Abstract
Background: Macronutrient intake varies substantially between individuals, and there is evidence that this variation is partly accounted for by genetic variants.
Objective: The objective of the study was to identify common genetic variants that are associated with macronutrient intake.
Design: We performed 2-stage genome-wide association (GWA) meta-analysis of macronutrient intake in populations of European descent. Macronutrients were assessed by using food-frequency questionnaires and analyzed as percentages of total energy consumption from total fat, protein, and carbohydrate. From the discovery GWA (n = 38,360), 35 independent loci associated with macronutrient intake at P < 5 × 10−6 were identified and taken forward to replication in 3 additional cohorts (n = 33,533) from the DietGen Consortium. For one locus, fat mass obesity-associated protein (FTO), cohorts with Illumina MetaboChip genotype data (n = 7724) provided additional replication data.
Results: A variant in the chromosome 19 locus (rs838145) was associated with higher carbohydrate (β ± SE: 0.25 ± 0.04%; P = 1.68 × 10−8) and lower fat (β ± SE: −0.21 ± 0.04%; P = 1.57 × 10−9) consumption. A candidate gene in this region, fibroblast growth factor 21 (FGF21), encodes a fibroblast growth factor involved in glucose and lipid metabolism. The variants in this locus were associated with circulating FGF21 protein concentrations (P < 0.05) but not mRNA concentrations in blood or brain. The body mass index (BMI)–increasing allele of the FTO variant (rs1421085) was associated with higher protein intake (β ± SE: 0.10 ± 0.02%; P = 9.96 × 10−10), independent of BMI (after adjustment for BMI, β ± SE: 0.08 ± 0.02%; P = 3.15 × 10−7).
Conclusion: Our results indicate that variants in genes involved in nutrient metabolism and obesity are associated with macronutrient consumption in humans. Trials related to this study were registered at clinicaltrials.gov as NCT00005131 (Atherosclerosis Risk in Communities), NCT00005133 (Cardiovascular Health Study), NCT00005136 (Family Heart Study), NCT00005121 (Framingham Heart Study), NCT00083369 (Genetic and Environmental Determinants of Triglycerides), NCT01331512 (InCHIANTI Study), and NCT00005487 (Multi-Ethnic Study of Atherosclerosis).
INTRODUCTION
Considerable variation in dietary choices exists across individuals. Human eating behavior is driven by many psychological and social factors, including culture, economics, and health beliefs. There is evidence from twin and family studies that consumption of major macronutrients has a genetic component, with estimated heritability ranging from 8% to 70% (1). Importantly, the specific genes that might underlie these associations have yet to be elucidated. The identification of genetic loci underlying macronutrient intake could provide insight into the biology of human dietary behaviors. Genome-wide linkage studies have identified several chromosomal regions for macronutrient intake (2–5). Some notable candidate genes within the linkage region include pro-opiomelanocortin (POMC), adiponectin (ADIPOQ), melanocortin receptor 4 (MC4R), and peroxisome proliferator-activated receptor γ (PPARγ). Furthermore, candidate gene association studies have focused primarily on genes involved in the central control of food intake, such as dopamine receptor (DRD2) (6, 7) and serotonin receptor (HTR2A) (8, 9), and the obesity genes fat mass obesity-associated protein (FTO) (10–13) and MC4R (11, 13, 14). Many of the results are inconsistent or lack replication to confirm the initial findings. The disparate findings between the studies may be a result of the differences in the study population, dietary assessment methods, or statistical methods used.
To identify common genetic variants associated with macronutrient intake, we conducted a meta-analysis of genome-wide association (GWA)5 analyses of carbohydrate, protein, and total fat intake by using data from 12 discovery cohorts (n = 38,360) from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Nutrition Working Group (15). The top signals from this analysis were combined in a joint analysis with data from a second macronutrient meta-analysis of data from 3 cohorts (n = 33,533) from the DietGen Consortium (16). For single nucleotide polymorphisms (SNPs) that were directly genotyped on the Illumina MetaboChip, data from an additional 3 cohorts (n = 7724) were used as replication data.
SUBJECTS AND METHODS
GWA cohorts
GWA was conducted in 37,537 subjects from the following 12 cohorts from the CHARGE Consortium Nutrition Working Group: the Atherosclerosis Risk in Communities Study; the Cardiovascular Health Study; the European Prospective Investigation into Cancer and Nutrition–Norfolk (EPIC-Norfolk); the Family Heart Study; the Fenland Study; the Framingham Heart Study; the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study; the Health, Aging, and Body Composition (Health ABC) Study; the InCHIANTI Study; the Multi-Ethnic Study of Atherosclerosis (MESA); the Rotterdam Study; and the Young Finns Study. Each cohort's study protocol was reviewed and approved by their respective institutional review board (see Supplemental Table S1 under “Supplemental data” in the online issue).
Stage 2 replication MetaboChip cohorts
Data from the Gene-Lifestyle interactions And Complex traits Involved in Elevated disease Risk (GLACIER) Study from Sweden, the Malmö Diet and Cancer Study from Sweden, and The Hellenic study of Interactions between Snps and Eating in Atherosclerosis Susceptibility (n = 7724) were used to replicate SNPs identified by stage 1 GWA that was genotyped by the MetaboChip.
Stage 2 replication GWA cohorts
A second GWA meta-analysis of macronutrient intake was conducted in parallel by the DietGen Consortium [n = 33,533 (16)]. The consortium is composed of 3 US population–based cohorts: the Health Professionals Follow-Up Study (17), the Nurses’ Health Study (18), and the Women's Genome Health Study (19). The DietGen Consortium performed GWA analysis by using the same methods as the CHARGE Consortium.
Assessment of macronutrient intake
Average dietary intake was assessed by using food-frequency questionnaires (FFQs). The type of FFQ used in each study varied slightly, with this variation generally designed to best capture the dietary habits of the population under study (see Supplemental Table S2 under “Supplemental data” in the online issue). Among these various CHARGE cohorts, 87% of the FFQs had been validated against other dietary assessment methods. On the basis of the responses to each FFQ and study-specific nutrient databases, usual nutrient consumption was calculated. The present analysis focused on the percentage of energy from total fat, protein, and carbohydrate intake.
Heritability estimates
In the Framingham Heart Study and the Family Heart Study, the heritability of macronutrient adjusted for age and sex was estimated by using the variance components method in Sequential Oligogenic Linkage Analysis Routines (SOLAR; Texas Biomedical Research Institute).
Genotyping and GWA analysis
Genome-wide genotyping was conducted by using Affymetrix or Illumina platforms. Each study performed quality control for genotyped SNPs on the basis of minor allele frequency, call rate, and departure from Hardy-Weinberg equilibrium (see Supplemental Table S3 under “Supplemental data” in the online issue). Phased haplotypes from HapMap CEU (build 35 or 36) were used to impute ∼2.5 million autosomal SNPs by using a Hidden Markov model algorithm implemented in MACH (20), IMPUTE (21), or BimBam (22). Study-specific GWA analyses were conducted for each macronutrient by using genotyped and imputed SNP dosages assuming an additive genetic model. The base model included age and sex for all studies and study-specific covariates (eg, study site) and population stratification principal components when applicable (model 1). In a second model (model 2), BMI was added to the covariates in model 1. SNPs with low minor allele frequency (<1%), low imputation quality (MACH: Rsq < 0.3; or IMPUTE: proper info < 0.4) were removed. The results from each study were combined in a fixed-effects meta-analysis with inverse variance weights by using METAL software (23). To account for population stratification, the association results from individual studies as well as meta-analyses were adjusted for genomic control.
To increase the power to detect significant associations, a joint analysis with results from the DietGen Consortium (n = 33,533) was conducted for 35 independent genomic regions containing SNPs with P < 10−6, a threshold that was selected on the basis of the deviation of observed P values from the expected (see Supplemental Figure S1 under “Supplemental data” in the online issue). The tag SNPs for each genomic region were selected by using HapMap CEU (release rel#23a NCBI B36) linkage disequilibrium structure with a cutoff of r2 > 0.5 within a 250-kb window. Of the top 35 independent loci, the top SNP in the FTO locus was evaluated in a second replication group (n = 7724) with MetaboChip data. The results from the 2 genome-wide meta-analyses and MetaboChip data were combined by using a fixed-effects inverse variance–weighted meta-analysis. For the joint analysis, genome-wide significance was considered at the Bonferroni-corrected threshold of P < 5 × 10−8.
Expression quantitative trait loci analysis
Proxy SNPs (D′ > 0.8 or r2 > 0.8) of the top SNPs (rs838145) on 19q13.33 were queried in expression SNP databases that included results from the following tissues: fresh lymphocytes (24, 25), fresh leukocytes (26), peripheral blood monocytes, fresh leukocytes (26), omental and subcutaneous adipose tissue (27, 28), whole-blood samples (27, 29), and liver (28, 30, 31).
Assessment of fibroblast growth factor 21 protein concentrations
In participants in the Baltimore Longitudinal Study on Aging, serum fibroblast growth factor 21 (FGF21) was measured with ELISA (Quantikine Human FGF-21 ELISA; R&D Systems) by using samples that were stored continuously at −70°C until the time of analysis of FGF21. The samples were analyzed at Johns Hopkins Hospital in the laboratory of one of the investigators (RDS). The intra- and interassay CVs were 2.8% and 7.0%, respectively. The assay has a minimum limit of detection of 4.6 pg/mL. No samples were below the lower limit of detection.
RESULTS
The mean macronutrient intake assessed through FFQs in the 12 discovery GWA studies were similar across studies and were, on average, 17 ± 1.3%, 33 ± 2.5%, and 48 ± 2.9% for protein, fat, and carbohydrate, respectively (see Supplemental Table S4 under “Supplemental data” in the online issue). Heritability estimates for protein, carbohydrate, and fat intake were similar in the 2 cohorts evaluated: 17 ± 0.02%, 20 ± 0.03%, and 20 ± 0.03%, respectively, in the Family Heart Study; 17 ± 0.02%, 20 ± 0.02%, and 23 ± 0.03%, respectively, in the Framingham Heart Study.
The meta-analysis of stage 1 discovery genome scans of macronutrient intake showed modest deviation from the expected distribution of P values under the null hypothesis, particularly for protein intake (see Supplemental Figure S1 under “Supplemental data” in the online issue). The genomic control values for the meta-analysis of carbohydrate, fat, and protein intake were 1.04, 1.04, and 1.06, respectively, for the model adjusted for age and sex (base model) and 1.04, 1.05, and 1.05, respectively, for the BMI-adjusted model. Genome-wide significant associations were observed with a locus in an ∼7-kb region on 4q28, near the mastermind-like 3 (MAML3) gene for protein intake (Table 1; see Supplemental Table S5 under “Supplemental data” in the online issue). The strongest association was observed for rs1350036 where the minor allele was associated with lower protein intake (βCHARGE ± SE: −0.14 ± 0.02%; P = 1.72 × 10−8). The signal remained significant after adjustment for BMI (βCHARGE ± SE: −0.15 ± 0.02%; P = 6.60 × 10−9). No genome-wide significant associations were observed for carbohydrate and fat intake.
TABLE 1.
Summary of GWA meta-analysis and joint analysis of top loci1
| SNP | Chr | Allele (effect/noneffect) | Allele frequency (effect) | CHARGE |
DietGen |
CHARGE + DietGen |
CHARGE + DietGen (BMI adjusted) |
||||
| β ± SE | P value | β ± SE | P value | β ± SE | P value | β ± SE | P value | ||||
| Carbohydrate | |||||||||||
| rs1667320 | 2 | G/T | 0.49 (0.01) | −0.31 ± 0.06 | 1.25 × 10−7 | 0.02 ± 0.06 | 0.8147 | −0.16 ± 0.04 | 0.0002 | −0.14 ± 0.04 | 0.0012 |
| rs2840445 | 8 | A/G | 0.27 (0.02) | −0.32 ± 0.06 | 7.55 × 10−7 | −0.10 ± 0.07 | 0.1781 | −0.22 ± 0.05 | 4.52 × 10−6 | −0.20 ± 0.05 | 5.78 × 10−5 |
| rs12122737 | 1 | A/C | 0.20 (0.02) | 0.35 ± 0.07 | 1.64 × 10−6 | −0.14 ± 0.08 | 0.0871 | 0.13 ± 0.05 | 0.0148 | 0.12 ± 0.05 | 0.0247 |
| rs12356048 | 10 | G/A | 0.32 (0.02) | 0.30 ± 0.06 | 2.44 × 10−6 | −0.10 ± 0.07 | 0.1669 | 0.12 ± 0.05 | 0.0108 | 0.08 ± 0.05 | 0.1034 |
| rs12912584 | 15 | A/G | 0.25 (0.02) | 0.31 ± 0.07 | 3.99 × 10−6 | −0.10 ± 0.07 | 0.1814 | 0.13 ± 0.05 | 0.0117 | 0.10 ± 0.05 | 0.0353 |
| rs1542608 | 7 | G/A | 0.18 (0.02) | 0.37 ± 0.08 | 4.23 × 10−6 | 0.00 ± 0.08 | 0.9857 | 0.19 ± 0.06 | 0.0008 | 0.17 ± 0.06 | 0.0035 |
| rs10508303 | 10 | A/C | 0.07 (0.02) | 0.60 ± 0.13 | 4.82 × 10−6 | −0.18 ± 0.13 | 0.1644 | 0.20 ± 0.09 | 0.0270 | 0.19 ± 0.09 | 0.0328 |
| rs1893113 | 6 | T/C | 0.50 (0.01) | 0.27 ± 0.06 | 5.19 × 10−6 | −0.01 ± 0.06 | 0.8810 | 0.14 ± 0.04 | 0.0011 | 0.12 ± 0.04 | 0.0045 |
| rs8019546 | 14 | A/G | 0.30 (0.01) | 0.29 ± 0.06 | 5.93 × 10−6 | 0.13 ± 0.07 | 0.0750 | 0.22 ± 0.05 | 5.01 × 10−6 | 0.19 ± 0.05 | 5.14 × 10−5 |
| rs838145 | 19 | G/A | 0.46 (0.04) | 0.27 ± 0.06 | 6.21 × 10−6 | 0.17 ± 0.07 | 0.0083 | 0.23 ± 0.04 | 3.13 × 10−7 | 0.25 ± 0.04 | 1.68 × 10−8 |
| rs1549309 | 5 | A/G | 0.17 (0.02) | 0.34 ± 0.07 | 9.50 × 10−6 | 0.18 ± 0.08 | 0.0350 | 0.27 ± 0.06 | 2.47 × 10−6 | 0.25 ± 0.06 | 1.59 × 10−5 |
| Protein | |||||||||||
| rs1350036 | 4 | T/C | 0.48 (0.03) | −0.14 ± 0.02 | 1.72 × 10−8 | −0.01 ± 0.02 | 0.7601 | −0.07 ± 0.02 | 3.49 × 10−5 | −0.07 ± 0.02 | 2.19 × 10−5 |
| rs1421085 | 16 | C/T | 0.42 (0.02) | 0.12 ± 0.03 | 1.53 × 10−6 | 0.10 ± 0.03 | 0.0002 | 0.11 ± 0.022 | 1.65 × 10−9 | 0.09 ± 0.023 | 4.80 × 10−7 |
| rs1191005 | 14 | T/C | 0.12 (0.01) | 0.19 ± 0.04 | 1.59 × 10−6 | −0.02 ± 0.04 | 0.5944 | 0.08 ± 0.03 | 0.0042 | 0.08 ± 0.03 | 0.0042 |
| rs9553939 | 13 | G/A | 0.43 (0.02) | −0.12 ± 0.03 | 3.60 × 10−6 | −0.02 ± 0.03 | 0.4968 | −0.07 ± 0.02 | 0.0002 | −0.07 ± 0.02 | 0.0003 |
| rs11640535 | 16 | A/G | 0.27 (0.03) | 0.14 ± 0.03 | 3.72 × 10−6 | 0.00 ± 0.03 | 0.8746 | 0.06 ± 0.02 | 0.0019 | 0.07 ± 0.02 | 0.0002 |
| rs16993715 | 21 | T/C | 0.04 (0.01) | 0.32 ± 0.07 | 8.17 × 10−6 | 0.05 ± 0.07 | 0.4621 | 0.18 ± 0.05 | 0.0003 | 0.17 ± 0.05 | 0.0007 |
| rs11699457 | 20 | A/G | 0.19 (0.01) | 0.14 ± 0.03 | 9.08 × 10−6 | 0.03 ± 0.03 | 0.3966 | 0.08 ± 0.02 | 0.0002 | 0.08 ± 0.02 | 0.0003 |
| rs7086704 | 10 | G/T | 0.25 (0.02) | −0.13 ± 0.03 | 9.61 × 10−6 | −0.01 ± 0.03 | 0.6798 | −0.07 ± 0.02 | 0.0008 | −0.05 ± 0.02 | 0.0091 |
| Fat | |||||||||||
| rs7625360 | 3 | C/A | 0.24 (0.01) | 0.30 ± 0.06 | 1.77 × 10−7 | 0.02 ± 0.06 | 0.7715 | 0.16 ± 0.04 | 9.78 × 10−5 | 0.14 ± 0.04 | 0.0006 |
| rs838145 | 19 | G/A | 0.46 (0.04) | −0.25 ± 0.05 | 3.43 × 10−7 | −0.19 ± 0.05 | 0.0002 | −0.22 ± 0.04 | 4.48 × 10−10 | −0.21 ± 0.04 | 1.57 × 10−9 |
| rs2609186 | 2 | A/G | 0.31 (0.05) | −0.25 ± 0.05 | 9.93 × 10−7 | −0.01 ± 0.05 | 0.8458 | −0.14 ± 0.04 | 0.0002 | −0.13 ± 0.04 | 0.0007 |
| rs7997717 | 13 | A/G | 0.05 (0.01) | −0.55 ± 0.11 | 2.99 × 10−6 | 0.03 ± 0.12 | 0.8167 | −0.27 ± 0.08 | 0.0014 | −0.28 ± 0.08 | 0.0009 |
| rs12156272 | 8 | C/A | 0.17 (0.01) | −0.28 ± 0.06 | 5.59 × 10−6 | −0.02 ± 0.06 | 0.7255 | −0.16 ± 0.04 | 0.0004 | −0.16 ± 0.04 | 0.0003 |
| rs3114727 | 7 | A/G | 0.40 (0.01) | 0.22 ± 0.05 | 5.76 × 10−6 | 0.03 ± 0.05 | 0.5503 | 0.13 ± 0.03 | 0.0002 | 0.13 ± 0.03 | 0.0001 |
| rs11725988 | 4 | G/A | 0.26 (0.02) | 0.26 ± 0.06 | 6.80 × 10−6 | 0.02 ± 0.06 | 0.7473 | 0.14 ± 0.04 | 0.0006 | 0.14 ± 0.04 | 0.0007 |
| rs4683038 | 3 | T/G | 0.27 (0.02) | 0.25 ± 0.05 | 7.78 × 10−6 | −0.02 ± 0.06 | 0.7704 | 0.12 ± 0.04 | 0.0026 | 0.11 ± 0.04 | 0.0052 |
| rs10152629 | 15 | A/G | 0.02 (0.01) | 0.80 ± 0.18 | 8.47 × 10−6 | −0.12 ± 0.19 | 0.5250 | 0.37 ± 0.13 | 0.0045 | 0.36 ± 0.13 | 0.0066 |
All analyses were adjusted for age, sex and study sex and study-specific covariates (eg, study site, population stratification principal components when applicable) (P < 10−6). CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; Chr, chromosome; GWA, genome-wide association; SNP, single nucleotide polymorphism.
Meta-analysis with 3 cohorts with Illumina MetaboChip data (n = 7724): 2β ± SE = 0.10 ± 0.02%, P = 9.96 × 10−10; 3β ± SE = 0.08 ± 0.02%, P = 3.15 × 10−7.
From stage 1 analysis, we took forward the SNPs that represented the 35 most significant loci (P < 10−6) in either the base model (Table 1) or the BMI-adjusted model (see Supplemental Table S5 under “Supplemental data” in the online issue). These SNPs were analyzed jointly with data from the DietGen Consortium and 3 studies with MetaboChip data for the FTO (rs1421085) locus because this SNP was directly genotyped on the chip. In joint meta-analyses, genome-wide significant associations were observed for rs1421085 in the FTO locus on chromosome 16 in the model 1 for protein intake (Table 1), where the BMI-increasing minor allele was associated with higher protein intake (βJoint ± SE: 0.10 ± 0.02%; P = 9.96 × 10−10). In the BMI-adjusted model, a small reduction in the strength of association was observed (βJoint ± SE: 0.08 ± 0.02%; P = 3.15 × 10−7).
Genome-wide significant associations were also observed on 19q13.33 for both fat and carbohydrate intake in the joint analysis (Table 1). The minor allele of rs838145 was associated with a higher percentage of energy intake from carbohydrate for the base model (βJoint ± SE: 0.23 ± 0.04%; P = 3.13 × 10−7) and for the BMI-adjusted model (βJoint ± SE: 0.25 ± 0.04%; P = 1.7 × 10−8) and a lower percentage of energy intake from fat for the base model (βJoint ± SE: −0.22 ± 0.04%; P = 4.48 × 10−10) and for the BMI-adjusted model (βJoint ± SE: −0.21 ± 0.04%; P = 1.57 × 10−9).
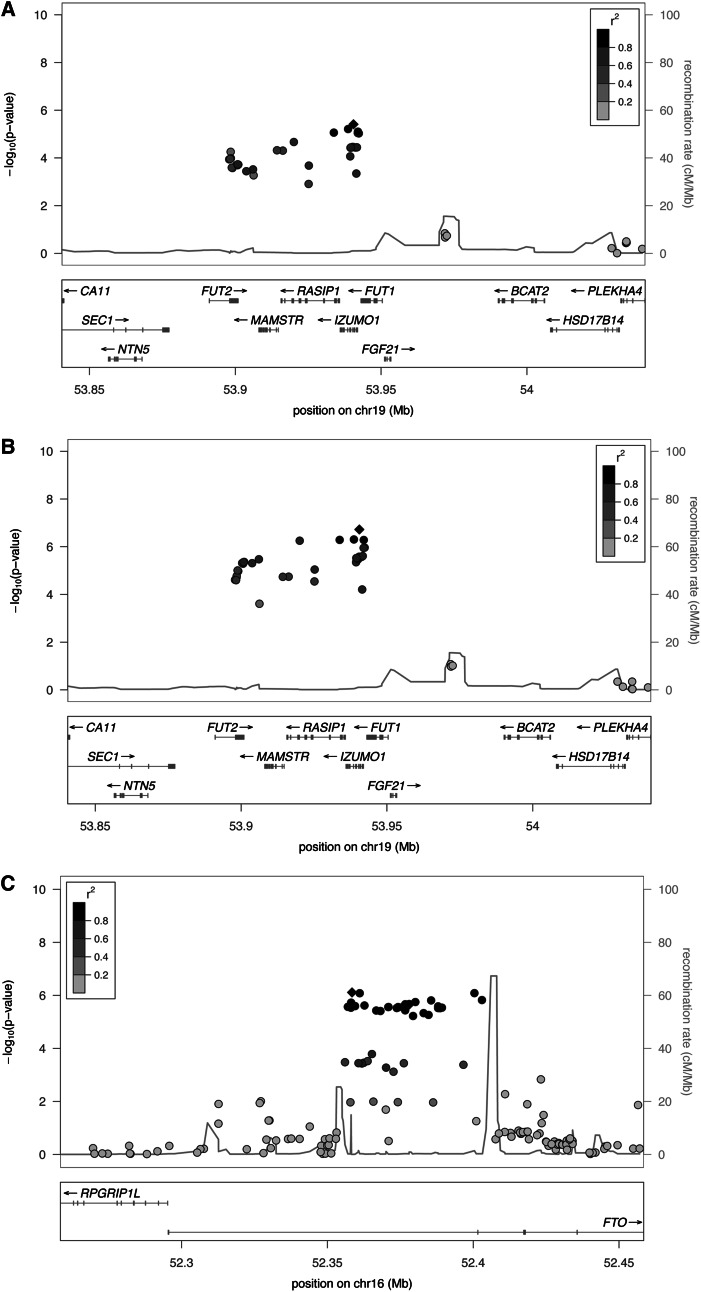
We identified FGF21 as a candidate gene in the 19q13.33 region, which encodes a hormone that regulates glucose and lipid metabolism (Figure 1, ). To investigate whether FGF21 could be the gene underlying this signal, we explored the association of rs838145 with the expression of the FGF21 gene and protein. First, we used an existing expression quantitative trait loci database of cis-gene expression with the use of data from genome-wide expression analysis in relevant tissues that express FGF21 including liver, adipose, and blood (see Supplemental Table S6 under “Supplemental data” in the online issue). Data show the expression of multiple genes in association with SNPs in high linkage disequilibrium with rs838145; however, the expression of FGF21 was not supported by the data. Second, to assess the association of rs838145 with circulating concentrations of FGF21 protein, we used plasma FGF21 concentrations in 377 individuals in the Baltimore Longitudinal Study on Aging. The minor allele of rs838145, which was associated with a higher percentage of energy from carbohydrate and a lower percentage of energy from fat, was significantly associated with higher FGF21 protein concentrations (β ± SE: 36 ± 17 pg/mL; P = 0.01), after adjustment for age and sex. This association remained significant after further adjusting the analyses for BMI (β ± SE: 32 ± 17 pg/mL; P = 0.02).
FIGURE 1.
Regional association plot of 19q13.33 and the FTO locus region. The panels show –log10 P values for SNPs that passed quality control from the stage 1 meta-analysis of intakes of carbohydrate (A), fat (B), and protein (C) as percentages of total energy adjusted for age, sex, and study-specific covariates (eg, study site, population-stratification principal components when applicable). The SNPs shown are those within 1 Mb of the index SNP: rs8183145 on chromosome 19 (A, B) and rs1421085 on chromosome 16 (C). The degree of linkage disequilibrium (r2) is shown from low (<0.2; light gray) to high (≥0.8; black). chr, chromosome; cM, centimorgan; Mb, mega base pair; SNP, single nucleotide polymorphism.
The SNP (rs1350036) in the 4q28 region that reached genome-wide significance in stage 1 for protein intake was not replicated in stage 2 and was not significant in the joint analysis for the base model (βJoint ± SE: −0.07 ± 0.02%; P = 3.49 × 10−5; Table 1) or in the BMI-adjusted model (βJoint ± SE: −0.07 ± 0.02%; P = 2.19 × 10−5).
DISCUSSION
In this study, we identified genome-wide significant associations of a locus on 19q13.33 for carbohydrate and fat intake, with reciprocal directions of association, and with the FTO locus for protein intake. FTO was one of the first BMI loci to be identified through GWA scans and has been consistently associated with obesity in various cohorts (32, 33). The association of variants in FTO with protein intake in our study was largely unchanged after adjustment for BMI, suggesting that this association is not mediated by BMI. The FTO SNP reported in this analysis (rs1421085) is in high linkage disequilibrium (r2 > 0.97) with SNPs showing the strongest association with BMI (rs993609, rs9930506) (32–34). Previous studies have investigated the association of FTO variants with food intake (35–37), hypothesizing an effect through the regulation of hunger and satiety (38). The focus of these studies was primarily on total energy intake rather than on specific macronutrients. Overall, the evidence was weak, but our findings highlight the importance of further investigation to clarify the interrelations of the FTO gene, macronutrient and energy intake, and body weight.
It is notable that, with the exception of FTO, other loci identified in previous linkage analyses (2–5) and candidate gene association studies (6–9, 11, 13, 14) of macronutrient intake were not among the top signals in our study. There have been 4 linkage studies of macronutrient intake, and although each study identified significant linkage regions, there were none that were consistent across the studies (2–5). It is possible that there are macronutrient loci that are specific to the families in each study. However, discrepancies may also be a result of differences in study population, methods for dietary assessment, or statistical methods used.
We report novel associations of a locus on 19q13.33 with higher carbohydrate and lower fat consumption, independent of BMI. One gene in this region with potential links to macronutrient intake was FGF21, which encodes a hormone produced primarily in the liver that is an important regulator of glucose and lipid metabolism (39, 40). In vitro, FGF21 promotes insulin-independent glucose uptake through the transcription of GLUT1 in rodent and human adipocytes (41). Pharmacologic doses of FGF21 improve glucose clearance and insulin sensitivity and lower plasma triglycerides and free fatty acids in diabetic and obese animal models (41–44). FGF21 also plays a critical role in regulating metabolic adaptation in nutritional states in which the primary source of energy is derived from fatty acids. During prolonged starvation or consumption of a ketogenic diet (high fat, low carbohydrate), FGF21 increases lipolysis in adipose tissue and increases ketogenesis and β-oxidation in the liver (45, 46). In humans, the role of FGF21 is not well defined. Some reports have linked FGF21 with BMI and other metabolic variables, but results are not consistent (47–49). In our study, we observed a significant association between the 19q13.33 variant with circulating protein concentrations of FGF21 but not with gene expression levels in liver, fat, and blood tissues. If FGF21 is the gene underlying the signal on 19q13.33, it is possible that there is a metabolic shift in glucose and fat metabolism, such that carriers of the minor allele preferentially consume more energy from carbohydrate and less from fat.
This study, along with the DietGen Consortium meta-analysis (16), is the largest GWA study of macronutrient intake in European ancestry cohorts. Interestingly, both studies identified associations with the locus on 19q13.33 but for different macronutrients. The DietGen Consortium observed a genome-wide significant association of 19q13.33 with protein intake, whereas we found associations with fat and carbohydrate intake. Although the reasons for these differences are unclear, it is possible that this locus is not specific to intake of any single macronutrient. Total dietary energy is derived from different macronutrients; therefore, when there is a change in the percentage of energy from one macronutrient, a change in one or both of the other macronutrients may occur in the opposite direction. Thus, the results of the 2 studies suggest that this locus is associated with dietary macronutrient composition rather than with the consumption of a specific macronutrient.
Although the associations reported in this study are significant, the effects are small in magnitude. These estimates may be underestimated because of measurement errors of macronutrient intake from FFQs. The FFQ is a commonly used dietary assessment tool in epidemiologic studies because it is feasible to use in large studies and accurately rank-orders individuals across the spectra of foods and nutrients; however, the validity of estimates varies across nutrients and populations. Of the cohorts involved in the CHARGE discovery meta-analysis, 10 of 12 used validated FFQs. Nevertheless, the clinical significance of differences in macronutrient intake of ∼0.2%, even when extended over a lifetime, is not clear. In this light, the size and scope of our study are important strengths to detect such a modest impact of genetics, and our findings are most relevant to elucidate the potential underlying biology of very complex human dietary behaviors. Given the complexity of the investigated trait, it is likely that other as yet unidentified loci, as well as other genetic differences such as copy number variants, contribute to macronutrient intake. Furthermore, replication of these findings in non-European cohorts will be needed to determine whether the identified loci are associated with macronutrient intake in other populations.
In conclusion, we report associations of a locus on chromosome 19 with carbohydrate and fat intake and of the FTO locus with protein intake. We also propose FGF21 as one strong candidate for the former association, and our findings support the need for further functional assessment and fine mapping of 19q13.33. Our results support a role of common genetic variants on macronutrient consumption in humans.
Acknowledgments
We thank Andrew Plump (Cardiovascular Diseases, Merck Research Laboratory, Rahway, NJ) for his contribution to the expression quantitative trait loci data collection.
The authors’ responsibilities were as follows—KEN, DSS, KTK, RJFL, IB, IBB, NGF, NJW, LAC, PWF, DKA, QL, DIC, SBK, SB, LF, JIR, JAN, AH, OHF, GD, JV, MKW, RO, and AJ: study concept and design; JAS, RDS, DS, DSS, K-TK, MFF, IBB, NGF, NJW, NMM, IJ, GH, EJD, DKA, DKH, SBK, SB, LF, JIR, FJAvR, AGU, JCW, MD, IPK, GD, VM, and VE: phenotype data collection; KEN, RJFL, IB, IBB, ZY, NGF, CL, NJW, FBH, JMO, KKL, YL, SBK, TT, AS, MO-M, AM, FJAvR, AGU, JCW, KS, PD, MKW, and AJ: genotyping and quality control; MCdOO, RNL, JHZ, RJFL, MKW, MFF, JL, JSN, NMM, LAC, FR, ACF-W, AYC, DKH, KKL, TT, ES, AM, JAN, FJAvR, SK, L-PL, and VE: data analysis; and MKW, JSN, LAC, ACF-W, TT, JAN, FJAvR, MCZ, and GD: writing of the manuscript. All authors contributed to the planning, execution, and interpretation of the submitted manuscript and read and approved the final manuscript. None of the authors had a conflict of interest.
Footnotes
Abbreviations used: CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology; FFQ, food-frequency questionnaire; FGF21, fibroblast growth factor 21; GWA, genome-wide association; SNP, single nucleotide polymorphism.
REFERENCES
- 1.Rankinen T, Bouchard C. Genetics of food intake and eating behavior phenotypes in humans. Annu Rev Nutr 2006;26:413–34 [DOI] [PubMed] [Google Scholar]
- 2.Collaku A, Rankinen T, Rice T, Leon AS, Rao DC, Skinner JS, Wilmore JH, Bouchard C. A genome-wide linkage scan for dietary energy and nutrient intakes: the Health, Risk Factors, Exercise Training, and Genetics (HERITAGE) Family Study. Am J Clin Nutr 2004;79:881–6 [DOI] [PubMed] [Google Scholar]
- 3.Choquette AC, Lemieux S, Tremblay A, Chagnon YC, Bouchard C, Vohl MC, Perusse L. Evidence of a quantitative trait locus for energy and macronutrient intakes on chromosome 3q27.3: the Quebec Family Study. Am J Clin Nutr 2008;88:1142–8 [DOI] [PubMed] [Google Scholar]
- 4.Cai G, Cole SA, Bastarrachea RA, Maccluer JW, Blangero J, Comuzzie AG. Quantitative trait locus determining dietary macronutrient intakes is located on human chromosome 2p22. Am J Clin Nutr 2004;80:1410–4 [DOI] [PubMed] [Google Scholar]
- 5.Cai G, Cole SA, Butte N, Bacino C, Diego V, Tan K, Goring HH, O'Rahilly S, Farooqi IS, Comuzzie AG. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obesity (Silver Spring) 2006;14:1596–604 [DOI] [PubMed] [Google Scholar]
- 6.Epstein LH, Wright SM, Paluch RA, Leddy JJ, Hawk LW, Jr, Jaroni JL, Saad FG, Crystal-Mansour S, Shields PG, Lerman C. Relation between food reinforcement and dopamine genotypes and its effect on food intake in smokers. Am J Clin Nutr 2004;80:82–8 [DOI] [PubMed] [Google Scholar]
- 7.Eny KM, Corey PN, El-Sohemy A. Dopamine D2 receptor genotype (C957T) and habitual consumption of sugars in a free-living population of men and women. J Nutrigenet Nutrigenomics 2009;2:235–42 [DOI] [PubMed] [Google Scholar]
- 8.Herbeth B, Aubry E, Fumeron F, Aubert R, Cailotto F, Siest G, Visvikis-Siest S. Polymorphism of the 5-HT2A receptor gene and food intakes in children and adolescents: the Stanislas Family Study. Am J Clin Nutr 2005;82:467–70 [DOI] [PubMed] [Google Scholar]
- 9.Aubert R, Betoulle D, Herbeth B, Siest G, Fumeron F. 5-HT2A receptor gene polymorphism is associated with food and alcohol intake in obese people. Int J Obes Relat Metab Disord 2000;24:920–4 [DOI] [PubMed] [Google Scholar]
- 10.Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr 2009;90:1418–25 [DOI] [PubMed] [Google Scholar]
- 11.Hasselbalch AL, Angquist L, Christiansen L, Heitmann BL, Kyvik KO, Sorensen TI. A variant in the fat mass and obesity-associated gene (FTO) and variants near the melanocortin-4 receptor gene (MC4R) do not influence dietary intake. J Nutr 2010;140:831–4 [DOI] [PubMed] [Google Scholar]
- 12.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med 2008;359:2558–66 [DOI] [PubMed] [Google Scholar]
- 13.Bauer F, Elbers CC, Adan RA, Loos RJ, Onland-Moret NC, Grobbee DE, van Vliet-Ostaptchouk JV, Wijmenga C, van der Schouw YT. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. Am J Clin Nutr 2009;90:951–9 [DOI] [PubMed] [Google Scholar]
- 14.Qi L, Kraft P, Hunter DJ, Hu FB. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet 2008;17:3502–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nettleton JA, McKeown NM, Kanoni S, Lemaitre RN, Hivert MF, Ngwa J, van Rooij FJ, Sonestedt E, Wojczynski MK, Ye Z, et al. Interactions of dietary whole grain intake with fasting glucose- and insulin-related genetic loci in individuals of European descent: a meta-analysis of 14 cohort studies. Diabetes Care 2010;33:2684–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu AY, Workalemahu T, Paynter NP, Rose LM, Giulianini F, Tanaka T, Ngwa JS, Qi Q, Curhan GC, Rimm EB, et al. Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet 2013;22:1895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, Stampfer MJ. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet 1991;338:464–8 [DOI] [PubMed] [Google Scholar]
- 18.Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer 2005;5:388–96 [DOI] [PubMed] [Google Scholar]
- 19.Ridker PM, Chasman DI, Zee RY, Parker A, Rose L, Cook NR, Buring JE. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy American women. Clin Chem 2008;54:249–55 [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006;S79:2290 [Google Scholar]
- 21.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13 [DOI] [PubMed] [Google Scholar]
- 22.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 2006;78:629–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nalls MA, Couper DJ, Tanaka T, van Rooij FJ, Chen MH, Smith AV, Toniolo D, Zakai NA, Yang Q, Greinacher A, et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet 2011;7:e1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Göring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 2007;39:1208–16 [DOI] [PubMed] [Google Scholar]
- 26.Idaghdour Y, Czika W, Shianna KV, Lee SH, Visscher PM, Martin HC, Miclaus K, Jadallah SJ, Goldstein DB, Wolfinger RD, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet 2010;42:62–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature 2008;452:423–8 [DOI] [PubMed] [Google Scholar]
- 28.Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, Beaulaurier J, Zhang B, Castro V, Zhu J, Sieberts SK, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res 2011;21:1008–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 2011;7:e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet 2011;7:e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol 2008;6:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 2007;3:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007;39:724–6 [DOI] [PubMed] [Google Scholar]
- 35.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 2008;16:1961–5 [DOI] [PubMed] [Google Scholar]
- 36.Timpson NJ, Emmett PM, Frayling TM, Rogers I, Hattersley AT, McCarthy MI, Davey Smith G. The fat mass- and obesity-associated locus and dietary intake in children. Am J Clin Nutr 2008;88:971–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haupt A, Thamer C, Staiger H, Tschritter O, Kirchhoff K, Machicao F, Haring HU, Stefan N, Fritsche A. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes 2009;117:194–7 [DOI] [PubMed] [Google Scholar]
- 38.Olszewski PK, Fredriksson R, Olszewska AM, Stephansson O, Alsio J, Radomska KJ, Levine AS, Schioth HB. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci 2009;10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kliewer SA, Mangelsdorf DJ. Fibroblast growth factor 21: from pharmacology to physiology. Am J Clin Nutr 2010;91(suppl):254S–7S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domouzoglou EM, Maratos-Flier E. Fibroblast growth factor 21 is a metabolic regulator that plays a role in the adaptation to ketosis. Am J Clin Nutr 2011;93(suppl):901S–5S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, et al. FGF-21 as a novel metabolic regulator. J Clin Invest 2005;115:1627–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarruf DA, Thaler JP, Morton GJ, German J, Fischer JD, Ogimoto K, Schwartz MW. Fibroblast growth factor 21 action in the brain increases energy expenditure and insulin sensitivity in obese rats. Diabetes 2010;59:1817–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharitonenkov A, Wroblewski VJ, Koester A, Chen YF, Clutinger CK, Tigno XT, Hansen BC, Shanafelt AB, Etgen GJ. The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology 2007;148:774–81 [DOI] [PubMed] [Google Scholar]
- 44.Fisher FM, Chui PC, Antonellis PJ, Bina HA, Kharitonenkov A, Flier JS, Maratos-Flier E. Obesity is a fibroblast growth factor 21 (FGF21)-resistant state. Diabetes 2010;59:2781–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 2007;5:426–37 [DOI] [PubMed] [Google Scholar]
- 46.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 2007;5:415–25 [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Yeung DC, Karpisek M, Stejskal D, Zhou ZG, Liu F, Wong RL, Chow WS, Tso AW, Lam KS, et al. Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes 2008;57:1246–53 [DOI] [PubMed] [Google Scholar]
- 48.Gälman C, Lundasen T, Kharitonenkov A, Bina HA, Eriksson M, Hafstrom I, Dahlin M, Amark P, Angelin B, Rudling M. The circulating metabolic regulator FGF21 is induced by prolonged fasting and PPARalpha activation in man. Cell Metab 2008;8:169–74 [DOI] [PubMed] [Google Scholar]
- 49.Tan BK, Hallschmid M, Adya R, Kern W, Lehnert H, Randeva HS. Fibroblast growth factor 21 (FGF21) in human cerebrospinal fluid: relationship with plasma FGF21 and body adiposity. Diabetes 2011;60:2758–62 [DOI] [PMC free article] [PubMed] [Google Scholar]