Summary
To improve replication fidelity, mismatch repair (MMR) must detect non-Watson-Crick base pairs and direct their repair to the nascent DNA strand. Eukaryotic MMR in vitro requires pre-existing strand discontinuities for initiation; consequently, it has been postulated that MMR in vivo initiates at Okazaki fragment termini in the lagging strand and at nicks generated in the leading strand by the mismatch-activated MLH1/PMS2 endonuclease. We now show that a single ribonucleotide in the vicinity of a mismatch can act as an initiation site for MMR in human cell extracts and that MMR activation in this system is dependent on RNase H2. As loss of RNase H2 in S.cerevisiae results in a mild MMR defect that is reflected in increased mutagenesis, MMR in vivo might also initiate at RNase H2-generated nicks. We therefore propose that ribonucleotides misincoporated during DNA replication serve as physiological markers of the nascent DNA strand.
Graphical Abstract
Highlights
► Ribonucleotides incorporated into eukaryotic DNA are not mismatch repair substrates ► Intermediates of rNMP processing can act as initiation sites for mismatch repair ► RNase H2 deficiency in S. cerevisiae decreases MMR efficiency ► rNMPs can act as markers of nascent DNA strands
Introduction
Correction of replication errors requires that MMR be targeted to the nascent DNA strand. In gram-negative bacteria such as Escherichia coli (E. coli), newly synthesized DNA is transiently unmethylated at adenines in d(GATC) sites, and this allows the MutS/MutL-activated MutH endonuclease to introduce a nick into the undermethylated nascent strand, where exonucleolytic degradation of the error-containing section commences (Jiricny, 2006). Gram-positive bacteria and eukaryotes do not use methylation in strand discrimination, and it was suggested that MMR may be directed to nascent DNA by strand discontinuities such as gaps between Okazaki fragments (Claverys and Lacks, 1986). This hypothesis was supported experimentally in extracts of human and D. melanogaster cells, where a single nick was shown to be necessary and sufficient to direct MMR to the discontinuous strand of a circular heteroduplex substrate carrying a single mismatch (Holmes et al., 1990; Thomas et al., 1991). The repair process could be directed by nicks situated either 3′ or 5′ from the misincorporated nucleotide. This was puzzling, because EXO1, the only exonuclease implicated in eukaryotic MMR, has an obligate 5′ → 3′ polarity. An answer to this puzzle came when the PMS2 subunit of MutLα (a heterodimer of MMR proteins MLH1 and PMS2) was shown to associate with PCNA and introduce additional nicks into the discontinuous strand that could be used as EXO1 loading sites (Pluciennik et al., 2010). This finding helped explain how MMR directs the repair process into the nascent DNA in both lagging and leading strands (Peña-Diaz and Jiricny, 2010). However, on the leading strand, the MutLα/PCNA complex would have to travel from the 3′ terminus toward and past the mismatch that could be several hundred nucleotides distant, and also the MMR system would have to compete with nucleosome loading behind the replication fork (Schöpf et al., 2012). Therefore, it was anticipated that MMR on the leading strand might be less efficient than on the lagging strand, where strand discontinuities are readily available. This was indeed shown to be the case (Nick McElhinny et al., 2010a), but because the difference was relatively small, it was postulated that MMR efficiency in the leading strand might be augmented by additional factors.
Recently, more than a million ribonucleotide monophosphates (rNMPs) were reported to be incorporated into mouse genomic DNA during replication (Hiller et al., 2012; Reijns et al., 2012), and a similar situation exists in Saccharomyces cerevisiae (S. cerevisiae), where the leading-strand polymerase (pol-ε) incorporates approximately four times more ribonucleotides into the nascent DNA than the lagging-strand enzyme (pol-δ): 1 rNMP/1,250 dNMPs versus 1/5,000, respectively (Nick McElhinny et al., 2010b, 2010c).
Ribonucleotides are removed from DNA by RNases H1 and H2, whereby the former enzyme processes stretches of more than three rNMPs such as those found at the 5′ termini of Okazaki fragments, while RNase H2 can incise the hybrid strand 5′ from even a single rNMP (Eder and Walder, 1991). The ribonucleotide(s) can then be removed by the flap endonuclease FEN1 and/or EXO1 (Rydberg and Game, 2002; Sparks et al., 2012). We wondered whether RNase H2-generated strand breaks arising during rNMP removal might be utilized by the MMR system as initiation sites for the exonucleolytic degradation of the error-containing strand. Here, we show that a single ribonucleotide in close proximity of a mismatch can act as an initiation site for MMR in cell extracts as well as in a reconstituted system, and that MMR activation in this scenario is dependent on RNase H2. As a consequence, loss of RNase H2 in S. cerevisiae leads to a mild defect in MMR and increased mutagenesis.
Results
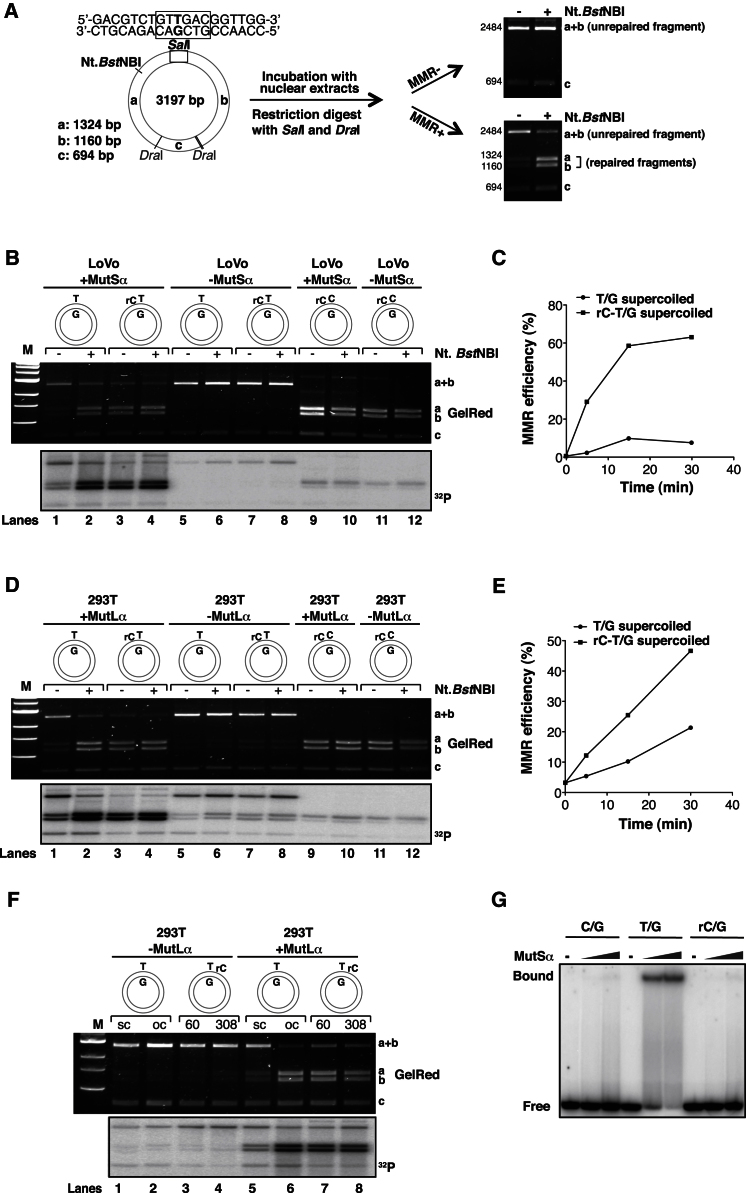
We first made use of an in vitro MMR assay (Baerenfaller et al., 2006; Modrich, 2006; Schanz et al., 2009), in which a phagemid substrate containing a T/G mismatch in a SalI restriction site is incubated with extracts of human cells. Because MMR requires a pre-existing strand discontinuity for initiation, the T/G mispair in a closed circular or supercoiled (sc) substrate is not repaired. However, introduction of an Nt.BstNBI-catalyzed nick 361 nucleotides 5′ from the mispaired T licenses MMR, which corrects the T/G mismatch to C/G and thus makes the phagemid susceptible to SalI digestion. In this system, MMR efficiency is estimated by digesting the phagemid recovered from the extracts with SalI and DraI, and quantitating the fraction of DNA in the 1,324 (band a) and 1,160 (band b) base-pair fragments, which are indicative of successful MMR (Figure 1A).
Figure 1.
A Single Ribonucleotide in a DNA Heteroduplex Acts as an Initiation Site for MMR
(A) Schematic representation of the in vitro MMR assay. In the absence of a nick, very little repair of the T/G mismatch takes place, and digestion of the phagemid DNA with SalI and DraI gives rise to 2,484 (a + b), 694 (c), and 19 bp fragments (the smallest fragment is not detectable on these 1% agarose gels stained with GelRed). Upon introduction of a nick at the Nt.BstNBI site, T/G-to-C/G repair restores the SalI site, such that the phagemid DNA is cut into 1,324 (a), 1,160 (b), 694 (c), and 19 bp fragments.
(B) The presence of a single cytidine (rC) 54 nucleotides 5′ from the mispaired T made the T/G substrate susceptible to MMR even in the absence of a Nt.BstNBI nick. The substrates were incubated with MutSα-deficient LoVo nuclear extracts (supplemented or not with recombinant MutSα), and repair efficiency was quantitated by estimating the percentage of DNA in bands a and b. Autoradiograph of the same gel (32P) showed that [α-32P]dCMP was incorporated preferentially into the repaired fragments a and b. The figure shows a representative result. The experiment was carried out in triplicate.
(C) Kinetic analysis performed with the supercoiled T/G and rC-T/G substrates in MutSα-complemented LoVo extracts.
(D) As in (B), but the substrates were incubated with MutLα-deficient extracts of 293TLα− cells supplemented or not with recombinant MutLα.
(E) Kinetic analysis performed with the supercoiled T/G and rC-T/G substrates in MutLα-complemented 293TLα− extracts.
(F) Position and distance of the ribocytidine does not affect MMR efficiency. Supercoiled (sc), Nt.BstNBI-nicked (oc) T/G phagemids, or T/G-rC substrates containing a single rC residue 60 or 308 nucleotides 3′ from the mispaired T, were incubated with 293TLα− cells supplemented or not with recombinant MutLα. Repair efficiency was quantitated by estimating the percentage of DNA in bands a and b. Autoradiograph of the same gel (32P) showed that [α-32P]dCMP was incorporated preferentially into the repaired fragments a and b. The figure shows a representative result.
(G) Band-shift assay (Cejka et al., 2005) using recombinant MutSα and oligonucleotides C/G, T/G, or rC/G. The figure shows an autoradiograph of a 6% native polyacrylamide gel. M, size marker (1 kb ladder, New England BioLabs).
Ribonucleotide-Dependent Mismatch Repair in Human Cell-free Extracts
We first generated a substrate containing a single rCMP residue (rC) 54 nucleotides 5′ from the mispaired T (rC-T/G), as well as a homoduplex (rC-C/G) control, and incubated them with extracts of LoVo cells, which lack the major mismatch recognition factor MSH2/MSH6 (MutSα) and are thus MMR deficient. As anticipated, none of the heteroduplex substrates were repaired (Figure 1B, lanes 5–8), but when the extract was supplemented with purified recombinant MutSα, ∼60% of the Nt.BstNBI-nicked T/G substrate was corrected to C/G within 30 min (Figure 1B, lane 2). The supercoiled T/G substrate remained largely uncorrected (Figure 1B, lane 1). Importantly, the supercoiled rC-T/G substrate was repaired nearly as efficiently as the nicked T/G phagemid in a reaction dependent on MutSα (Figure 1B, lane 3, compare with lane 2), and the efficiency of repair was only slightly improved by Nt.BstNBI pretreatment (Figure 1B, lane 4). Kinetic analysis (Figure 1C) further confirmed that a single ribonucleotide in the supercoiled T/G heteroduplex was sufficient to convert a MMR-refractory substrate into a MMR-susceptible one in this system.
We repeated the above experiments with MMR-deficient 293TLα− extracts (293T) that lack MutLα (Cejka et al., 2003). When these extracts were supplemented with purified recombinant MutLα, the supercoiled rC-T/G substrate was repaired almost as efficiently as the nicked rC-T/G or T/G heteroduplexes (Figure 1D, lanes 2–4). As observed in previous studies (Constantin et al., 2005; Schanz et al., 2009), small amounts of repair were detected also in the absence of added MutLα, as this factor is not absolutely required for 5′ → 3′ MMR (Figure 1D, lanes 5–8). When compared with MutSα-supplemented LoVo extracts (Figure 1B, lane 1; and Figure 1C), the supercoiled T/G substrate was repaired to a greater extent in the MutLα-supplemented 293T extracts (Figure 1D, lane 1; and Figure 1E). This is most likely due to the endonuclease activity of MutLα, which can introduce random nicks with low efficiency also into either strand of a supercoiled substrate (Pluciennik et al., 2010) and trigger noncanonical MMR (Peña-Diaz et al., 2012).
By carrying out the in vitro MMR reactions in the presence of [α-32P]dCTP, we saw only background levels of radioactivity incorporated into the T/G or rC-T/G substrates incubated with LoVo extracts in the absence of added MutSα (Figure 1B, lanes 5–8 in lower panel). Similarly, the rC-C/G control substrate remained largely unlabeled even when the extracts were supplemented with MutSα (lanes 9–12 in lower panel). This shows that the mere presence of the ribonucleotide in a substrate does not trigger long-patch DNA synthesis, irrespective of whether the extracts are MMR deficient or proficient.
Identical experiments carried out in the MMR-deficient extracts of 293TLα− cells yielded similar results, even though the background levels of [α-32P]dCMP incorporation into the T/G and rC-T/G substrates were somewhat higher (Figure 1D, lanes 5–8 in lower panel; compare with Figure 1B, lanes 5–8 in lower panel). This is due to residual 5′ → 3′ MMR that can proceed independently of MutLα, and that initiates at rare random nicks in the closed circular phagemids. Notably, the a + b fragments contained proportionally more radioactive nucleotide than the cleaved fragments a and b. This indicates that most of the excision/resynthesis events failed to reach the mispaired T and therefore convert the T/G mispair to C/G, which would have restored the SalI site.
In contrast to 5′ → 3′ repair, MutLα is indispensable for 3′ → 5′ MMR (Kadyrov et al., 2006). To learn whether similar criteria apply also to ribonucleotide-directed MMR, we first constructed a substrate that contained the rCMP residue 60 or 308 nucleotides 3′ from the mispaired T (T/G-rC). When incubated with 293TLα− extracts that are devoid of MutLα, no repair of the control T/G phagemid was detected, irrespective of whether the substrate was supercoiled or nicked (Figure 1F, lanes 1 and 2, respectively). This result differed from that obtained with the 5′-nicked substrate (Figure 1D, lane 6) and confirms the requirement for MutLα in 3′ nick-directed MMR (Pluciennik et al., 2010). Similarly, only background levels of [α-32P]dCMP were incorporated into the T/G-rC substrates (Figure 1F, lanes 3 and 4). However, when the extracts were supplemented with purified recombinant MutLα, the closed circular T/G-rC substrates were repaired almost as efficiently as the nicked T/G heteroduplex (Figure 1F, compare lane 6 with lane 7 or lane 8).
Taken together, the above results demonstrate that strand discontinuities arising during processing of ribonucleotides in DNA can be hijacked by the MMR machinery for initiation of mismatch-activated excision, even over a distance of more than 300 nucleotides. Moreover, the finding that only background levels of [α-32P]dCMP were incorporated into the rC-C/G phagemid (Figure 1B, lanes 9–12) showed that removal of the ribonucleotide, presumably by an RNaseH2-dependent mechanism (Sparks et al., 2012), involved only short-patch (fewer than ∼30 nucleotides) repair synthesis. The mere presence of the ribonucleotide in these substrates was not sufficient to trigger long-patch repair synthesis, be it MMR independent or MMR dependent. The lack of activation of the MMR process by the rCMP was further strengthened by the finding that purified recombinant human MutSα did not bind the rC/G “mispair” in an electrophoretic mobility-shift assay (Figure 1G), similarly to S. cerevisiae MutSα, which bound even an rG/T mispair with only very low affinity (Clark et al., 2011).
RNase H2, but Not H1, Can Initiate Mismatch-Dependent Strand Degradation in a Reconstituted In Vitro System
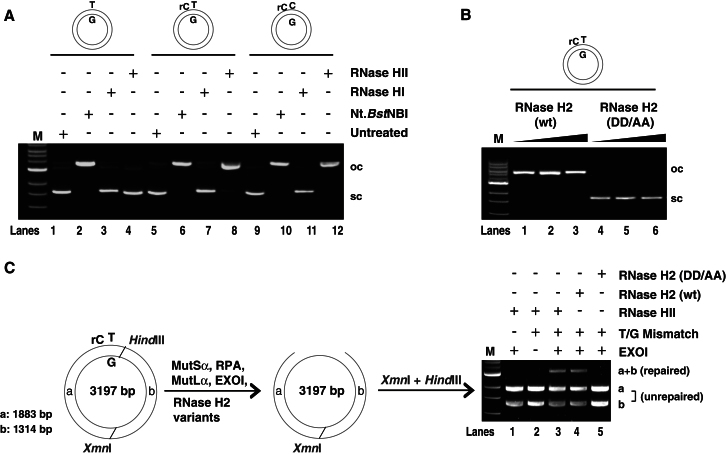
We wanted to learn which enzyme was responsible for incising the heteroduplexes in the above in vitro MMR assays. Since ribonucleotide removal from DNA is initiated by RNase H, we tested the activity of RNases H1 and H2 on our substrate. Incubation of the rC-T/G and rC-C/G substrates with bacterial RNase HII or with human RNase H2 led to the formation of open-circular forms of the phagemids, whereas those incubated with bacterial RNase HI or a catalytically inactive version of human RNase H2 (D34A/D169A or DD/AA) remained supercoiled (Figures 2A and 2B). This result agrees with work from other laboratories, which reported that only RNase H2 can incise RNA/DNA hybrids containing single ribonucleotides (Cerritelli and Crouch, 2009; Reijns et al., 2012; Sparks et al., 2012).
Figure 2.
RNase H2-Mediated Processing of a Single Ribonucleotide in the Heteroduplex Substrate Provides MMR with an Initiation Site In Vitro
(A and B) The indicated supercoiled (sc) substrates were incubated with (A) purified recombinant RNase HI or HII or with the nickase Nt.BstNBI or (B) increasing concentrations of human recombinant RNase H2 wild-type (WT) or the catalytically inactive (DD/AA) mutant. Only RNase HII, Nt.BstNBI, and RNase H2 WT generated open circular (oc) form, which confirms that only RNase H type 2 ribonuclease can incise DNA substrates containing a single ribonucleotide.
(C) Schematic representation of mismatch-dependent strand-degradation reaction, using the indicated purified recombinant proteins. Phagemid molecules containing MMR-generated single-stranded gaps around the mismatch (indicative of efficient strand degradation) are resistant to cleavage with HindIII and are only linearized with XmnI (a + b). Fully double-stranded (unrepaired) molecules are cleaved with both enzymes into fragments a and b. M, size marker (1 kb ladder, New England BioLabs).
We wanted to confirm our findings by reconstituting the ribonucleotide-dependent MMR reaction from individual purified proteins. Using a substrate containing a nick within ∼1 kb of the mispair, the minimal 5′-to-3′ MMR system could be reconstituted from MutSα, MutLα, RPA, and EXO1 (Genschel and Modrich, 2003). In that system, mismatch-dependent strand degradation gave rise to a single-stranded gap spanning the distance between the nick and ∼150 nucleotides past the mismatch. The appearance of the single-stranded gap could be monitored by restriction enzyme digestion, because the substrate became refractory to cleavage by enzymes with recognition sites within the gap. We decided to adopt this approach but substitute the T/G substrate with the rC-C/G or rC-T/G phagemids, which can be digested with HindIII and XmnI to give rise to fragments a and b of 1,883 and 1,314 bp, respectively. The HindIII recognition sequence is situated ten base pairs from the T/G mismatch and is thus an ideal indicator of single-stranded gap formation in this region. As shown in Figure 2C, incubation of the rC-C/G control substrate with MutSα, MutLα, RPA, EXO1, and bacterial RNase HII, followed by digestion with HindIII and XmnI, yielded only the fragments a and b (lane 1). A similar result was obtained with the rC-T/G substrate when EXO1 was omitted (lane 2). In contrast, incubation of the rC-T/G substrate with MutSα, MutLα, RPA, EXO1, and bacterial RNase HII (lane 3) or human RNase H2 (lane 4) gave rise, in addition to fragments a and b, also to fragment a + b, which is indicative of single-strand gap formation. Addition of the RNase H2 DD/AA mutant (lane 5) yielded no a + b fragment. Collectively, these data provided formal proof that mismatch- and MutSα/MutLα-activated EXO1 could load at RNase H2-catalyzed strand breaks and initiate the strand-degradation process in spite of the fact that the 5′ terminus of the nick is a ribonucleotide.
RNase H2-Mediated MMR in Human and Mouse Nuclear Extracts
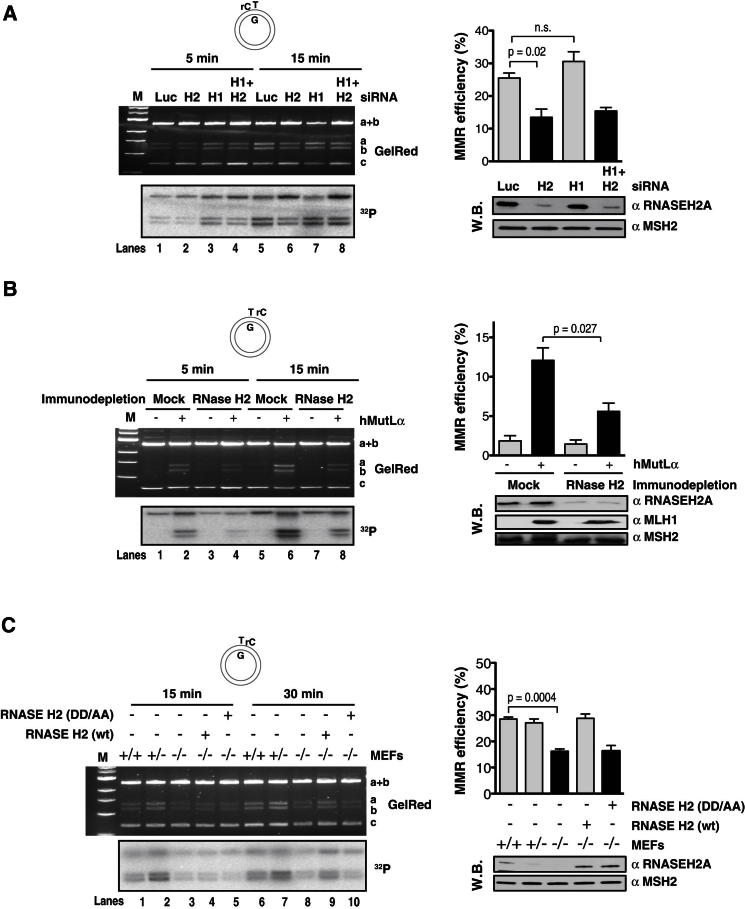
We next set out to confirm that the MMR process seen in nuclear extracts was also dependent on RNase H2. In the first experiment, we knocked down RNase H1, RNASEH2A, or both in 293 cells by siRNA (Figure 3A, bottom right panel). The knockdown of RNASEH2A decreased repair efficiency of the rC-T/G substrate to ∼50% of that seen in extracts of cells treated with siLUC (Figure 3A, compare lanes 5 and 6), while knockdown of RNase H1 had no apparent effect (Figure 3A, lanes 3 and 7). We next immunodepleted RNase H2 from MutLα-deficient 293T nuclear extracts, using a polyclonal antibody raised against the heterotrimer (Figure 3A, bottom right). As anticipated, no repair activity was observed in the absence of MutLα in either mock- or RNase H2-depleted extracts (Figure 3B, lanes 1, 3, 5, and 7), whereas addition of recombinant MutLα successfully rescued MMR in the mock-depleted extracts (lanes 2 and 6). In contrast, MutLα rescued MMR only weakly in the RNase H2-depleted extracts (lanes 4 and 8). Finally, we employed nuclear extracts derived from RNase H2 WT (+/+), heterozygous (+/−), and homozygous null (−/−) mouse fibroblasts (Reijns et al., 2012) and performed MMR assays with the T/G-rC substrate (Figure 3C, bottom right). As shown in Figure 3C, there was no significant difference in repair efficiency between WT and heterozygous fibroblasts (compare lanes 2 and 7 with lanes 1 and 6). However, RNase H2 deficiency resulted in a substantial decrease in repair efficiency when compared to WT extracts (compare lanes 3 and 8 with lanes 1 and 6). Importantly, this MMR defect could be completely rescued by the addition of recombinant human WT RNase H2 (lane 9), but not the DD/AA mutant (lane 10). Our data therefore show that RNase H2 is the key nuclease responsible for MMR initiation in the rC-T/G substrate in human and mouse cell extracts.
Figure 3.
RNase H2-Activated MMR in Human and Mouse Nuclear Extracts
(A) RNASEH2A knockdown decreases MMR activity on the rC-T/G heteroduplex. (Left panels) Efficiency of the mismatch repair reaction at 5 and 15 min time points in extracts of 293 cells transiently transfected with the indicated siRNAs. The figure shows an agarose gel image (GelRed) and autoradiograph (32P) of a representative experiment. (Top right) Plot of data from the 15 min time points of three independent experiments, with error bars representing standard deviation from the mean. (Bottom right) Western blot showing siRNA-mediated knockdown efficiency of RNASEH2A in 293 cells. Luc, siRNA against luciferase control; H1, H2a, H1+H2a, siRNAs against RNase H1, H2a, or both.
(B) Immunodepletion of RNase H2 decreases MMR activity on the T/G-rC heteroduplex. (Left panels) MMR assay carried out in extracts of 293T cells immunodepleted of RNase H2. The figure shows an agarose gel image (GelRed) and autoradiograph (32P) of 5 and 15 min time points of a representative experiment. (Top right) Plot of data from the 15 min time points of three independent experiments, with error bars representing standard deviation from the mean. (Bottom right) Immunodepletion of RNase H2 and supplementation of 293T extracts with recombinant MutLα. This western blot shows the efficiency of the immunodepletion procedure and the amounts of recombinant MLH1 relative to endogenous MSH2 levels.
(C) Extracts of RNase H2 knockout mouse embryonal fibroblasts display decreased MMR activity on the T/G-rC heteroduplex at the 15 and 30 min time points. (Left panel) An agarose gel image (GelRed) and autoradiograph (32P) of a representative experiment. (Top right) Plot of data from the 30 min time points of three independent experiments, with error bars representing standard deviation from the mean. (Bottom right) Western blot showing the amount of RNASEH2A in the MEFs as well as the amount of recombinant RNase H2 protein added to restore the MMR defect. MSH2 visualized with an anti-MSH2 antibody served as the loading control in the above experiments. M, size marker (1 kb ladder, New England BioLabs).
Loss of RNase H2 in S. cerevisiae Impairs MMR in the Leading Strand
In the above experiments we have demonstrated that RNase H2 can direct MMR to the ribonucleotide-containing strand and thus facilitate strand discrimination in MMR assays using cell extracts. We next wanted to test whether RNase H2 performs this function also in vivo. To this point, we made use of an assay in which the fidelity of leading- and lagging-strand replication can be monitored by the reversion rate of a mutant ura3-29 allele at the agp1 locus of a S. cerevisiae ogg1Δ strain. In this genetic background, reversion of the ura3-29 point mutation was previously shown to be caused by MMR dysfunction (Pavlov et al., 2003), whereby the loss of MSH2 increased lagging-strand (OR2 in Table 1) mutagenesis nearly 7-fold, as compared to only 3-fold in the leading strand (OR1 in Table 1). This difference was proposed to be caused by the higher MMR efficiency in the lagging strand that is linked to the ready availability of initiation sites provided by Okazaki fragment termini (Nick McElhinny et al., 2010a). The loss of RNH201 (the catalytic subunit of RNase H2 in yeast) led to an ∼1.5-fold increase in mutation rates in both leading and lagging strands in this system (Clark et al., 2011; Kim et al., 2011; Nick McElhinny et al., 2010b). We now show that the rnh201Δ mutation failed to further increase the mutation rate in the MMR-deficient background (rnh201Δ msh2Δ in Table 1). Moreover, the relative contribution of RNase H2 toward replication fidelity was higher on the leading strand (24% of the total MMR activity) and amounted to twice that seen on the lagging strand (10% of the total MMR activity). In addition, DNA sequencing revealed that 100% of the mutants displayed C-to-A transversions in the leading strand, the signature phenotype of a MMR defect in this reporter system (Pavlov et al., 2003). Taken together, the data indicate that the increased mutagenesis detected in the rnh201Δ mutant is likely linked to diminished MMR efficiency.
Table 1.
Reversion Rates of a Mutant ura3-29 Allele at the agp1 Locus of a S. cerevisiae ogg1Δ Strain
| Strain | ura3-29 Orientation | Mutation Rate × 10−7 | 95% Confidence Limits | Fold Increase Relative to WT | Residual MMR Activity (%) |
|---|---|---|---|---|---|
| Wild-type | LD | 3.95 | (3.25–4.4) | 1 | 100 |
| LG | 1.01 | (0.69–1.25) | 1 | 100 | |
| rnh201Δ | LD | 5.86 | (4.91–8.47) | 1.48 | 76 |
| LG | 1.64 | (1.41–1.93) | 1.62 | 90 | |
| msh2Δ | LD | 12.05 | (9.89–13.25) | 3.05 | 0 |
| LG | 6.95 | (5.97–8.14) | 6.89 | 0 | |
| msh2Δrnh201Δ | LD | 11.75 | (10.41–12.61) | 2.98 | 4 |
| LG | 7.16 | (5.28–9.12) | 7.09 | 2 |
LD represents the leading strand and LG the lagging strand. Data from one representative experiment are shown. Fold increase was calculated relative to the wild type (WT) strain. The residual MMR activity was calculated using the following formula: [1 − (mutation rate of mutant strain − mutation rate of WT)/(mutation rate of msh2Δ strain − mutation rate of WT)] × 100, as described previously (Pavlov et al., 2003).
A key hallmark of MMR deficiency is instability of the so-called microsatellites—sequence elements consisting of repeats of mono-, di-, or trinucleotides. These motifs are prone to strand misalignments during replication, which give rise to insertion/deletion loops (IDLs). Because IDLs are repaired predominantly by MMR, cells lacking this repair pathway display microsatellite instability (MSI), which can be measured in suitable reporter assays (Strand et al., 1993). We decided to make use of this phenomenon to determine the effect of RNase H2 deficiency on MMR efficiency. In our assay, a T9 repeat was introduced in-frame into the URA3 coding sequence, and the construct was integrated into the ADE2 locus in the S. cerevisiae genome. Alterations other than ±3 nucleotides in the length of the T9 repeat inactivate the URA3 gene and render the cells resistant to 5-fluoroorotic acid (FOA). As anticipated, the T9URA3 marker was highly unstable in the MMR-deficient msh2 or mlh1 strains; we observed a 64- and 52-fold increase in mutation rates relative to WT, respectively (Table 2). Importantly, in the absence of RNH201, we observed an overall 4-fold increase in mutation rates relative to WT, whereas no significant increase was observed when RNH201 was deleted in the MMR-deficient background. Sequencing of a 242 bp region of URA3 flanking the T9 repeat in 50 FOA-resistant rnh201Δ clones revealed an 8.5-fold increase in the rate of −2 deletions compared to WT, which are most likely due to mutagenic processing of ribonucleotides in DNA catalyzed by topoisomerase I (Kim et al., 2011). Importantly, we also detected a 7.4-fold increase in the rate of −1 deletions in the T9 repeat of the rnh201Δ strain compared to WT. This is a key hallmark of MMR deficiency, as clearly substantiated by the finding that all mutations identified in the msh2Δ and rnh201Δmsh2Δ strains were −1 deletions in this motif. Taken together, the above results show that the eukaryotic MMR system can and does use intermediates of processing of misincorporated ribonucleotides as additional repair initiation sites.
Table 2.
Mutation Rates within the T9URA3 Cassette in the Indicated S. cerevisiae Strains
| Genotype | Overall Mutation Rate × 10−7 | Fold Increase Relative to WT | −1 Deletion Rate at the T9 Repeat × 10−7 | Fold Increase Relative to WT |
|---|---|---|---|---|
| Wild-type | 0.24 (0.17 − 0.36) a | 1 | 0.015 (3/45)b | 1 |
| rnh201Δ | 1.02 (0.79 − 1.55)a | 4.3 | 0.12 (6/50)b | 7.3 |
| msh2Δ | 15.4 (11.77 − 19.62)a | 64.2 | 15.4 (31/31)b | 917 |
| msh2Δrnh201Δ | 15.21 (13 − 19.07)a | 63.4 | 15.21 (28/28)b | 905 |
| mlh1Δ | 12.57 (10.04 −19.92)a | 52.4 | ND | ND |
| mlh1Δrnh201Δ | 13.53 (10.33 − 17.31)a | 56.4 | ND | ND |
The data were obtained from 32-50 independent cultures of each strain. Fold increases were calculated relative to the wild type strain. Mutation rates and confidence levels were calculated by the method of Lea and Coulson (Lea and Coulson, 1949). The −1 deletion rates at the T9 repeat were calculated from the formula [del rate = (nr. of sequenced mutants with −1 deletions inT9/total number of sequenced mutants) x overall mutation rate]. ND, not determined.
Numbers in parentheses represent 95% confidence levels.
Numbers in parentheses represent the number of colonies with −1 deletions versus the number of sequenced colonies (see the Experimental Procedures).
Discussion
How organisms lacking MutH homologs direct MMR to the continuous leading stand has puzzled researchers for decades. The requirement for the endonucleolytic activity of MutL or its eukaryotic orthologs (Kadyrov et al., 2006) showed that this process was dependent on the introduction of discontinuities where EXO1 can load, but it was only with the characterization of an association between MutLα and PCNA (Pluciennik et al., 2010) that the molecular mechanism of the process could be understood (Peña-Diaz and Jiricny, 2010). Given the strong mutator phenotype of cells lacking the MutLα endonuclease, it must be assumed that this enzymatic activity represents the key determinant of strand directionality in MMR, similarly to MutH in E. coli (Zell and Fritz, 1987). However, DNA polymerases incorporate several noncanonical nucleotides into nascent DNA, such as deoxyuridine (Andersen et al., 2005), 8-oxo-deoxyguanosine (Maki and Sekiguchi, 1992), and ribonucleotides (Nick McElhinny et al., 2010b; Reijns et al., 2012). Because metabolism of these nucleotides involves excision, we wondered whether the strand breaks generated during their removal might be deployed by the MMR system for initiation of excision in cases where a misincorporated nucleotide is in the vicinity. We decided to investigate the link between MMR and the processing of ribonucleotides, because their abundance in nascent DNA is high (Reijns et al., 2012)—particularly in the leading strand, due to the fact that they are inefficiently excised by the proofreading activity of pol ε (Williams et al., 2012). Moreover, the loss of RNase H1/2 gives rise to higher mutation rates and genomic instability (Clark et al., 2011; Lazzaro et al., 2012; Reijns et al., 2012), and, although it was proposed that the mutagenicity of ribonucleotides might be caused by their misincorporation opposite a noncomplementary template nucleotide (Clark et al., 2011; Shen et al., 2012), this possibility appeared to us unlikely, given that the bases of ribonucleotides form perfect hydrogen bonds with DNA bases. We now show that the presence of ribonucleotides in nascent DNA and their processing by RNase H2 increases MMR efficiency, most likely through providing the system with additional strand breaks that can be utilized as entry sites for EXO1 (Figure 4). The contribution of this pathway to MMR fidelity is small, because it requires that the ribonucleotide and the mispair are within less than ∼1 kb of each other. However, if the MMR system also makes use of intermediates of processing of other nascent strand markers such as deoxyuridine and 8-oxo-deoxyguanosine, then this mechanism may play a substantially more important part in the initiation of MMR than first thought.
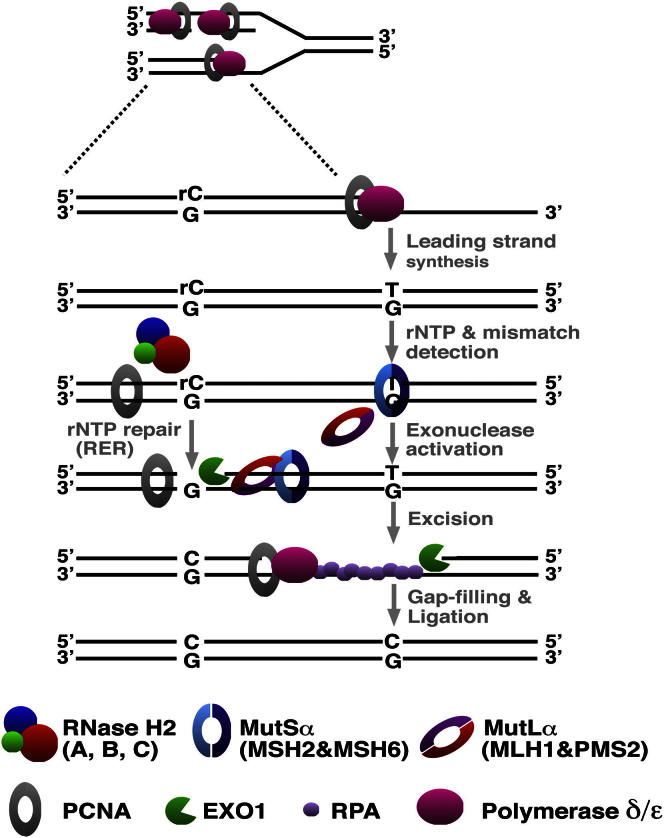
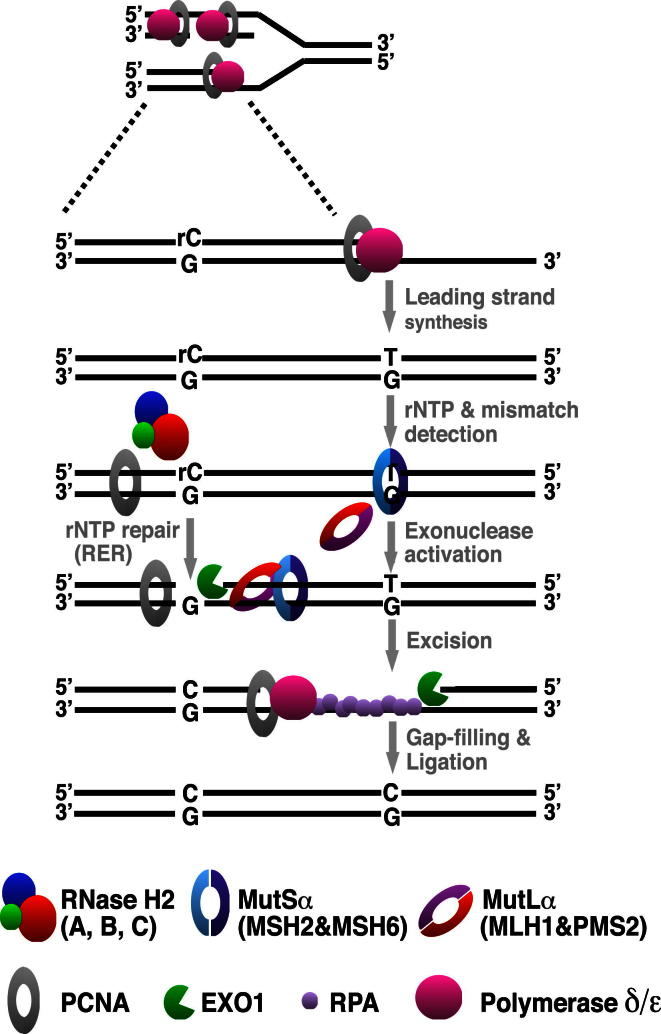
Figure 4.
Putative Mechanism of Ribonucleotide-Mediated Strand Discrimination during Eukaryotic MMR
Replicative polymerases such as polymerase epsilon (pol ε) erroneously incorporate single ribonucleotides (1 rNMP per 1,250 dNMPs) into the nascent DNA strand. RNase H2 cleaves the DNA 5′ from the rNMP. In the event that the polymerase generates a mispair in the vicinity, the mismatch-activated MMR proteins can “hijack” the transient RNase H2-mediated nick as a loading site for EXO1, which then degrades the error-containing strand.
Given that the presence of ribonucleotides in DNA is linked to genomic instability (Clark et al., 2011; Lazzaro et al., 2012; Reijns et al., 2012), it might appear surprising that replicative polymerases have not evolved to exclude them. Our findings suggest that the presence of rNMPs, possibly together with other markers of nascent DNA, may have been tolerated during evolution (Andersen et al., 2005; Russo et al., 2004), because they aid metabolic processes that need to distinguish between the parental and daughter DNA strands.
Experimental Procedures
Substrates, Nuclear Extracts, and MMR Assays
The substrates were generated as described previously (Baerenfaller et al., 2006). Briefly, the hetero- and homoduplexes were constructed by primer extension using the oligonucleotides listed below as primers and the single-stranded phagemid DNA as template. Depending on the orientation of the ribonucleotide and the nick, two different ssDNA templates were used (pRichi-350topSalI creates 3′ substrates, and pRichi-2850topSalI creates 5′ substrates). After primer extension, ligation, and isolation of the desired supercoiled heteroduplex substrates on CsCl gradients, MMR assays were carried out as described (Baerenfaller et al., 2006).
Unless otherwise specified, the MMR reactions were carried out with 100 ng DNA substrate and 100 μg nuclear extracts in a total volume of 25 μl in a buffer containing 20 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 110 mM KCl, 1 mM glutathione, 50 μg/ml BSA, 100 μM dNTPs, and, where indicated, 2 μCi of [α-32P]dCTP. The reactions were incubated at 37°C for 30 min. For time course experiments, 8 μl aliquots were withdrawn at the indicated time points. The reaction was stopped by adding an equal volume of 2× stop solution containing 1 mM EDTA, 3% SDS, and 5 mg/ml Proteinase K. The samples were incubated at 45°C for 30 min, purified on Mini-Clean columns (QIAGEN), and subjected to restriction digests. The digested DNA was resolved on 1% agarose gels. Nuclear extracts used in this study were prepared from HEK293, LoVo, 293TLα−, and mouse embryonal fibroblasts obtained from RNase H2, +/−, −/− mice as indicated in the figures.
Primers
All primers were obtained from Microsynth (Balgach, Switzerland). The sequences are indicated below. The SalI restriction site (GTCGAC) is underlined. The mispaired residue is highlighted in bold. The position of the ribocytidine is indicated in lower case.
T/G-SalI primer is as follows: 5′-CCAGACGTCTGTTGACGTTGGGAAGCTTGAG-3′. 5′ rC-T/G primer is as follows: 5′-GAATTGTAATAcGAACACTATAGGGCGAATTGGCGGCCGCGATCTGATCAGATCCAGACGTCTGTTGACGTTGGGAAGCTTGAG-3′. 5′-rC-C/G primer is as follows: 5’GAATTGTAATAcGAACACTATAGG-3′. 3′ rC-T/G-60 primer is as follows: 5′-CCAGACGTCTGTTGACGTTGGGAAGCTTGAGTATTCTATAGTGTCACCTAAATAGCTTGGCGTAATCATGGTcATAGCTGTTTCCTGTGTG-3′. 3′ rC-C/G primer is as follows: 5′-GCGTAATCATGGTCATAGCTGTTTCC-3′. 3′ rC-T/G-308 primer is as follows: 5′-GCTTCCTCGCTCACTGAGTCGCTGcGCTCGGTCGTTC-3′.
Antibodies and Recombinant Proteins
The RNASEH2A antibody for western blots (rabbit polyclonal, GeneTex) was used at a dilution of 1:1,000, MSH2 (mouse monoclonal, BD Transduction Laboratories) was used at a dilution of 1:1,000, and MLH1 (mouse monoclonal, Oncogene) was used at a dilution of 1:1,000. The RNase H2 (sheep polyclonal) antibody used for immunodepletion was generated by A.P.J. Recombinant MutSα and MutLα were expressed and purified in our laboratory. RNase HI and HII were obtained from New England BioLabs.
Band-Shift Assay
This was performed essentially as described (Cejka et al., 2005). The oligonucleotide heteroduplexes were created by annealing 5′ radiolabelled (∗) oligonucleotide 5′CTCAAGCTTCCCAACGTCGACAGACGTCTGG3′ with the following unlabeled oligonucleotides: T/G mismatch, 5′-CCAGACGTCTGTTGACGTTGGGAAGCTTGAG-3′; C/G match, 5′-CCAGACGTCTGTCGACGTTGGGAAGCTTGAG-3′; and rC/G mismatch, 5′- CCAGACGTCTGTcGACGTTGGGAAGCTTGAG 3′, where boldface nucleotides define the residues creating the mismatches. The position of the ribocytidine is indicated in lowercase. The binding reaction mixtures contained 40 fmol oligonucleotide duplex T/G∗, C/G∗, or rC/G∗ and MSH2/MSH6 (100 or 250 ng). Protein-bound substrates were separated from free probes by electrophoresis on a 6% native polyacrylamide gel eluted with TAE.
In Vitro Nicking Assay
Supercoiled DNA substrates (100 ng) were incubated with 1 U of Nt.BstNBI, RNase HI, or RNase HII in buffers recommended by the manufacturers. Nicking assays with recombinant human RNase H2 (WT or the catalytically inactive DD/AA mutant, 0.1, 1, and 10 μM) were carried out with 100 ng of supercoiled rC-T/G substrate in a buffer containing 20 mM Tris.HCl (pH 7.6), 5 mM MgCl2, 110 mM KCl, 1 mM glutathione, and 50 μg/ml BSA. The substrates were incubated at 37°C for 45 min. The products were then loaded on a 1% agarose gel and visualized with GelRed.
Mismatch-Dependent Strand Degradation Assays
The experiments were performed essentially as described (Genschel and Modrich, 2003) with some modifications. Briefly, 100 ng of the homo-/heteroduplex was treated with 1 U of bacterial RNase HII or 10 μmole human RNase H2 (WT or DD/AA) in a reaction containing the recombinant MMR proteins (150 ng MutSα, 100 ng MutLα, 70 ng RPA, and 1.6 ng of EXO1 in 20 mM Tris.HCl [pH 7.6], 1 mM glutathione, 5 mM MgCl2, 0.05 mg/ml BSA, 3 mM ATP, 100 mM KCl). Mismatch-provoked excision reactions were allowed to proceed for 7 min at 37°C, following which the samples were digested with HindIII and XmnI at 37°C overnight and analyzed on 1% agarose gels containing GelRed.
siRNA and Knockdown Experiments
293 cells were transfected with 40 pmol of siRNA against luciferase (siLuc), RNase H1, RNase H2a, or both H1 and H2a, using RNaiMax (Invitrogen) as per the manufacturer’s protocol. For each knockdown, six 15 cm dishes were transfected. The following day, the plates were trypsinized and scaled up into 20 15 cm dishes. Forty-eight hours posttransfection, the nuclear extracts were prepared as described above. The siRNAs used in this study were as follows: siLuc, 5′-CGUACGCGGAAUACUUCGA-3′; siRNase H1, 5′-GAAGACAAGUGCAGGGAAA-3′; and siRNase H2A, 5′-GGACUUGGAUACUGAUUAU-3′.
RNase H2-Immunodepletion of Nuclear Extracts
Protein A/G beads (SantaCruz) were washed twice with binding buffer (30 mM HEPES.KOH [pH 7.5], 7 mM MgCl2) and incubated with the RNase H2 antibody at 4°C for 3 hr (10 μl of the serum was used to bind 25 μl of the bead slurry). They were then washed thrice with the binding buffer and subsequently used to immunodeplete the nuclear extracts. For 150 μg of nuclear extracts, 10 μl of antibody-preadsorbed beads were used. Mock-depleted nuclear extracts were obtained by incubating with the beads alone. The immunodepletion was carried out for 30 min at 4°C, and the MMR assays were performed immediately.
In Vivo Mutagenesis Assays
Strand-specific mutation rates in S. cerevisiae ogg1Δ strains were measured using the ura3-29 marker as described previously (Pavlov et al., 2003). The RNH201 and MSH2 deletions were generated by standard genetic procedures. Mutation rates were obtained by fluctuation tests using 9–20 independent cultures. The 95% confidence limits for the median and the differences between mutation frequencies using the Mann-Whitney nonparametric criterion were determined as described (Dixon and Massey, 1983).
In the second assay, we used a reporter cassette containing a (T)9 repeat within the URA3 open reading frame that had been introduced into the ADE2 locus by standard genetic techniques. Mutation rates were calculated from fluctuation analysis with 32–50 independent cultures using the Lea-Coulson method of median (Lea and Coulson, 1949), based on the appearance of mutants resistant to 5-FOA. The fluctuation analysis calculator (FALCOR) software was used to calculate the mutation rate and confidence levels (Hall et al., 2009). The strains were derivatives of FF18733/FF18734 (Cejka et al., 2005).
DNA Sequencing
Genomic DNA from ten URA+ clones per S. cerevisiae ogg1Δ strain was purified using an YDER Kit (http://www.piercenet.com/browse.cfm?fldID=06010499). The URA3 region containing the ura3-29 mutation was amplified by PCR using the primers 5′-GAACGTGCTGCTACTCATCC-3′ and 5′-CATTCTGCTATTCTGTATAC-3′. The PCR product was sequenced by Cogentech (http://www.cogentech.it/) using the primer 5′-TAGTTGAAGCATTAGGTCCC-3′.
Mutations flanking the T9 repeat were identified by sequencing a 500 bp fragment covering the first 242 bp of the URA3 coding sequence (including the T9 repeat), amplified by PCR using primers 5′-GCATTGGATGGTGGTAACG-3′ and 5′-GGAACGACAGTACCCTCATAAC-3′. PCRs were carried out using ReddyMix PCR Master Mix (Thermo Scientific) and 10 ng of yeast genomic DNA (95°C 5 min; then three cycles of 94°C 30 s, 65°C 30 s, and 72°C 45 s; then three cycles of 94°C 30 s, 62°C 30 s, and 72°C 45 s; then three cycles of 94°C 30 s, 59°C 30 s, and 72°C 45 s; then 35 cycles of 94°C 30 s, 56°C 30 s, and 72°C 45 s; then 72°C for 10 min). Purified PCR amplification products from 28–50 independent clones were sequenced using dye-terminator chemistry and electrophoresed on an ABI 3730 capillary sequencer (Applied Biosystems). Sequencing data were analyzed using Sequencher 4.8 (Gene Codes).
Acknowledgments
The authors thank Kalpana Surendranath and Anja Saxer for recombinant MutSα and MutLα, Murat Aykut for sharing reagents for the yeast work, Ailsa Revuelta for technical support, Stefano Ferrari for recombinant EXO1, Pavel Janscak for recombinant RPA, Thomas A. Kunkel for yeast strains, and Luca Gianfranceschi for help with statistical analyses. The work was supported by Swiss National Science Foundation grants 310030A-118158 and 310030B-133123, and by European Community’s Seventh Framework Programme (FP7/2007-2013) grant agreement number HEALTH-F4-2008-223545 to J.J.; by grants from AIRC and MIUR to M.M.-F.; and from MIUR (FIRB RBFR10S3UQ) to F.L. The financial support of Telethon-Italy (grant number GGP11003) is also gratefully acknowledged. A.P.J. and M.A.R. were supported by a MRC Senior Clinical Fellowship. J.J., M.M.-F., M.M.G., P.C., and P.P. conceived the study and devised the experiments; M.M.G. designed and M.A.-B. constructed all the substrates generated in this study. M.M.G. carried out the biochemical experiments; F.L. and M.M.-F. carried out the strand-specific genetic studies; M.M.G., with the help of M.O.-P. and P.C., carried out the T9URA3 genetic studies. M.O.-P. performed the band-shift assay and assisted in the preparation of nuclear extracts and substrates. M.A.R. and A.P.J. carried out the sequencing of the T9 repeats and provided the RNase H2 polyclonal antibody and the RNase H2 knockout MEFs; J.J., M.M.-F., and M.M.G. wrote the manuscript.
References
- Andersen S., Heine T., Sneve R., König I., Krokan H.E., Epe B., Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K., Fischer F., Jiricny J. Characterization of the “mismatch repairosome” and its role in the processing of modified nucleosides in vitro. Methods Enzymol. 2006;408:285–303. doi: 10.1016/S0076-6879(06)08018-9. [DOI] [PubMed] [Google Scholar]
- Cejka P., Stojic L., Mojas N., Russell A.M., Heinimann K., Cannavó E., di Pietro M., Marra G., Jiricny J. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Mojas N., Gillet L., Schär P., Jiricny J. Homologous recombination rescues mismatch-repair-dependent cytotoxicity of S(N)1-type methylating agents in S. cerevisiae. Curr. Biol. 2005;15:1395–1400. doi: 10.1016/j.cub.2005.07.032. [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.B., Lujan S.A., Kissling G.E., Kunkel T.A. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε. DNA Repair (Amst.) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J.P., Lacks S.A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol. Rev. 1986;50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin N., Dzantiev L., Kadyrov F.A., Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon W.J., Massey F.J., Jr. McGraw-Hill; New York: 1983. Introduction to Statistical Analysis. [Google Scholar]
- Eder P.S., Walder J.A. Ribonuclease H from K562 human erythroleukemia cells. Purification, characterization, and substrate specificity. J. Biol. Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- Genschel J., Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- Hall B.M., Ma C.X., Liang P., Singh K.K. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller B., Achleitner M., Glage S., Naumann R., Behrendt R., Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J., Jr., Clark S., Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl. Acad. Sci. USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kadyrov F.A., Dzantiev L., Constantin N., Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kim N., Huang S.N., Williams J.S., Li Y.C., Clark A.B., Cho J.E., Kunkel T.A., Pommier Y., Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F., Novarina D., Amara F., Watt D.L., Stone J.E., Costanzo V., Burgers P.M., Kunkel T.A., Plevani P., Muzi-Falconi M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D.E., Coulson C.A. The distribution of the numbers of mutants in bacterial population. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Kissling G.E., Kunkel T.A. Differential correction of lagging-strand replication errors made by DNA polymerases alpha and delta. Proc. Natl. Acad. Sci. USA. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundström E.B., Johansson E., Chabes A., Kunkel T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundström E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov Y.I., Mian I.M., Kunkel T.A. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Peña-Diaz J., Jiricny J. PCNA and MutLα: partners in crime in triplet repeat expansion? Proc. Natl. Acad. Sci. USA. 2010;107:16409–16410. doi: 10.1073/pnas.1011692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Diaz J., Bregenhorn S., Ghodgaonkar M., Follonier C., Artola-Borán M., Castor D., Lopes M., Sartori A.A., Jiricny J. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 2012;47:669–680. doi: 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Pluciennik A., Dzantiev L., Iyer R.R., Constantin N., Kadyrov F.A., Modrich P. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. USA. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Leitch A., Keighren M., Kilanowski F. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo M.T., Blasi M.F., Chiera F., Fortini P., Degan P., Macpherson P., Furuichi M., Nakabeppu Y., Karran P., Aquilina G., Bignami M. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol. Cell. Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B., Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanz S., Castor D., Fischer F., Jiricny J. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc. Natl. Acad. Sci. USA. 2009;106:5593–5598. doi: 10.1073/pnas.0901726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöpf B., Bregenhorn S., Quivy J.P., Kadyrov F.A., Almouzni G., Jiricny J. Interplay between mismatch repair and chromatin assembly. Proc. Natl. Acad. Sci. USA. 2012;109:1895–1900. doi: 10.1073/pnas.1106696109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Koh K.D., Weiss B., Storici F. Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat. Struct. Mol. Biol. 2012;19:98–104. doi: 10.1038/nsmb.2176. [DOI] [PubMed] [Google Scholar]
- Sparks J.L., Chon H., Cerritelli S.M., Kunkel T.A., Johansson E., Crouch R.J., Burgers P.M. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand M., Prolla T.A., Liskay R.M., Petes T.D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Thomas D.C., Roberts J.D., Kunkel T.A. Heteroduplex repair in extracts of human HeLa cells. J. Biol. Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- Williams J.S., Clausen A.R., Nick McElhinny S.A., Watts B.E., Johansson E., Kunkel T.A. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ϵ. DNA Repair (Amst.) 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R., Fritz H.J. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987;6:1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]