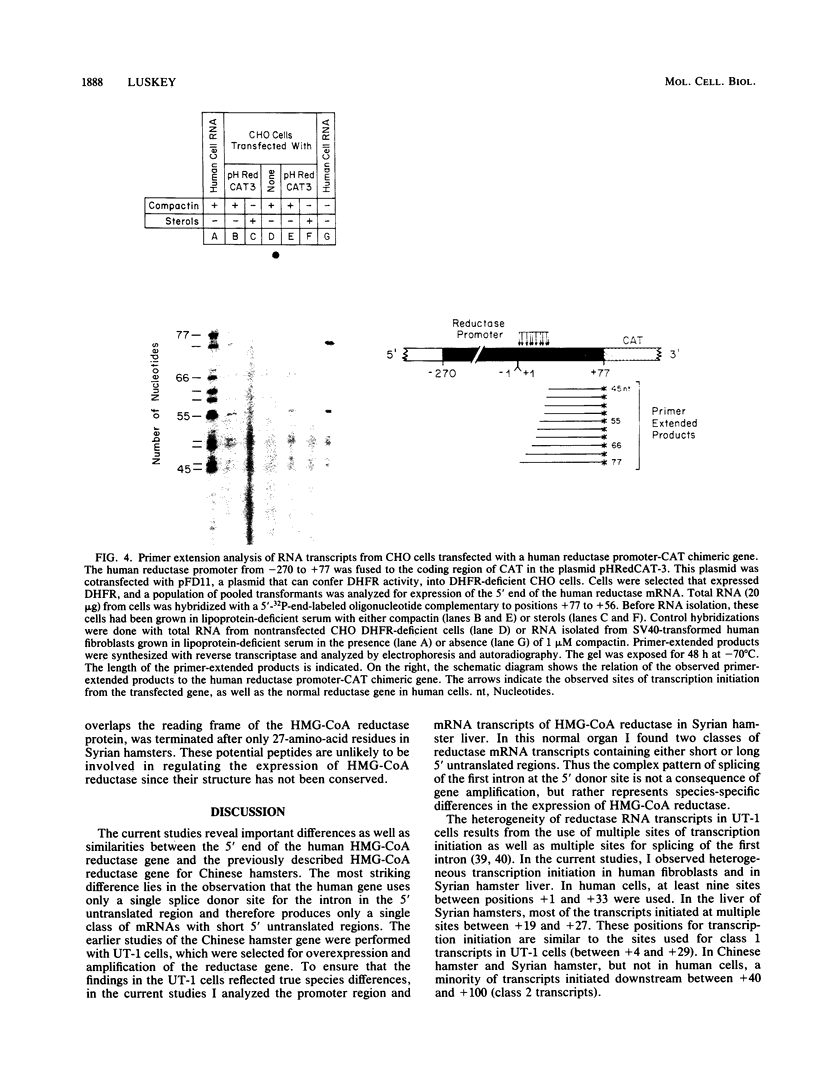
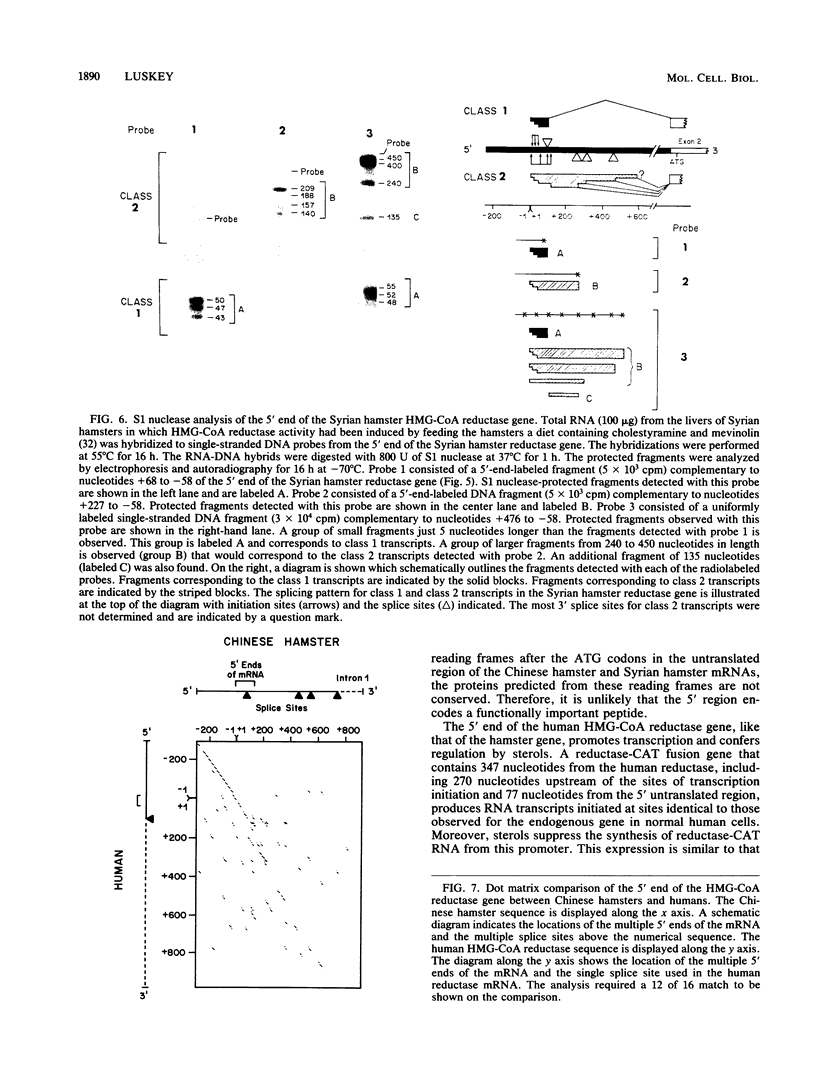
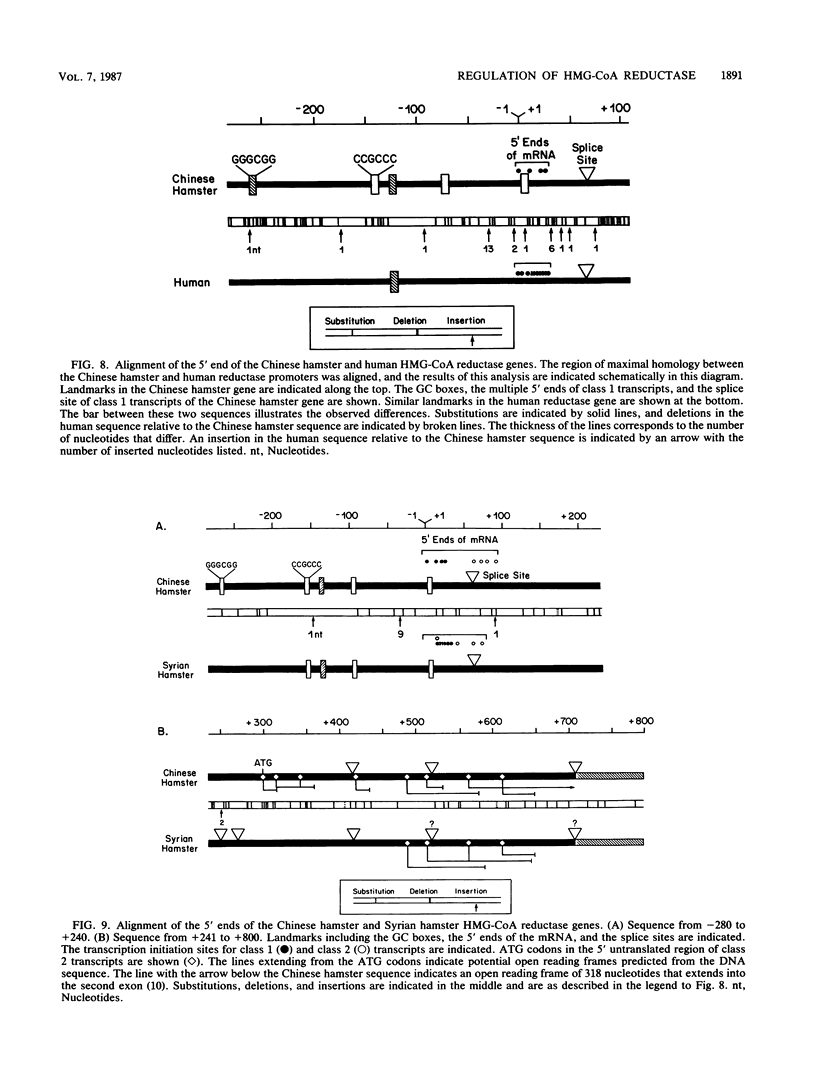
Regulation of the expression of 3-hydroxy-3-methyglutaryl coenzyme A (HMG-CoA) reductase is a critical step in controlling cholesterol synthesis. Previous studies in cultured Chinese hamster ovary cells have shown that HMG-CoA reductase is transcribed from a cholesterol-regulated promoter to yield a heterogeneous collection of mRNAs with 5' untranslated regions of 68 to 670 nucleotides in length. Synthesis of these molecules is initiated at multiple sites, and multiple donor sites are used to excise an intron in the 5' untranslated region. In the current paper, I report that human HMG-CoA reductase gene resembles the Chinese hamster gene in having multiple sites of transcription initiation that are subject to suppression by cholesterol. The human gene differs from the hamster gene in that a single donor splice site is used to excise the intron in the 5' untranslated region. All of the resulting RNAs have short 5' untranslated regions of 68 to 100 nucleotides. This difference in the splicing pattern of the first intron is species specific and not a peculiarity of cultured cells in that HMG-CoA reductase mRNAs from Syrian hamster livers resemble those of the cultured Chinese hamster ovary cells. Comparison of the DNA sequences of the HMG-CoA reductase promoters from three different species--humans, Syrian hamsters, and Chinese hamsters--shows a highly conserved region of 179 nucleotides that extends from 220 to 42 nucleotides upstream of the transcription initiation sites. This region is 88% identical between the human and Chinese hamster promoter. When fused to the coding region of the Escherichia coli chloramphenicol acetyltransferase gene, this highly conserved region of the reductase gene directs the cholesterol-regulated expression of chloramphenicol acetyltransferase in transfected hamster cells, further indicating the interspecies conservation of the regulatory elements.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basler K., Oesch B., Scott M., Westaway D., Wälchli M., Groth D. F., McKinley M. P., Prusiner S. B., Weissmann C. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 1986 Aug 1;46(3):417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Chang T. Y., Limanek J. S. Regulation of cytosolic acetoacetyl coenzyme A thiolase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and mevalonate kinase by low density lipoprotein and by 25-hydroxycholesterol in Chinese hamster ovary cells. J Biol Chem. 1980 Aug 25;255(16):7787–7795. [PubMed] [Google Scholar]
- Chen M. J., Shimada T., Moulton A. D., Cline A., Humphries R. K., Maizel J., Nienhuis A. W. The functional human dihydrofolate reductase gene. J Biol Chem. 1984 Mar 25;259(6):3933–3943. [PubMed] [Google Scholar]
- Chin D. J., Gil G., Faust J. R., Goldstein J. L., Brown M. S., Luskey K. L. Sterols accelerate degradation of hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase encoded by a constitutively expressed cDNA. Mol Cell Biol. 1985 Apr;5(4):634–641. doi: 10.1128/mcb.5.4.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984 Apr 12;308(5960):613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Faust J. R., MacDonald R. J., Brown M. S., Goldstein J. L. Molecular cloning of 3-hydroxy-3-methylglutaryl coenzyme a reductase and evidence for regulation of its mRNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7704–7708. doi: 10.1073/pnas.79.24.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. F., Fogelman A. M., Edwards P. A. Transcriptional regulation of the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene in rat liver. J Biol Chem. 1985 Nov 15;260(26):14363–14367. [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J Biol Chem. 1983 Jun 25;258(12):7272–7275. [PubMed] [Google Scholar]
- Endo A., Kuroda M., Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976 Dec 31;72(2):323–326. doi: 10.1016/0014-5793(76)80996-9. [DOI] [PubMed] [Google Scholar]
- Faust J. R., Luskey K. L., Chin D. J., Goldstein J. L., Brown M. S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5205–5209. doi: 10.1073/pnas.79.17.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Gil G., Brown M. S., Goldstein J. L. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster. II. Isolation of the gene and characterization of the 5' flanking region. J Biol Chem. 1986 Mar 15;261(8):3717–3724. [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985 May;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Basu S. K., Brown M. S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeman E. C., Endo A., Simoni R. D. Effects of compactin on the levels of 3-hydroxy-3-methylglutaryl coenzyme A reductase in compactin-resistant C100 and wild-type cells. Arch Biochem Biophys. 1984 Aug 1;232(2):549–561. doi: 10.1016/0003-9861(84)90572-1. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Moores J. C., David D., Respess J. G., Jolly D. J., Friedmann T. The organization of the human HPRT gene. Nucleic Acids Res. 1986 Apr 11;14(7):3103–3118. doi: 10.1093/nar/14.7.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lawn R. M., Fritsch E. F., Parker R. C., Blake G., Maniatis T. The isolation and characterization of linked delta- and beta-globin genes from a cloned library of human DNA. Cell. 1978 Dec;15(4):1157–1174. doi: 10.1016/0092-8674(78)90043-0. [DOI] [PubMed] [Google Scholar]
- Lindgren V., Luskey K. L., Russell D. W., Francke U. Human genes involved in cholesterol metabolism: chromosomal mapping of the loci for the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase with cDNA probes. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8567–8571. doi: 10.1073/pnas.82.24.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskey K. L., Faust J. R., Chin D. J., Brown M. S., Goldstein J. L. Amplification of the gene for 3-hydroxy-3-methylglutaryl coenzyme A reductase, but not for the 53-kDa protein, in UT-1 cells. J Biol Chem. 1983 Jul 10;258(13):8462–8469. [PubMed] [Google Scholar]
- Luskey K. L., Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J Biol Chem. 1985 Aug 25;260(18):10271–10277. [PubMed] [Google Scholar]
- Ma P. T., Gil G., Südhof T. C., Bilheimer D. W., Goldstein J. L., Brown M. S. Mevinolin, an inhibitor of cholesterol synthesis, induces mRNA for low density lipoprotein receptor in livers of hamsters and rabbits. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8370–8374. doi: 10.1073/pnas.83.21.8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. W., McEwan C., McKie A. B., Reid A. M. Expression of the mouse HPRT gene: deletional analysis of the promoter region of an X-chromosome linked housekeeping gene. Cell. 1986 Jan 31;44(2):319–328. doi: 10.1016/0092-8674(86)90766-x. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne T. F., Goldstein J. L., Brown M. S. 5' end of HMG CoA reductase gene contains sequences responsible for cholesterol-mediated inhibition of transcription. Cell. 1985 Aug;42(1):203–212. doi: 10.1016/s0092-8674(85)80116-1. [DOI] [PubMed] [Google Scholar]
- Patel P. I., Framson P. E., Caskey C. T., Chinault A. C. Fine structure of the human hypoxanthine phosphoribosyltransferase gene. Mol Cell Biol. 1986 Feb;6(2):393–403. doi: 10.1128/mcb.6.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Goldstein J. L., Brown M. S. Multiple mRNAs for 3-hydroxy-3-methylglutaryl coenzyme A reductase determined by multiple transcription initiation sites and intron splicing sites in the 5'-untranslated region. J Biol Chem. 1985 Aug 25;260(18):10369–10377. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Isolation and expression of an altered mouse dihydrofolate reductase cDNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2495–2499. doi: 10.1073/pnas.80.9.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J., Keith D. H., Tani K., Simmer R. L., Shively L., Lindsay S., Yoshida A., Riggs A. D. Sequence of the promoter region of the gene for human X-linked 3-phosphoglycerate kinase. Gene. 1984 Dec;32(3):409–417. doi: 10.1016/0378-1119(84)90016-7. [DOI] [PubMed] [Google Scholar]
- Skalnik D. G., Simoni R. D. The nucleotide sequence of Syrian hamster HMG-CoA reductase cDNA. DNA. 1985 Dec;4(6):439–444. doi: 10.1089/dna.1985.4.439. [DOI] [PubMed] [Google Scholar]
- Südhof T. C., Goldstein J. L., Brown M. S., Russell D. W. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985 May 17;228(4701):815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Chasin L. A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]