Abstract
Purpose of review
In recent years we have seen remarkable progress in our understanding of the disease mechanism underlying facioscapulohumeral muscular dystrophy (FSHD). The purpose of this review is to provide a comprehensive overview of our current understanding of the disease mechanism and to discuss the observations supporting the possibility of a developmental defect in this disorder.
Recent findings
In the majority of cases FSHD is caused by contraction of the D4Z4 repeat array (FSHD1). This results in local chromatin relaxation and stable expression of the DUX4 retrogene in skeletal muscle, but only when a polymorphic DUX4 polyadenylation signal is present. In some cases (FSHD2), D4Z4 chromatin relaxation and stable DUX4 expression occurs in the absence of D4Z4 array contraction. DUX4 is a germline transcription factor and its expression in skeletal muscle leads to activation of early stem cell and germline programs and transcriptional activation of retroelements.
Summary
Recent studies have provided a plausible disease mechanism for FSHD where FSHD results from inappropriate expression of the germline transcription factor DUX4. The genes regulated by DUX4 suggest several mechanisms of muscle damage, and provide potential biomarkers and therapeutic targets that should be investigated in future studies.
Keywords: Facioscapulohumeral, muscular dystrophy, DUX4, germline, retrotransposon
Introduction
With an incidence between 1:15,000 and 1:20,000 FSHD is the third most common myopathy [1-3]. Patients suffer from a progressive and irreversible weakness of the facial, shoulder and upper arm muscles. With disease progression, other muscles may also become affected. Interestingly, muscle weakness in FSHD is often asymmetric. Symptomatic non-muscular disease manifestations are rare but can include sensorineural deafness, retinovasculopathy and intellectual disability. Pain and fatigue is a frequent complaint [4].
Despite the typical onset in the late teens, there are several clinical observations that support the hypothesis that FSHD can be considered a developmental disorder. First, in about 10% of patients, the disease onset is much earlier, with symptoms occasionally occurring at birth [5]. In some of these patients there is a congenital absence of specific muscle groups [6]. Second, the incidence of pectus excavatum independent from muscle weakness is 1,000-fold increased compared to the general population. Finally, the asymmetric, often seemingly random, muscle weakness may point to a developmental aspect to this disorder [7].
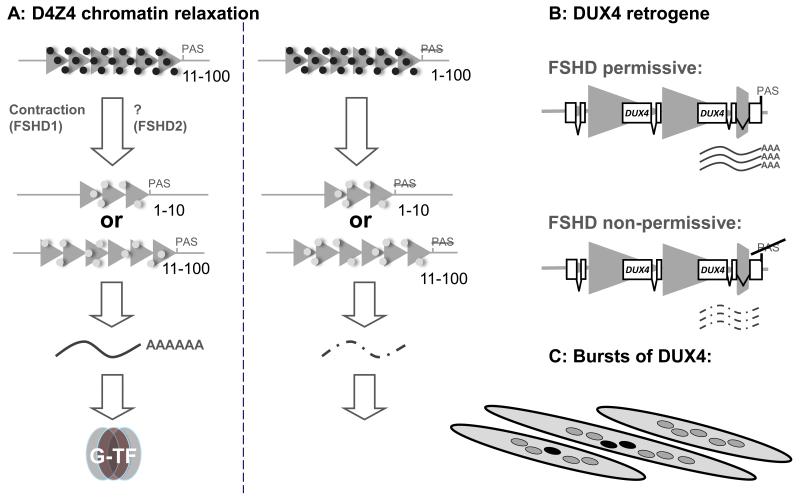
FSHD is caused by chromatin relaxation of the polymorphic D4Z4 macrosatellite repeat array on chromosome 4 in a repeat array contraction-dependent (FSHD1) or contraction-independent (FSHD2) fashion (Fig. 1) [8]. Normally the D4Z4 repeat array located on the tip of the long arm of chromosome 4 contains 11-100 D4Z4 units of 3.3kb each. Patients with FSHD1 have one chromosome with an array of 1-10 units [9;10]. Contraction of D4Z4 needs to occur on a specific genetic background of chromosome 4 to cause FSHD as not all contractions on chromosome 4 are pathogenic [11-14]. Contraction of a highly similar repeat structure on chromosome 10 is normally also not associated with disease [15-18].
Figure 1.
(Epi)genetic mechanism of FSHD1 and FSHD2. A) Normally the D4Z4 repeat (triangles) on DUX4 polyadenylation site containing (PAS) and non-containing genetic backgrounds varies between 11-100 units and contains heterochromatic modifications (black circles). By D4Z4 contraction to 1-10 units in FSHD1, or by an unknown mechanism in FSHD2, the chromatin structure of the D4Z4 repeat becomes relaxed (light grey circles) leading to the production of DUX4 mRNA and protein (G-TF: germline transcription factor). Alleles that do not contain a PAS also show chromatin relaxation but fail to produce stable DUX4 mRNA and protein. B) FSHD permissive alleles contain an extra exon for DUX4 immediately distal to the D4Z4 repeat array that provides this retrogene with a PAS. Non-permissive fail to stabilize DUX4 mRNA in the absence of this exon/PAS signal. C) In myotube cultures of FSHD1 and FSHD2 patients, a variegated pattern of DUX4 immunoreactive nuclei (black nuclei) can be observed.
Genetic backgrounds of chromosome 4 on which contractions lead to FSHD facilitate the production of the double homeobox protein DUX4 in skeletal muscle [19;20]. Each unit of the array contains a copy of the DUX4 retrogene but transcripts that originate from this retrogene are unstable in the absence of a polyandenylation signal. In the context of a genetic background of chromosome 4 that is permissive for FSHD, and when the chromatin structure is relaxed, the distal copy of DUX4 is spliced to an additional exon that provides a polyadenylation signal and results in transcript stabilization leading to variegated expression of DUX4 protein in a small subset of myonuclei [19-24].
A small but significant group of patients show D4Z4 chromatin relaxation and somatic derepression of DUX4 in the absence of repeat contraction and leading to a clinical and molecular phenotype indistinguishable from FSHD1. These FSHD2 patients also carry at least one chromosome 4 with a FSHD-permissive genetic background and produce DUX4 in their muscle [25-27].
Chromatin Structure and Bidirectional Transcription
Macrosatellite repeats represent a class of very large repeat structures in our genome. They are often associated with bidirectional transcription [20;28-31] and the tandem repeat structure of these arrays contributes to the regulatory consequences of bidirectional transcription. These include the possibility of stearic effects from colliding polymerases [32], or the production of sense and antisense transcripts that may mediate post-transcriptional [33;34] or transcriptional gene silencing through heterochromatin formation [33;35]. These activities serve to inhibit further transcription, and paradoxically inhibit production of the antisense molecules themselves suggesting that maintenance levels of transcription are occurring even from silenced loci to maintain suppression. DUX4 transcriptional suppression likely involves both initiation and maintenance phases and can be modeled using pluripotent stem cells and retroviral vectors encoding D4Z4 promoter sequences and fluorescent reporters.
The use of human embryonic stem (ES) cells and induced pluripotent stem (IPS) cells allows an examination of CpG methylation and chromatin structure before and after transcriptional silencing has occurred. Thus, transcriptionally inactive D4Z4 arrays from normal individuals become actively transcribed after the induction of pluripotency [20].
Active transcription in pluripotent cells is associated with histone signatures typical of transcribed sequences [20;36] including a relative depletion of histones containing repressive modifications (Histone 3 lysine 9 trimethylation: H3K9me3) and an enrichment of modifications typical of active transcription (H3K4me2). As ES and IPS cells are induced to differentiate, the D4Z4 region becomes populated by histones containing repressive marks and depleted of those with H3K4me2. IPS cells from FSHD-affected individuals with shortened D4Z4 arrays also demonstrate active DUX4 transcription with histone modifications typical of transcribed sequences, but unlike D4Z4 arrays in normal individuals the modifications remain unchanged as these cells are induced to differentiate suggesting that failure of suppression during differentiation is an important mechanism of FSHD-related pathology [20].
In addition to the generation of antisense transcripts that may be involved in silencing in early development, bidirectional transcription may also be important for maintenance of D4Z4 repression in somatic cells. Indeed, in addition to several coding and non-coding transcripts recently identified at the D4Z4 units, si/miRNA-sized small RNAs were shown to be expressed from the D4Z4 arrays in control and FSHD muscles [20;21;37;38] suggesting that deregulation of bidirectional transcription or small RNA-processing may play a role in chromatin derepression observed in FSHD.
First reports on the altered chromatin structure of the 4q D4Z4 region in FSHD came from the studies of DNA methylation [39]. While normally D4Z4 repeats show ~50% CpG methylation at the sites investigated, in FSHD chromosomes D4Z4 methylation levels were reduced by a factor of 2 [27]. Interestingly, FSHD2 patients without detectable deletion of D4Z4 also showed significant hypomethylation of D4Z4 [25;27;40]. Different from FSHD1 patients where hypomethylation is restricted to the disease chromosome, in FSHD2 D4Z4 repeats on chromosomes 4q and 10q are hypomethylated. D4Z4 DNA hypomethylation is also observed in unaffected carriers indicating that loss of DNA methylation may be necessary but not sufficient for development of disease [25;27].
In addition to DNA hypomethylation, specific loss of H3K9me3, a repressive chromatin mark associated with heterochromatin formation, in D4Z4 was reported for FSHD chromosomes accompanied by secondary loss of HP1γ and cohesin binding, all suggestive of a more relaxed chromatin structure of D4Z4 in disease chromosomes [36]. Although, chromatin immunoprecipitation (ChIP) analysis of D4Z4 for H3K27, another repressive chromatin mark associated with Polycomb silencing, did not reveal any changes between control and FSHD muscle cells, levels of H3K27me3 associated with contacted D4Z4 repeats were significantly reduced when analyzed by 3D-fluorescence and in situ hybridization (3D-FISH) [41]. Consistently, levels of the histone H3K27 methyl-transferase EZH2, a component of the Polycomb repressive complex 2, at the D4Z4 repeats were shown to be reduced in FSHD muscle cells compared to controls. Conversely, ASH1L, a Trithorax group protein that is associated with transcriptionally active chromatin, was found to be specifically recruited to D4Z4 in FSHD cells but not in controls [37]. Notably, ASH1L recruitment was dependent on expression of a non-coding transcript mapped to the proximal D4Z4 unit [37].
Several DNA binding proteins, such as YY1 and CTCF, have been also shown to be associated with D4Z4. While YY1 normally binds D4Z4 as part of a repressor complex [42], CTCF has been recently reported to bind to the D4Z4 repeat in FSHD chromosomes and function as a Lamin A/C-dependent chromatin insulator and a boundary element [43]. Consistent with the presence of a chromatin boundary in the region, a distinct differentially methylated chromatin domain has been identified in the proximal D4Z4 unit and adjacent p13E-11 sequence [44]. However, in addition to protecting D4Z4 from epigenetic silencing by surrounding heterochromatic regions, CTCF may also prevent spreading of DNA methylation and limit RNA-mediated heterochromatin formation at D4Z4 as it was observed at other genomic loci [45] and therefore keep the D4Z4 chromatin open.
Since CTCF has also been shown to tether DNA to nuclear periphery and mediate chromatin loop formation [45;46], the increased CTCF binding to D4Z4 in FSHD may result in altered nuclear and chromatin organization. Indeed, the CTCF binding site in a D4Z4 unit has been shown to behave as a CTCF- and Lamin A-dependent perinuclear positioning element [43]. Finally, growing evidence suggests that the higher order chromatin architecture and the chromatin domain structure seem to be also affected in FSHD [41;47;48].
Although the exact mechanisms of chromatin changes in FSHD are still largely unknown, the recent findings seem to point to a unified model for both D4Z4 contraction-dependent (FSHD1) and contraction-independent (FSHD2) FSHD, where a failure in chromatin compaction during development of the normally heterochromatic D4Z4 repeat arrays on the disease permissive chromosomes results in inappropriate DUX4 expression.
However, even when D4Z4 arrays are packaged as euchromatin, only a small subset of myonuclei expresses DUX4 in culture [20]. A potentially related observation is the fact that FSHD-affected individuals have relatively normal muscle strength in their childhood and even after development of symptoms, muscle groups with normal strength are found adjacent to severely affected muscles [49;50] suggesting that additional regulatory mechanisms modulate DUX4 production. Bi-directional transcription in FSHD-patient cells reveals active transcription from short D4Z4 arrays with a predilection for the antisense direction [31]. A careful analysis of this activity using a bidirectional transcriptional reporter allowed the initiation site of antisense transcripts to be mapped, and identification of promoter sequences important for modulating the balance between sense and antisense transcription. D4Z4 contains two distinct promoters that initiate transcription in opposite directions and elimination of the start site of antisense transcripts causes sense transcription to predominate, suggesting that active antisense transcription modulates the production of sense transcripts (and thus DUX4 protein) even when arrays have a euchromatin configuration [31]. Antisense transcripts overlay an important DUX4 transcriptional enhancer [47], a situation previously shown to suppress enhancer activity and modulate transcription in the sense direction [35]. Thus bidirectional transcription may have dual roles in FSHD pathogenesis, one in early development where array suppression is established normally, and when this doesn’t occur, antisense transcription modulates DUX4 sense transcription in somatic cells and may be partly responsible for the sporadic expression pattern seen in myotube cultures.
Consequences of D4Z4 relaxation and DUX4 expression
DUX4 is a transcription factor of the double-homeobox family and is normally expressed in the human germline [20]. In the male germline, the spermatogonia and primary spermatocytes express DUX4, whereas the more mature spermatids do not. Although the role of DUX4 in normal germline biology is not known, when mis-expressed in human muscle cells, DUX4 activates the expression of many genes that are normally expressed in the germline [51], essentially activating a germline transcriptional program in postmitotic skeletal muscle cells.
The genes induced by DUX4 in skeletal muscle suggest several possible mechanisms of cellular damage in response to DUX4 expression [51]. DUX4-induced apoptosis of muscle cells might be, at least in part, secondary to the activation of a germline mitotic program that is incompatible with a postmitotic skeletal muscle cell. In addition, DUX4 activates the expression of numerous genes involved in atrophy and protein degradation, which might contribute to muscle deterioration.
One interesting possible mechanism of damage in response to DUX4 is the potential of germline genes to induce an immune response when expressed in somatic tissues. The germline is an immune privileged site, similar to the brain, and proteins expressed exclusively in the germline are recognized as foreign antigens when mis-expressed in non-germline tissues. This has been particularly well studied in cancer immunology because many cancers mis-express germline genes and induce an immune response that can have therapeutic benefit. Collectively, these germline genes mis-expressed by cancers have been called cancer testis antigens [52]. Several of the genes activated by DUX4 when it is expressed in skeletal muscle belong to this class of cancer testis antigens, and some of these gene products can be detected in FSHD muscle biopsies. FSHD muscle pathology is characterized by an infiltration of CD4 and CD8 T-cells [53;54] so it is plausible that this represents an immune response to germline antigens resulting in immune-mediated damage to FSHD skeletal muscle.
In addition to potentially stimulating an immune response to mis-expressed germline proteins, DUX4 induces the expression of human beta-defensin 3 (DEFB103A/B) [51]. The normal function of this secreted peptide is not fully known, but it has been shown to both block the innate immune response and to be an antagonistic ligand for the CXCR4 receptor [55-57]. Blocking the CXCR4 receptor prevents muscle regeneration and, therefore, this could have a direct role in preventing normal muscle regeneration in FSHD [58;59].
Finally, DUX4 activates the expression of several groups of endogenous retrotransposons [51]. The consequence of this is unknown, nor is it known whether this represents a function of DUX4 in the germline.
In summary, the genes regulated by DUX4 suggest several mechanisms of muscle damage when it is expressed in FSHD muscle. In addition, these genes provide a group of potential biomarkers that can be used as markers of disease activity and progression.
Conclusion
Over the last few years remarkable progress in understanding the molecular pathophysiology of FSHD has been made. Although many questions still remain unanswered and will require additional investigation, the prevailing mechanism from recent studies is that FSHD is caused by incomplete suppression of the DUX4 retrogene in skeletal muscle. Being a germline transcription factor, somatic expression of DUX4 releases a complex pattern of inappropriate gene expression in FSHD muscle, including activation of early stem cell and germline programs.
However, although DUX4 mis-expression should be considered the most likely cause for FSHD, many observations have not yet been adequately explained. We have only started to connect the molecular findings to the phenotype but have not yet explained the variability in onset and penetrance, nor the asymmetric nature of the disease. We do not yet fully understand the cause and consequence of the variegated pattern of bursts of DUX4 expression in tissue culture: does this represent a stochastic or triggered activation and, does this expression pattern explain the variability in onset and progression, and perhaps even the often asymmetric presentation of the disease? An alternative and perhaps equally likely explanation is that other genetic and environmental factors contribute to these clinical observations. Indeed, the existence of a separate mechanism for D4Z4 chromatin relaxation (FSHD2) strongly suggests that other factors must be involved in this disease.
From a therapeutic point of view, an important question that remains to be answered is which nuclei in skeletal muscle express DUX4 and the timing of DUX4 expression. Studies to interfere with DUX4 expression by RNA interference using siRNAs, shRNAs or AONs are ongoing [60;61], but without knowledge about the spatiotemporal distribution of DUX4 expression in the muscle and during development the efficacies of such therapies are difficult to predict. Future studies should therefore focus on these issues.
Key points.
FSHD is caused by D4Z4 repeat array contraction-dependent (FSHD1) or – independent (FSHD2) chromatin relaxation leading to transcriptional derepression of DUX4 in skeletal muscle;
The clinical presentation of FSHD is linked to a polymorphic DUX4 polyadenylation signal immediately distal to the D4Z4 repeat array;
The germline transcription factor DUX4 releases a complex pattern of inappropriate gene expression in FSHD muscle, including activation of early stem cell and germline programs;
DUX4 also induces the expression of several classes of retrotransposons
Acknowledgements
The authors thank all patients and family members for their long term commitment to our studies. Our work is supported by grants from the NIH (NINDS P01NS069539; CTSA UL1RR024160; NIAMS R01AR045203; NHGRI HG005608 and HG006493), the MDA (217596), the Geraldi Norton and Eklund family foundation, the FSH Society, The Friends of FSH Research, and the Stichting FSHD.
References
- 1.Flanigan KM, Coffeen CM, Sexton L, et al. Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul Disord. 2001;11:525–529. doi: 10.1016/s0960-8966(01)00201-2. [DOI] [PubMed] [Google Scholar]
- 2.Mostacciuolo ML, Pastorello E, Vazza G, et al. Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin Genet. 2009;75:550–555. doi: 10.1111/j.1399-0004.2009.01158.x. [DOI] [PubMed] [Google Scholar]
- 3.Padberg GW, Frants RR, Brouwer OF, et al. Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve. 1995;2:S81–S84. [PubMed] [Google Scholar]
- 4*.Statland JM, Tawil R. Facioscapulohumeral muscular dystrophy: molecular pathological advances and future directions. Curr Opin Neurol. 2011;24:423–428. doi: 10.1097/WCO.0b013e32834959af. A comprehensive review outlining the clinical features and pathogenic mechanism of FSHD
- 5.Brouwer OF, Padberg GW, Bakker E, et al. Early onset facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S67–S72. [PubMed] [Google Scholar]
- 6.Tyler FH, Stephens FH. Studies of disorders in muscle II: clinical manifestations and inheritance of facioscapulohumeral dystrophy in a large family. Ann Int Med. 1950;640:660. doi: 10.7326/0003-4819-32-4-640. [DOI] [PubMed] [Google Scholar]
- 7.Padberg GW. Thesis. Leiden University; Leiden, The Netherlands: 1982. Facioscapulohumeral disease. [Google Scholar]
- 8.van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Deutekom JC, Bakker E, Lemmers RJ, et al. Evidence for subtelomeric exchange of 3. 3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet. 1996;5:1997–2003. doi: 10.1093/hmg/5.12.1997. [DOI] [PubMed] [Google Scholar]
- 10.Wijmenga C, Hewitt JE, Sandkuijl LA, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 11.Lemmers RJ, de Kievit P, Sandkuijl L, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 12.Lemmers RJ, Wohlgemuth M, Frants RR, et al. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scionti I, Greco F, Ricci G, et al. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy. Am J Hum Genet. 2012;90:628–635. doi: 10.1016/j.ajhg.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas NS, Wiseman K, Spurlock G, et al. A large patient study confirming that facioscapulohumeral muscular dystrophy (FSHD) disease expression is almost exclusively associated with an FSHD locus located on a 4qA-defined 4qter subtelomere. J Med Genet. 2007;44:215–218. doi: 10.1136/jmg.2006.042804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakker E, Wijmenga C, Vossen RH, et al. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve. 1995;2:39–44. [PubMed] [Google Scholar]
- 16.Deidda G, Cacurri S, Grisanti P, et al. Physical mapping evidence for a duplicated region on chromosome 10 qter showing high homology with the Facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur J Hum Genet. 1995;3:155–167. doi: 10.1159/000472291. [DOI] [PubMed] [Google Scholar]
- 17.Lemmers RJL, de Kievit P, van Geel M, et al. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol. 2001;50:816–9. doi: 10.1002/ana.10057. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Forner J, Fournet S, et al. Improved characterization of FSHD mutations. Ann Genet. 2001;44:105–10. doi: 10.1016/s0003-3995(01)01075-9. [DOI] [PubMed] [Google Scholar]
- 19.Lemmers RJ, van der Vliet PJ, Klooster R, et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snider L, Geng LN, Lemmers RJLF, et al. Facioscapulohumeral Dystrophy: Incomplete Suppression of a Retrotransposed Gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixit M, Ansseau E, Tassin A, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabriels J, Beckers MC, Ding H, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 23.Hewitt JE, Lyle R, Clark LN, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 24.Snider L, Asawachaicharn A, Tyler AE, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Greef JC, Lemmers RJ, van Engelen BG, et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 26.de Greef JC, Lemmers RJ, Camano P, et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Overveld PG, Lemmers RJ, Sandkuijl LA, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 28.Chadwick BP. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18:1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh Y, Miyamoto N, Okada T, et al. The RS447 human megasatellite tandem repetitive sequence encodes a novel deubiquitinating enzyme with a functional promoter. Genomics. 2000;67:291–300. doi: 10.1006/geno.2000.6261. [DOI] [PubMed] [Google Scholar]
- 30.Tremblay DC, Alexander G, Jr., Moseley S, et al. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics. 2010;11:632. doi: 10.1186/1471-2164-11-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Block GJ, Petek LM, Narayanan D, et al. Asymmetric bidirectional transcription from the FSHD-causing D4Z4 array modulates DUX4 production. PLoS One. 2012;7:e35532. doi: 10.1371/journal.pone.0035532. In this study the authors show that D4Z4 contains two promoters that initiate sense and antisense transcription within the array, with the antisense transcription predominating. This study offers insight in some aspects of the chromatin regulation of D4Z4.
- 32.Hongay CF, Grisafi PL, Galitski T, et al. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 33.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 34.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanno T, Bucher E, Daxinger L, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 36.Zeng W, de Greef JC, Chen YY, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Cabianca DS, Casa V, Bodega B, et al. A Long ncRNA Links Copy Number Variation to a Polycomb/Trithorax Epigenetic Switch in FSHD Muscular Dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. In this study the authors identify a chromatin-associated long non-coding RNA that is involved in the derepression of the FSHD locus and the recuitment the Trithorax group protein Ash1L.
- 38.Kowaljow V, Marcowycz A, Ansseau E, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 39.de Greef JC, Frants RR, van der Maarel SM. Epigenetic mechanisms of facioscapulohumeral muscular dystrophy. Mutat Res. 2008;647:94–102. doi: 10.1016/j.mrfmmm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Greef JC, Wohlgemuth M, Chan OA, et al. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- 41.Bodega B, Ramirez GD, Grasser F, et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabellini D, Green M, Tupler R. Inappropriate Gene Activation in FSHD. A Repressor Complex Binds a Chromosomal Repeat Deleted in Dystrophic Muscle. Cell. 2002;110:339–248. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 43.Ottaviani A, Rival-Gervier S, Boussouar A, et al. The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet. 2009;5:e1000394. doi: 10.1371/journal.pgen.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X, Tsumagari K, Sowden J, et al. DNaseI hypersensitivity at gene-poor, FSH dystrophy-linked 4q35. 2. Nucleic Acids Res. 2009;37:7381–7393. doi: 10.1093/nar/gkp833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 46.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 47.Petrov A, Allinne J, Pirozhkova I, et al. A nuclear matrix attachment site in the 4q35 locus has an enhancer-blocking activity in vivo: implications for the facio-scapulo-humeral dystrophy. Genome Res. 2008;18:39–45. doi: 10.1101/gr.6620908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pirozhkova I, Petrov A, Dmitriev P, et al. A functional role for 4qA/B in the structural rearrangement of the 4q35 region and in the regulation of FRG1 and ANT1 in facioscapulohumeral dystrophy. PLoS One. 2008;3:e3389. doi: 10.1371/journal.pone.0003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedman SD, Poliachik SL, Carter GT, et al. The magnetic resonance imaging spectrum of facioscapulohumeral muscular dystrophy. Muscle Nerve. 2012;45:500–506. doi: 10.1002/mus.22342. [DOI] [PubMed] [Google Scholar]
- 50.Kan HE, Scheenen TW, Wohlgemuth M, et al. Quantitative MR imaging of individual muscle involvement in facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2009;19:357–362. doi: 10.1016/j.nmd.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 51**.Geng LN, Yao Z, Snider L, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. In this study the authors explore the consequences of DUX4 expression in skeletal muscle and show that it leads to the activation of early stem cell and germline programs.
- 52.Akers SN, Odunsi K, Karpf AR. Regulation of cancer germline antigen gene expression: implications for cancer immunotherapy. Future Oncol. 2010;6:717–732. doi: 10.2217/fon.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arahata K, Ishihara T, Fukunaga H, et al. Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve. 1995;2:56–66. [PubMed] [Google Scholar]
- 54*.Frisullo G, Frusciante R, Nociti V, et al. CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol. 2011;31:155–166. doi: 10.1007/s10875-010-9474-6. By a combination of MRI studies and immunological examinations in this study the autors provide evidence that circulating activated immune cells contribute to disease progression.
- 55.Semple F, Webb S, Li HN, et al. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur J Immunol. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Semple F, MacPherson H, Webb S, et al. Human beta-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur J Immunol. 2011;41:3291–3300. doi: 10.1002/eji.201141648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Z, Dubyak GR, Lederman MM, et al. Cutting edge: human beta defensin 3--a novel antagonist of the HIV-1 coreceptor CXCR4. J Immunol. 2006;177:782–786. doi: 10.4049/jimmunol.177.2.782. [DOI] [PubMed] [Google Scholar]
- 58.Melchionna R, Di CA, De MR, et al. Induction of myogenic differentiation by SDF-1 via CXCR4 and CXCR7 receptors. Muscle Nerve. 2010;41:828–835. doi: 10.1002/mus.21611. [DOI] [PubMed] [Google Scholar]
- 59.Griffin CA, Apponi LH, Long KK, et al. Chemokine expression and control of muscle cell migration during myogenesis. J Cell Sci. 2010;123:3052–3060. doi: 10.1242/jcs.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanderplanck C, Ansseau E, Charron S, et al. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61*.Wallace LM, Liu J, Domire JS, et al. RNA Interference Inhibits DUX4-induced Muscle Toxicity In Vivo: Implications for a Targeted FSHD Therapy. Mol Ther. 2012 doi: 10.1038/mt.2012.68. In this study the authors provide proof-of-concept that suppression of DUX4 expression by RNA interference in skeletal muscle may have therapeutic benefit.