Abstract
Alzheimer's disease (AD) is a neurodegenerative disease concomitant with grey and white matter damages. However, the interrelationship of volumetric changes between grey and white matter remains poorly understood in AD. Using joint independent component analysis, this study identified joint grey and white matter volume reductions based on structural magnetic resonance imaging data to construct the covariant networks in twelve AD patients and fourteen normal controls (NC). We found that three networks showed significant volume reductions in joint grey–white matter sources in AD patients, including (1) frontal/parietal/temporal-superior longitudinal fasciculus/corpus callosum, (2) temporal/parietal/occipital-frontal/occipital, and (3) temporal-precentral/postcentral. The corresponding expression scores distinguished AD patients from NC with 85.7%, 100% and 85.7% sensitivity for joint sources 1, 2 and 3, respectively; 75.0%, 66.7% and 75.0% specificity for joint sources 1, 2 and 3, respectively. Furthermore, the combined source of three significant joint sources best predicted the AD/NC group membership with 92.9% sensitivity and 83.3% specificity. Our findings revealed joint grey and white matter loss in AD patients, and these results can help elucidate the mechanism of grey and white matter reductions in the development of AD.
Keywords: Alzheimer's disease, Joint source, Joint independent component analysis, Structural MRI, Voxel-based morphometry
1. Introduction
Alzheimer's disease (AD) is a well-known neurodegenerative disease. Numerous structural magnetic resonance imaging (sMRI) studies reported widespread grey matter (GM) reductions; more specifically, the medial temporal structures are involved primarily at the initial phase of the disease, and the parietal and frontal lobes are affected as the disease advances [13,20,21,28]. The pathological processes in AD also include white matter (WM) alterations [8,10,11,24]. A recent study reported AD-associated volume reductions in many gyral WM, including the parahippocampal, inferior parietal and middle frontal regions [24].
WM volume changes are most likely related to GM atrophy that produces cognitive impairment in AD patients [7,26]. However, many sMRI studies investigated GM or WM separately [6,17]. As a result, the interrelationship of volumetric changes between GM and WM remains poorly understood in AD. Thus, it is imperative to document the joint contributions of both GM and WM using sMRI.
Voxel-based morphometry (VBM) is an objective technique [3–5] and has been widely utilized to detect differences in brain tissue in various populations [7,17,21]. However, most reported VBM findings are based on a univariate approach. The univariate VBM can detect brain abnormalities related to AD [7,13,21], but it is unable to identify the anatomical interrelationships in those brain areas. Multivariate methods consider multiple voxels simultaneously based on the view that the overall brain structure is interconnected and covaried as a network [2,9,18,19,30]. Moreover, multivariate methods emphasize on the covariance information of imaging data and generate interrelated morphological features and a network pattern representation. Again, multivariate approaches may capture the subtle changes and have greater sensitivity in examining the regional differences [2,9]. Recently, Xu et al. proposed a multivariate method referred to as joint source based morphometry (jSBM). Unlike VBM, jSBM using joint independent component analysis (jICA) grounds the statistical outcome of “source” by including a set of regions which carry similar covariance information and exhibit inter-subject and group differences [30]. Xu et al. applied jICA to detect differences in GM and WM in schizophrenia, suggesting that jICA is a potentially efficient approach to examine brain anatomical changes due to a specific disease [30].
Using multivariate jICA, this study identified joint GM and WM volume reductions in sMRI data to construct the covariant networks in AD patients and normal controls (NC). In addition, a multiple linear regression model was performed with the components that significantly discriminated AD patients from NC to attain the integrated network. We expect that combining multiple sources will best predict the AD/NC group membership.
2. Materials and methods
2.1. Participants
Twelve AD patients (mean age: 71.8 ± 6.7 [rang 58–81], 5 males) with a mean Mini-Mental State Examination (MMSE) score of 18.3 ± 11.7 [range 12–23], and 14 NC (mean age: 70.4 ± 3.5 [range 61–82], 6 males) with a mean MMSE of 28.5 ± 0.6 [27–29], were recruited from the memory clinic at Xuanwu Hospital of Capital Medical University in Beijing, China. Written informed consent was obtained from each participant. This study was approved by the Medical Research Ethics Committee of Xuanwu Hospital. The diagnosis of probable AD was made according to the NINCDS-ADRDA criteria. NC had no cognitive complaints and did not have neurological or psychiatric disorders. The AD group did not significantly differ from the NC in sex ratio or age (t(24) = 0.57, p = 0.573), but had significantly lower MMSE scores (t(24) = −10.93, p = 4.23E − 11).
2.2. Structural MRI scanning
For each participant, 3D sMRI data were acquired using a MPRAGE sequence at 3.0 T (TR = 1900 ms, TE = 2.2 ms, TI = 900 ms, FA = 9°, FOV = 224 mm × 256 mm, matrix size = 448 × 512 and voxel size = 0.5 mm × 0.5 mm × 1 mm).
2.3. Preprocessing of structural MRI Data
The preprocessing of sMRI data was performed using the VBM8 Toolbox (http://dbm.neuro.uni-jena.de/vbm8) implemented in SPM8 (http://www.fil.ion.ucl.ac.uk/spm). The two main outcomes of the VBM8 procedure are segmentation and normalization. The segmentation approach applied adaptive Maximum a posterior [23] and partial volume estimation [29] to segment the native space images into a rigid-body aligned GM and WM images in the Montreal Neurological Institute (MNI) space. Subsequently, two denoising methods, including a spatially adaptive nonlocal means denoising filter [22] and a classical Markov random field approach, were utilized to improve the segmentation [23]. We employed a diffeomorphic anatomical registration using exponential Lie algebra (DARTEL) protocol for normalization [4]. DARTEL utilizes the large deformation parameterized by a single constant velocity field to generate diffeomorphic and invertible deformations. The template creation and image registration were iteratively implemented, and the brain tissue maps for each subject were warped to new templates at each iteration. The initial registration worked on the average templates from the rigidly aligned images of all subjects, separately for GM and WM. The flow field encoding the warping information was generated for each subject, and finally GM and WM tissue maps were deformed to their own increasingly crisp average templates and further normalized to MNI space. Afterwards, the registered GM and WM partitions were separately multiplied by the Jacobian determinants from the deformations to preserve the total amount of tissue. Lastly, the grey and white maps were smoothed separately with a 12 mm Gaussian kernel.
2.4. Joint ICA and statistical analysis
The jICA was implemented using the Fusion ICA toolbox (http://icatb.sourceforge.net). Akaike's information criterion (AIC) was used to determine the number of joint sources [1]. The SPM default brain mask was used to exclude the non-brain voxels separately for the registered and smoothed GM and WM partitions. The initial data matrix was formed by combining horizontally GM and WM data (a row vector) for each subject. Spatial ICA using an infomax algorithm decomposed the initial data matrix (subjects by voxels; the number of voxels was the sum of the dimension of GM and WM maps) into a mixing matrix (subjects by sources) and a source matrix (sources by voxels) [30]. Each column of the mixing matrix (ICA weights) indicates the degree to which each subject expresses the joint source differences between two groups. The source matrix comprises of a GM source matrix and a WM one. Each row of the source matrix represents a joint source that includes both GM and WM maps that share the same covariance patterns. Such joint sources are maximally independent spatial components.
The jICA used the mixing coefficients for statistical inferences. A two sample t-test was performed on each column of the mixing matrix to detect AD/NC group differences at a statistical threshold of p ≤ 0.05. The rows of the source matrix corresponding to the significant set of mixing coefficients were converted to Z-scores with unit standard deviation. The significant joint sources were reshaped into two 3D maps that individually depicted GM and WM with a threshold of Z ≥ 3.0 to reflect the related brain regions involved in the joint source networks. Such statistical tests are free of multiple comparison correction for multivariate jICA. Furthermore, the significant mixing coefficients were entered into a multiple linear regression model. Based on the multiple-regression results, the AD-related integrated network pattern was created as a linear combination of the corresponding components.
3. Results
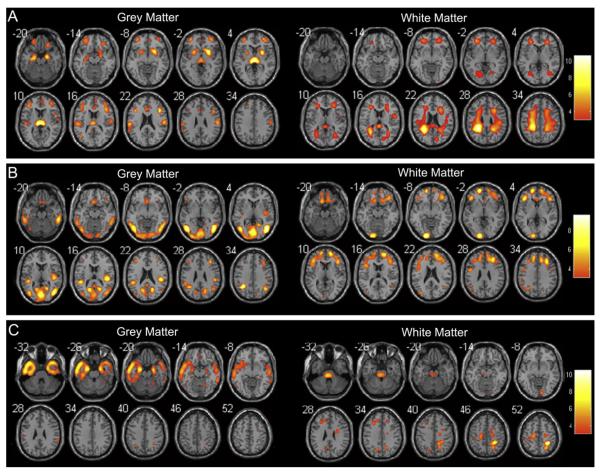
Six joint sources were extracted according to AIC. Among those six sources, three of them expressed significant differences in the AD/NC group relationship. The positive weighted coefficient in the joint source maps illustrated decreases in GM and WM volumes in AD patients (Fig. 1).
Fig. 1.
Significance maps illustrating covariant decreases in grey (left) and white matter volumes (right) in AD patients compared with NC for joint sources 1 (A), 2 (B) and 3 (C). Left of the plane is the left of brain. The colour bar represents Z-scores.
For joint source 1, GM volume reductions were detected mainly in the middle/inferior/superior frontal gyrus, supramarginal gyrus, anterior cingulate gyrus, hippocampus and parahippocampal gyrus, whereas WM volume reductions in the superior longitudinal fasciculus (SLF), corpus callosum (CC) and corona radiata in AD patients (Fig. 1A). Table 1 shows the specific localization information for three sources.
Table 1.
Locations of joint grey and white matter volume reductions in AD patients compared with NC for joint sources 1–3.
| Brain regions | Peak coordinates (X, Y, Z) | Z | Cluster size (mm3) | ||
|---|---|---|---|---|---|
| Joint source 1, grey matter | |||||
| L middle frontal gyrus | −36 | 49 | −2 | 5.18 | 9494 |
| R middle frontal gyrus | 40 | 26 | 19 | 6.34 | 9812 |
| L inferior frontal gyrus | −39 | 21 | 20 | 5.75 | 4182 |
| R inferior frontal gyrus | 42 | 21 | 20 | 7.53 | 6686 |
| L superior frontal gyrus | −33 | 51 | −3 | 5.04 | 3304 |
| R superior frontal gyrus | 21 | 33 | 44 | 4.96 | 5920 |
| L thalamus/putamen | −6 | −20 | 8 | 10.20 | 7989 |
| R thalamus/putamen | 6 | −19 | 8 | 9.59 | 10,223 |
| L supramarginal gyrus | −58 | −22 | 22 | 6.06 | 2774 |
| R supramarginal gyrus | 58 | −16 | 24 | 5.89 | 2015 |
| L anterior cingulate gyrus | −3 | 26 | 19 | 4.49 | 1603 |
| R anterior cingulate gyrus | 7 | 34 | 15 | 3.98 | 1991 |
| L hippocampus/parahippocampal gyrus | −21 | −11 | −17 | 6.29 | 5947 |
| R hippocampus/parahippocampal gyrus | 22 | −10 | −17 | 6.75 | 3287 |
| Joint source 1, white matter | |||||
| L superior longitudinal fasciculus | −31 | −39 | 28 | 12.85 | 45,914 |
| R superior longitudinal fasciculus | 30 | −42 | 30 | 7.01 | 40,352 |
| Splenium of corpus callosum | −18 | −36 | 29 | 10.63 | 7945 |
| Body of corpus callosum | −18 | −28 | 30 | 9.07 | 3959 |
| Genu of corpus callosum | 21 | 20 | 21 | 5.67 | 405 |
| L posterior corona radiata | −27 | −38 | 28 | 13.41 | 3581 |
| R posterior corona radiata | 22 | −44 | 30 | 8.38 | 3210 |
| L superior corona radiata | −24 | −25 | 34 | 8.75 | 6669 |
| R superior corona radiata | 25 | 14 | 26 | 6.71 | 4961 |
| L anterior corona radiata | −24 | 39 | −2 | 6.20 | 4023 |
| R anterior corona radiata | 25 | 19 | 25 | 7.21 | 4145 |
| Joint source 2, grey matter | |||||
| L inferior temporal gyrus | −46 | −67 | −2 | 8.04 | 9349 |
| R inferior temporal gyrus | 46 | −68 | 1 | 8.15 | 15,174 |
| L middle temporal gyrus | −48 | −67 | 3 | 8.33 | 4796 |
| R middle temporal gyrus | 46 | −68 | 8 | 9.61 | 9474 |
| L superior temporal gyrus | −45 | −36 | 17 | 6.70 | 3564 |
| R superior temporal gyrus | 45 | −25 | 16 | 7.46 | 2720 |
| L angular gyrus | −34 | −49 | 36 | 9.43 | 2589 |
| R angular/supramarginal gyrus | 42 | −58 | 36 | 6.46 | 6652 |
| L inferior parietal lobule | −36 | −49 | 36 | 9.32 | 1829 |
| L cuneus | −15 | −63 | 21 | 6.55 | 4820 |
| R cuneus/precuneus | 19 | −60 | 21 | 6.71 | 6706 |
| L inferior/middle occipital gyrus | −45 | −67 | 2 | 8.87 | 16,835 |
| R inferior/middle occipital gyrus | 46 | −70 | 8 | 9.61 | 7702 |
| Joint source 2, white matter | |||||
| L superior frontal gyrus | −18 | 58 | 3 | 4.99 | 19,295 |
| R superior frontal gyrus | 18 | 31 | 37 | 3.83 | 14,475 |
| L middle frontal gyrus | −30 | 41 | 12 | 4.48 | 7044 |
| R middle frontal gyrus | 36 | 20 | 43 | 5.81 | 17,456 |
| L inferior frontal gyrus | −42 | 35 | −2 | 5.57 | 14,084 |
| R inferior frontal gyrus | 42 | 24 | 10 | 5.15 | 11,573 |
| L inferior/middle occipital gyrus | −15 | −93 | −2 | 9.43 | 7121 |
| L inferior parietal lobule | −27 | −58 | 40 | 5.89 | 5812 |
| Joint source 3, grey matter | |||||
| L inferior temporal gyrus | −36 | 3 | −35 | 10.45 | 14,438 |
| R inferior temporal gyrus | 39 | 6 | −36 | 7.34 | 9484 |
| L superior temporal gyrus | −52 | 4 | −12 | 5.63 | 13,784 |
| R superior temporal gyrus | 59 | −12 | −7 | 4.87 | 10,287 |
| L middle temporal gyrus | −34 | 1 | −37 | 10.15 | 5306 |
| R middle temporal gyrus | 39 | 13 | −31 | 7.47 | 6929 |
| L hippocampus/parahippocampal gyrus | −22 | −7 | −19 | 8.53 | 6311 |
| R hippocampus/parahippocampal gyrus | 24 | −5 | −19 | 7.05 | 7276 |
| L amygdala | −22 | −5 | −19 | 8.55 | 1502 |
| R amygdala | 24 | −4 | −19 | 6.90 | 1164 |
| Joint source 3, white matter | |||||
| L precentral gyrus | −18 | −16 | 49 | 5.57 | 5414 |
| R precentral/postcentral gyrus | 30 | −42 | 53 | 10.58 | 18,771 |
| L middle cerebellar peduncle | −22 | −58 | −32 | 9.24 | 4847 |
| L middle frontal gyrus | −28 | 23 | 32 | 4.35 | 4138 |
L, left; R, right; coordinates in Talairach space.
For joint source 2, AD patients showed reduced GM volumes mainly in the inferior/middle/superior temporal gyrus, supramarginal gyrus, inferior parietal lobule (IPL), precuneus and inferior/middle occipital gyrus as well as reduced WM volumes in the superior/middle/inferior frontal gyrus, inferior occipital gyrus and IPL (Fig. 1B).
For joint source 3, less GM was found in the inferior/superior/middle temporal gyrus, hippocampus and parahippocampal gyrus, whereas less WM was found mainly in the precentral/postcentral gyrus in AD patients (Fig. 1C).
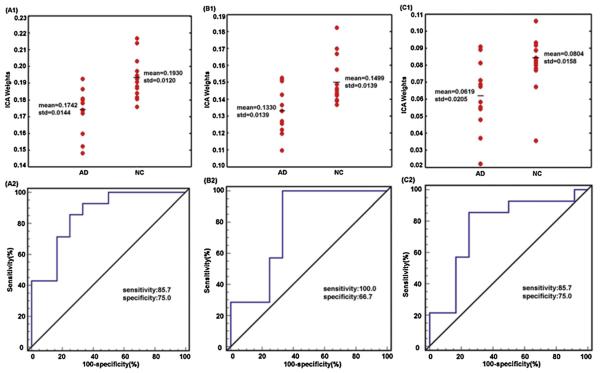
The expression of covariance patterns was significantly lower in AD patients than NC for joint source 1 (t(24) = −3.64, p = 6.48E − 4) (Fig. 2A1), joint source 2 (t(24) = −3.09, p = 0.0025) (Fig. 2B1) and joint source 3 (t(24) = −2.60, p = 0.0079) (Fig. 2C1). The receiver operating curve (ROC) analysis determined discrimination with 85.7% sensitivity and 75.0% specificity (area under curve = 0.857, SE = 0.074, p = 0.002) for joint source 1 (Fig. 2A2), 100% sensitivity and 66.7% specificity (area under curve = 0.786, SE = 0.099, p = 0.014) for joint source 2 (Fig. 2B2), and 85.7% sensitivity and 75.0% specificity (area under curve = 0.768, SE = 0.100, p = 0.021) for joint source 3 (Fig. 2C2).
Fig. 2.
The effects of discrimination from ICA weights in AD patients and NC for joint sources 1–3. The above panel shows the scatterplot of ICA weights for joint sources 1 (A1), 2 (B1) and 3 (C1). The below panel shows the ROC of the discriminability for joint sources 1 (A2), 2 (B2) and 3 (C2).
For the combined source from significant joint sources 1–3, the brain regions with decreased volumes in AD broadly involved these three sources but mainly involved sources 1 and 2, such as GM in the middle frontal gyrus, putamen, thalamus, hippocampus and parahippocampal gyrus, and WM in the SLF, corona radiata, CC and the superior/middle/inferior frontal gyrus. Analysis of the combined source and joint sources 1–3 showed that the overlapping regions ratios were 38.3%, 34.1% and 17.7% for GM and 57.2%, 36.9% and 10.7% for WM. The expression of covariance patterns was significantly lower in AD patients compared with NC (t(24) = −4.89, p = 2.73E − 5) (Fig. 3). ROC analysis demonstrated the discrimination with 92.9% sensitivity and 83.3% specificity (area under curve = 0.917, SE = 0.057, p = 3.18E − 4) (Fig. 3).
Fig. 3.
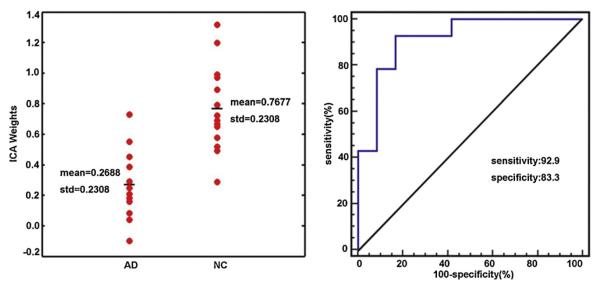
The effects of discrimination from ICA weights in AD patients and NC for the combined source. The left panel shows the scatterplot of ICA weights. The right panel shows the ROC of the discriminability.
4. Discussion
This study performed jICA analysis of sMRI data to capture AD-related network patterns. AD patients showed volume reductions in both GM and WM in three significant joint sources, and they had lower expression scores of covariance patterns compared with NC. In addition, ROC analysis determined the discrimination with high sensitivity and specificity. Furthermore, the combined source constructed from three sources best predicted AD/NC group membership.
For joint source 1, the expression of covariance patterns was the most significant among three sources, with the maximum discriminability between AD patients and NC (Fig. 2A1 and A2). Joint source 1 revealed GM reductions mainly in the frontal, parietal and temporal areas (Fig. 1A), similar to previous studies [6,17,21,28]. Interestingly, AD patients exhibited regions of GM atrophy in joint source 1, including the middle/superior frontal gyrus, cingulate gyrus, supramarginal gyrus, hippocampus and parahippocampal gyrus from the default mode network (DMN) investigated in resting-state studies [14]. Greicius et al. found that DMN activity distinguished AD from healthy ageing and confirmed that resting-state functional connectivity reflected structural connectivity [15,16]. In addition, Seely et al. found concordance between AD-affected atrophy patterns and intrinsic connectivity networks corresponding to the DMN [25]. He et al. also reported correlated variations among several “default” regions in AD [19]. The GM alterations in joint source 1 indicate that these regions possibly may be in the same structural network embodying episodic memory and executive function impairment in AD.
The majority of WM alterations in joint source 1 involved the bilateral SLF and CC (Fig. 1A), in accordance with sMRI and diffusion tensor imaging studies [8,11,17,27]. The SLF is an anterior–posterior longitudinal fibre that connects the frontal, parietal, temporal and occipital cortices. A reduction in this tract suggests that the connection of the frontal, parietal and temporal GM in joint source 1 was interrupted [27]. Volume or fractional anisotropy reductions in CC were associated with neuronal loss in AD; for example, the splenium of CC was found to be responsible for neuronal loss involving parieto-temporal neocortical regions and the anterior callosal portions of the prefrontal regions [8,11].
For joint source 2, the expression of covariance patterns showed a higher discriminability, and ROC analysis had a sensitivity of 100% (Fig. 2B1 and B2). Joint source 2 showed GM loss in the temporal, parietal and occipital lobes (Fig. 1B), congruent with previous reports [17,20,21,28]. GM atrophy patterns could explain the differential involvement of the parietal regions in the development of AD [20]; furthermore, our results support the idea that the IPL and precuneus are likely the first regions to show atrophy within the parietal lobe [20]. Correspondingly, WM loss occurred in the IPL within joint source 2.
Previous neuroimaging research indicated that WM integrity damage due to normal ageing and AD started mainly in the late myelinating regions such as the frontal lobes [24,27]. Salat et al. found that the inferior parietal and rostral middle frontal WM regions showed the largest AD-associated volume reductions [24]. Similar to Salat et al. study, joint source 2 demonstrated apparent degeneration in the bilateral superior/inferior/middle frontal gyri WM, possibly contributing to the deterioration of the executive functional network in AD. In addition, our data showed reductions in both GM and WM in the inferior/middle occipital gyri. These results, however, are in contrast to the early sMRI studies that reported relatively spared changes in the occipital lobe [21,28].
For joint source 3, the significance level of the covariance patterns expression was the lowest; however, joint source 3 did have a significant discriminability with reliable sensitivity and specificity (Fig. 2C1 and C2). GM atrophy of the temporal lobe and its medial regions (Fig. 1C), is one of the most concurrent AD-associated findings in sMRI studies [6,13,17,21,26]. Volume decreases in the hippocampus and parahippocampal gyrus are considered to be the most valuable sMRI biomarker of AD pathology for prediction and early diagnosis [12].
Stoub et al. found significant WM volume decreases in the parahippocampal gyrus in individuals at risk for AD [26]. In contrast, we detected WM reductions in the inferior longitudinal fasciculi in our recent report using univariate VBM [17], and we report now a lack of related WM changes in the temporal lobe and its medial structures in joint source 3. A possible explanation is that the myelin breakdown of WM had already occurred in our AD patients; subsequently, there may have been timing differences relative to the adjacent GM reductions in the temporal lobe. Joint source 3 consisted of WM changes mainly in the precentral/postcentral gyrus in AD patients. These findings seem to be discrepant with the literature [7,27]. Our findings should be further investigated in longitudinal sMRI studies.
The combined source represented three joint sources but mainly consisted of sources 1 and 2. As we expected, the combined source had the highest statistical power of distinguishing AD patients from NC and had the optimal degree of sensitivity and specificity compared with each joint source (Fig. 3). It is worth noting that there are some overlapping regions among the various joint sources. For example, the hippocampus and parahippocampal gyrus are in both joint sources 1 and 3, and the supramarginal gyrus and genu of CC are in both joint sources 1 and 2. These three joint sources are spatially independent, but this does not mean that they cannot overlap [30]. The overlapping brain regions may indicate their distinctive relevance to specific functional networks.
Multivariate techniques have been increasingly utilized to elucidate neural networks in different populations [2,9,18,19,25,30]. In contrast to univariate approaches such as VBM, multivariate jICA integrates the joint information and simultaneously identifies GM and WM changes across the whole brain while the univariate approaches can only examine differences in peak voxels; moreover, the jICA mixing coefficients need no multiple comparison correction while the statistics based on univariate approaches do.
Using jICA, we have shown a comprehensive network pattern of GM and WM reductions in AD patients. These network-based findings contribute to the understanding of the mechanism behind brain tissue changes in the development of AD. Although the sample size was relatively small in our study, the results suggest that jICA can detect group differences with reliable statistical power. It should be mentioned that jICA brings out the covariation patterns of GM and WM, but it could not tell us what the specific relationship among various brain regions was. It is necessary to further explore the linear or nonlinear associations among core AD-affected brain regions using other multivariate methods, such as Bayesian network.
Highlights
-
▶
We performed joint independent component analysis (jICA) to structural MRI data.
-
▶
Three joint grey-white matter sources showed significant volume reductions in AD.
-
▶
The combined source best predicted the AD/NC group membership.
Acknowledgements
This work was supported by the National Natural Science Foundation (NNSF), China (81000603), Key Program of NNSF, China (60931003), the Fundamental Research Funds for the Central Universities, China, the National Institute of Mental Health, US (RO1MH57899), the National Institute on Aging, US (9R01AG031581-10, P30AG19610, k23AG24062), and the State of Arizona.
References
- [1].Akaike H. A new look at statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- [2].Alexander GE, Bergfield KL, Chen K, Reiman EM, Hanson KD, Lin L, Bandy D, Caselli RJ, Moeller JR. Gray matter network associated with risk for Alzheimer's disease in young to middle-aged adults. Neurobiology of Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.01.014. http://dx.doi.org/10.1016/j.neurobiolaging.2012.01.014. [DOI] [PMC free article] [PubMed]
- [3].Ashburner J, Friston KJ. Voxel-based morphometry – the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- [4].Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- [5].Ashburner J, Friston KJ. Diffeomorphic registration using geodesic shooting and Gauss–Newton optimization. NeuroImage. 2011;55:954–967. doi: 10.1016/j.neuroimage.2010.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Balthazar ML, Yasuda CL, Pereira FR, Pedro T, Damasceno BP, Cendes F. Differences in grey and white matter atrophy in amnestic mild cognitive impairment and mild Alzheimer's disease. European Journal of Neurology. 2009;16:468–474. doi: 10.1111/j.1468-1331.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- [7].Baxter LC, Sparks DL, Johnson SC, Lenoski B, Lopez JE, Connor DJ, Sabbagh MN. Relationship of cognitive measures and gray and white matter in Alzheimer's disease. Journal of Alzheimer's Disease. 2006;9:253–260. doi: 10.3233/jad-2006-9304. [DOI] [PubMed] [Google Scholar]
- [8].Bosch B, Arenaza-Urquijo EM, Rami L, Sala-Llonch R, Junqué C, Solé-Padullés C, Pena-Gómez C, Bargalló N, Molinuevo JL, Bartrés-Faz D. Multiple DTI index analysis in normal aging, amnestic MCI and AD: relationship with neuropsychological performance. Neurobiology of Aging. 2012;33:61–74. doi: 10.1016/j.neurobiolaging.2010.02.004. [DOI] [PubMed] [Google Scholar]
- [9].Brickman AM, Habeck C, Zarahn E, Flynn J, Stern Y. Structural MRI covariance patterns associated with normal aging and neuropsychological functioning. Neurobiology of Aging. 2007;28:284–295. doi: 10.1016/j.neurobiolaging.2005.12.016. [DOI] [PubMed] [Google Scholar]
- [10].Canu E, Frisoni GB, Agosta F, Pievani M, Bonetti M, Filippi M. Early and late onset Alzheimer's disease patients have distinct patterns of white matter damage. Neurobiology of Aging. 2012;33:1023–1033. doi: 10.1016/j.neurobiolaging.2010.09.021. [DOI] [PubMed] [Google Scholar]
- [11].Chaim TM, Duran FLS, Uchida RR, Périco CAM, de Castro CC, Busatto GF. Volumetric reduction of the corpus callosum in Alzheimer's disease in vivo as assessed with voxel-based morphometry. Psychiatry Research: Neuroimaging. 2007;154:59–68. doi: 10.1016/j.pscychresns.2006.04.003. [DOI] [PubMed] [Google Scholar]
- [12].Ewers M, Sperling RA, Klunk WE, Weiner MW, Hampel H. Neuroimaging markers for the prediction and early diagnosis of Alzheimer's disease dementia. Trends in Neurosciences. 2011;34:430–442. doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Good CD, Scahill RI, Fox NC, Ashburner J, Friston KJ, Chan D, Crum WR, Rossor MN, Frackowiak RS. Automatic differentiation of anatomical patterns in the human brain: validation with studies of degenerative dementias. NeuroImage. 2002;17:29–46. doi: 10.1006/nimg.2002.1202. [DOI] [PubMed] [Google Scholar]
- [14].Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Greicius MD, Srivastava G, Reiss AL, Menon V. Default mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cerebral Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo XJ, Chen CS, Chen KW, Jin Z, Peng DL, Yao L. Voxel-based assessment of gray and white matter volumes in Alzheimer's disease. Neuroscience Letters. 2010;468:146–150. doi: 10.1016/j.neulet.2009.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Habeck C, Stern Y. Multivariate data analysis for neuroimaging data: overview and application to Alzheimer's disease. Cell Biochemistry and Biophysics. 2010;58:53–67. doi: 10.1007/s12013-010-9093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].He Y, Chen Zh., Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. Journal of Neuroscience. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jacobs HIL, Van Boxtel MP, Uylings HB, Gronenschild EH, Verhey FR, Jolles J. Atrophy of the parietal lobe in preclinical dementia. Brain and Cognition. 2011;75:154–163. doi: 10.1016/j.bandc.2010.11.003. [DOI] [PubMed] [Google Scholar]
- [21].Karas GB, Burton EJ, Rombouts SA, van Schijndel RA, O'Brien JT, Scheltens P, McKeith IG, Williams D, Ballard C, Barkhof F. A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel-based morphometry. NeuroImage. 2003;18:895–907. doi: 10.1016/s1053-8119(03)00041-7. [DOI] [PubMed] [Google Scholar]
- [22].Manjón JV, Coupé P, Martí-Bonmatí L, Collins DL, Robles M. Adaptive non local means denoising of MR images with spatially varying noise levels. Journal of Magnetic Resonance Imaging. 2010;31:192–203. doi: 10.1002/jmri.22003. [DOI] [PubMed] [Google Scholar]
- [23].Rajapakse JC, Giedd JN, Rapoport JL. Statistical approach to segmentation of single-channel cerebral MR images. IEEE Transactions on Medical Imaging. 1997;16:176–186. doi: 10.1109/42.563663. [DOI] [PubMed] [Google Scholar]
- [24].Salat DH, Greve DN, Pacheco JL, Quinn BT, Helmer KG, Buckner RL, Fischl B. Regional white matter volume differences in nondemented aging and Alzheimer's disease. NeuroImage. 2009;44:1247–1258. doi: 10.1016/j.neuroimage.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Seely WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stoub TR, deToledo-Morrell L, Stebbins GT, Leurgans S, Bennett DA, Shah RC. Hippocampal disconnection contributes to memory dysfunction in individuals at risk for Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10041–10045. doi: 10.1073/pnas.0603414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stricker NH, Schweinsburg BC, Delano-Wood L, Wierenga CE, Bangen KJ, Haaland KY, Frank LR, Salmon DP, Bondi MW. Decreased white matter integrity in late-myelinating fiber pathways in Alzheimer's disease supports retrogenesis. NeuroImage. 2009;45:10–16. doi: 10.1016/j.neuroimage.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer S, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer's disease. Journal of Neuroscience. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tohka J, Zijdenbos A, Evans A. Fast and robust parameter estimation for statistical partial volume models in brain MRI. NeuroImage. 2004;23:84–97. doi: 10.1016/j.neuroimage.2004.05.007. [DOI] [PubMed] [Google Scholar]
- [30].Xu L, Calhoun VD. Joint source based morphometry identifies linked gray and white matter group differences. NeuroImage. 2009;44:777–789. doi: 10.1016/j.neuroimage.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]