Abstract
AIM: To identify the downstream regulated genes of GAEC1 oncogene in esophageal squamous cell carcinoma and their clinicopathological significance.
METHODS: The anti-proliferative effect of knocking down the expression of GAEC1 oncogene was studied by using the RNA interference (RNAi) approach through transfecting the GAEC1-overexpressed esophageal carcinoma cell line KYSE150 with the pSilencer vector cloned with a GAEC1-targeted sequence, followed by MTS cell proliferation assay and cell cycle analysis using flow cytometry. RNA was then extracted from the parental, pSilencer-GAEC1-targeted sequence transfected and pSilencer negative control vector transfected KYSE150 cells for further analysis of different patterns in gene expression. Genes differentially expressed with suppressed GAEC1 expression were then determined using Human Genome U133 Plus 2.0 cDNA microarray analysis by comparing with the parental cells and normalized with the pSilencer negative control vector transfected cells. The most prominently regulated genes were then studied by immunohistochemical staining using tissue microarrays to determine their clinicopathological correlations in esophageal squamous cell carcinoma by statistical analyses.
RESULTS: The RNAi approach of knocking down gene expression showed the effective suppression of GAEC1 expression in esophageal squamous cell carcinoma cell line KYSE150 that resulted in the inhibition of cell proliferation and increase of apoptotic population. cDNA microarray analysis for identifying differentially expressed genes detected the greatest levels of downregulation of calpain 10 (CAPN10) and upregulation of trinucleotide repeat containing 6C (TNRC6C) transcripts when GAEC1 expression was suppressed. At the tissue level, the high level expression of calpain 10 protein was significantly associated with longer patient survival (month) of esophageal squamous cell carcinoma compared to the patients with low level of calpain 10 expression (37.73 ± 16.33 vs 12.62 ± 12.44, P = 0.032). No significant correction was observed among the TNRC6C protein expression level and the clinocopathologcial features of esophageal squamous cell carcinoma.
CONCLUSION: GAEC1 regulates the expression of CAPN10 and TNRC6C downstream. Calpain 10 expression is a potential prognostic marker in patients with esophageal squamous cell carcinoma.
Keywords: Esophageal squamous cell carcinoma, Oncogene, RNA interference, Calpain 10, Tissue microarray
INTRODUCTION
Esophageal squamous cell carcinoma (ESCC) has a multifactorial etiology which involves environmental and/or genetic factors[1,2]. The incidence of ESCC also shows marked variation in its geographic distribution and occurs at relatively high frequency in Asian regions including China[3]. Current modalities of therapy for this disease offer relatively poor survival and cure rates[4], thus more investigations at the molecular level are essential for a better understanding the molecular pathogenesis of this disease and for making further improvements in diagnosis and treatment of ESCC.
Gene amplification and overexpression have been suggested as the major genomic aberrations involved in the pathogenesis of ESCC[5,6]. We previously employed the method of comparative DNA fingerprinting using inter-simple sequence repeat polymerase chain reaction (ISSR-PCR) and revealed that amplifications or deletions of chromosomal sequences are common events in both the preneoplastic lesions and carcinomas[7]. An analysis of the frequency of amplification or loss of individual ISSR-PCR profile bands led to the identification of a novel expressed sequence tag database entry of a cDNA clone from a chromosome 7 placental cDNA library[7,8]. Moreover, the ISSR-PCR fragment also showed 98% homology to a Homo sapiens chromosome 7 P1-derived artificial chromosome clone (approximately 125 kb) which has been mapped to chromosome band 7q22[9]. The amplification of chromosomal segment 7q22 has been implicated in many types of cancer. Reported examples include ESCC[10], breast carcinoma[11], pancreatic carcinoma[12], renal-cell carcinomas[13] and T-cell leukemia[14]. Thus, further investigation on the newly identified ESCC-related genomic and expressed sequences mapped to chromosomal region 7q22 can be a fruitful approach for identifying new candidate genes crucial to the disease. We subsequently identified and characterized the role of a novel oncogene GAEC1 which is located at 7q22 region, encodes a nuclear protein and shows a high frequency of gene amplification and overexpression in ESCC cell lines and primary tumors[15], as well as in colorectal adenocarcinoma[16]. Overexpression of GAEC1 in 3T3 mouse fibroblasts caused increased cell proliferation, foci formation and colony formation in soft agar, comparable to H-ras overexpression. Further, injection of GAEC1-transfected 3T3 cells into athymic nude mice formed undifferentiated sarcoma in vivo, providing the first evidence about the oncogenic nature of GAEC1[15]. An increased GAEC1 DNA copy number was also reported in 79% of colorectal adenocarcinomas and the copy numbers were significantly different among colorectal adenocarcinomas, adenomas, and non-neoplastic colorectal tissues[16].
In this report, GAEC1 was further characterized by identifying the downstream partners using cDNA microarray analysis on GAEC1-suppressed human esophageal carcinoma cell line KYSE150 which shows GAEC1 overexpression. The prominently downstream-regulated genes were then studied by immunohistochemistry on a tissue microarray (TMA) of ESCC to determine their clinicopathological significance.
MATERIALS AND METHODS
ESCC specimens and cell lines
One hundred and thirty-two paired non-tumor and tumor fresh tissue samples were collected after esophagectomy with patients’ consent at the Department of Surgery, Queen Mary Hospital, Hong Kong from 2001 to 2006. They were collected consecutively from esophagectomy specimens performed on patients who had received no prior treatment directed to the primary ESCC. The histopathological features were reported by specialist pathologists of the Department of Pathology, Queen Mary Hospital, Hong Kong. The clinicopathological parameters of the patients were collected prospectively and they included age, gender, tumor-node-metastasis pathological stages and histological grades. The actuarial survival rate of the patients was calculated from the date of surgical resection of the ESCC to the date of death or last follow-up. Management was by a pre-agreed standardized multidisciplinary protocol supervised by a senior specialist surgeon. The ESCC cell line KYSE150 is of Japanese origin. It was purchased from DSMZ (Braunschweig, Germany) and cultured as described[17]. The non-tumor esophageal epithelial cell line NE1 was used as the control to confirm the overexpression of GAEC1 in KYSE150 and was cultured as previously described[18].
Preparation of small interfering RNA expression vector
A vector based RNA interference (RNAi) approach was used for suppressing the expression of GAEC1 in KYSE150 ESCC cells. The pSilencer2.1-U6 neo vector (Ambion) was used to express the siRNA which is specific for targeting GAEC1 expression. The pSilencer2.1-U6 neo Negative Control vector (Ambion) was used as the negative control which expressed a hairpin small interfering RNA (siRNA) with limited homology to any known sequences in the human, mouse and rat genomes. The siRNA target sequence of GAEC1 and the insert sequence were determined by the programs siRNA Target Finder and Insert Design Tool for the pSilencer™ Vectors (Ambion). The top strand of the insert sequence (P3-4) is 5’-GATCCGAAGTGGCTTCTGGATTAATTCAAGAGATTAATCCAGAAGCCACTTCTTTTTTGGAAA-3’ and the bottom strand is 5’-AGCTTTTCCAAAAAAGAAGTGGCTTCTGGATTAATCTCTTGAATTAATCCAGAAGCCACTTCG-3’. The top and bottom strands were annealed and cloned into the pSilencer2.1-U6 neo vector according to the manufacturer’s instruction. The vectors were transfected into the KYSE150 cells as previously described[15] using FuGene HD (Roche Diagnostics GmbH) with G418 selection.
RNA extraction and reverse transcription-polymerase chain reaction analysis
RNA was extracted from the parental, pSilencer-P3-4 and pSilencer-negative control vectors transfected KYSE150 cells using the RNeasy mini Kit (Qiagen) after 2 mo selection under G418. About 2 μg DNA-free RNA from each sample was used for the multiplex semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis with β-actin as the internal control to show the expression level of GAEC1 as previously described[15]. Densitometry analysis was performed to compare the intensity of the PCR products after agarose gel electrophoresis under UV using Quantity-One program (Bio-Rad).
Cell proliferation assay
The cell proliferation of the parental, pSilencer-P3-4 and pSilencer-negative control vectors transfected KYSE150 cells was determined by MTS assay using the CellTiter 96 Aqueous One Solution (Promega) as previously described[15].
Cell cycle analysis
The parental, pSilencer-P3-4 and pSilencer-negative control vectors transfected KYSE150 cells were resuspended in 500 L 1 × phosphate buffered saline and fixed with 500 L 70% ethanol. The cells were then suspended in 1 mL PI (20 μg/mL)/Triton X-100 (0.1% v/v) staining solution with RNase A (200 μg/mL) and then analyzed by the BD FACSCalibur flow cytometer. Different fractions of cell cycles were analyzed using the Modfit LT software (Verity Software House).
cDNA microarray analysis
The differentially expressed genes of the pSilencer-P3-4 vector transfected KYSE150 cells with suppressed GAEC1 expression were identified using cDNA microarray analysis by making comparisons between the parental cells, pSilencer-negative control vectors transfected cells, and pcDNA3.1-GAEC1 transfected cells with GAEC1 overexpression[15]. The cDNA microarray analysis and the associated quality control using Human Genome U133 Plus 2.0 arrays (Affymetrix) were performed in the Genome Research Centre of the University of Hong Kong according to the Affymetrix’s protocol. Briefly, total RNA was extracted from 2 × 106 cells of each sample using RNeasy mini Kit (Qiagen). The RNA integrity was measured by the ratio of 28S/18S ribosomal RNA using Agilent 2100 Bioanalyzer. One microgram total RNA from each source was then reverse transcribed to the first-stranded cDNA by using oligo-dT linked-T7 RNA polymerase promoter sequence and the double-stranded cDNA was synthesized by using RT Kit (Invitrogen). The biotin labelled-cRNA was produced by in vitro transcription kit (Invitrogen) and purified by RNeasy mini columns (Qiagen). About 15 μg denatured cRNA was hybridized to each Human Genome U133 Plus 2.0 array (Affymetrix) and then stained with a streptavidin-phycoerythrin conjugate and the signals were detected with GeneArray scanner (Agilent). The microarray signals were analyzed by using Agilent Genespring GX and Affymetrix GeneChip Operating Softwares. The signal of each differentially expressed gene in the pSilencer-P3-4 transfected cells was determined by comparing with the parental cells and normalized with the pSilencer-negative control vector transfected cells. The threshold level of the corresponding up- or down-regulated genes with transfected pcDNA3.1-GAEC1 vector is ≥ 2 folds.
Tissue microarray and immunohistochemical staining
A TMA containing the 132 paired non-tumor esophageal epithelia and ESCC specimens were constructed as described previously[19]. The archival paraffin-embedded ESCC tissues were used under the ethical guidelines in the Department of Pathology of The University of Hong Kong. Immunohisto-chemical staining on the TMA sections was performed using the calpain 10 (0.03 mg/mL; Sigma-Aldrich) rabbit polyclonal antibody and TNRC6C (1 mg/mL; Abnova) mouse monoclonal antibody using the previously described methodology[19]. The dilution factors for the calpain 10 and TNRC6C antibodies were 1:50 and 1:150 respectively. The percentage of tumor cells positively stained formed the basis of grading as follows: Grade 0: less than 5%, Grade I: 5% to less than 25%, Grade II: 25% to less than 50% and Grade III: more than 50%. For each tissue sample, the tissue core with the highest grade was selected for subsequent statistical analysis. The high expression group combined those tumors with Grade II or III, and the low expression group combined those tumors with Grade 0 or I.
Statistical analysis
The Student’s t test was used to evaluate the statistical significance of the differences in calpain 10 and TNRC6C expression between tumor and non-tumor tissues. The χ2 test and t test were used to examine the statistical significance of the correlations between calpain 10 and TNRC6C expression with clinicopathological parameters. Kaplan-Meier plots and Cox multi-variant analysis were produced for overall patient survival, and statistical significance was evaluated by using Wilcoxon’s signed-rank test. Statistical analysis were performed using SPSS Ver. 20.0 (SPSS, Chicago, IL, United States). Differences were considered statistically significant when the relevant P values were < 0.05.
RESULTS
The overexpression of GAEC1 in KYSE150 over NE1 was confirmed by multiplex semi-quantitative RT-PCR and densitometry analysis (Figure 1A). The expression level of GAEC1 in pSilencer-P3-4 transfected KYSE150 cells was also determined by comparing with the parental and pSilencer-negative control vector transfected cells using densitometry measurement. The results indicated that the pSilencer-P3-4 transfected KYSE150 cells showed a down-regulation of GAEC1 expression compared with the parental cells and the control. The comparison of the band intensities among the samples by densitometry measurement showed that the GAEC1 expression level was down-regulated in the pSilencer-P3-4 transfected cells by about three folds (Figure 1B).
Figure 1.
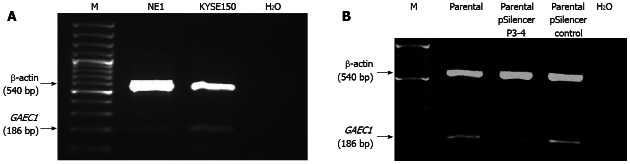
Expression level of GAEC1 in KYSE150 cells. A: Multiplex reverse transcription-polymerase chain reaction (RT-PCR) analysis showed the overexpression of GAEC1 in KYSE150 compared with the non-tumor esophageal epithelial cell line NE1; B: Multiplex RT-PCR analysis demonstrated the down-regulation of GAEC1 expression in KYSE150 cells transfected with pSilencer P3-4 vector compared with the parental cells and those transfected with pSilencer control vector. The amount of RNA in each lane was normalized with the amplification of β-actin. M: 100 bp ladder marker; H2O: Water control.
To assess the effect on cell proliferation with suppressed GAEC1 expression, a comparison was made between the MTS activities generated from the parental cells and the cells transfected with the pSilencer-negative control vector. The results indicated that the KYSE150 cells with down-regulated GAEC1 showed an obvious reduction in proliferation rate compared with the parental and control-vector transfected cells (Figure 2). Further analysis on the cell cycle related changes using flow cytometry demonstrated an approximately 50% increase in apoptotic population with suppressed GAEC1 expression, compared with the parental and control-vector transfected cells (Figure 3).
Figure 2.
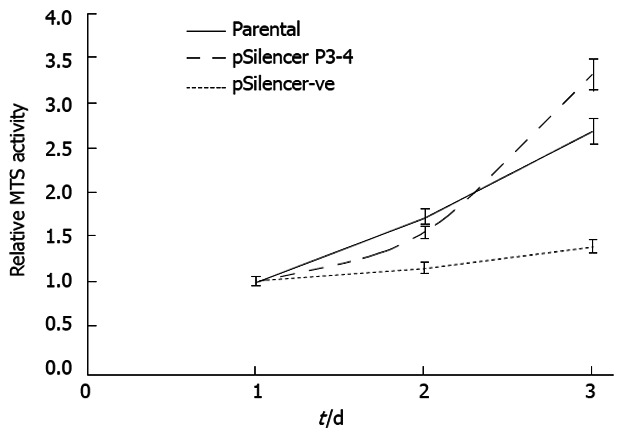
MTS cell proliferation assays for esophageal squamous cell carcinoma cell line KYSE150. Cells were transfected with pSilencer vector cloned with the P3-4 sequence (pSilencer P3-4) or control vector (pSilencer-ve). MTS assays were then performed every 24 h for 3 d on each type of transfected cells and the parental cells. The respective MTS activities on each day were compared with the corresponding activities of day 1. Representative data from 3 independent experiments are shown.
Figure 3.
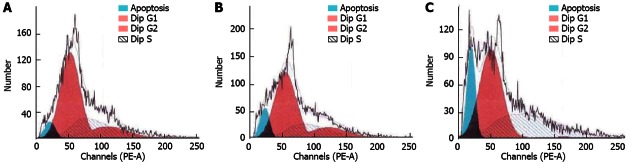
Flow cytometry analyses for KYSE150 cells. KYSE150 transfected with pSilencer cloned with P3-4 sequence demonstrated an increased apoptotic population by approximately 50% (C) compared with the parental cells (A) and cells transfected with pSilencer-ve control vector (B).
To identify the downstream candidate genes which are regulated by the suppressed GAEC1 expression, cDNA microarray analysis was performed using the Human Genome U133 Plus 2.0 array (Affymetrix) which comprises of more than 47000 transcripts and variants in each chip. The results of the identified lists of more than 5-fold down-regulated (total 10 genes) and more than 3-fold up-regulated (total 9 genes) targets were shown in Table 1 respectively. All the listed genes were selected based on more than 2-fold expression signals of the corresponding up- or down-regulation of the respective genes when the cells were transfected with the pcDNA3.1-GAEC1 vector and no significant fold change was detected with transfected pSilencer-negative control vector compared with parental cells. With suppressed GAEC1 expression, calpain 10 (CAPN10) was identified to have the highest level (> 15 folds) of down-regulation (Table 1), while trinucleotide repeat containing 6C (TNRC6C) was shown to have the highest level (> 7 folds) of up-regulation (Table 1). These two GAEC1-regulated target genes with the greatest changes in expression level were followed up by the immunohistochemical analysis using the ESCC tissue microarray.
Table 1.
List of more than 5-fold and 3-fold down-regulated genes induced by stable GAEC1 knowndown in KYSE150 cells compared with the parental cells
| Probe set ID | Gene title | Down-regulation with transfected pSilencer P3-4 | Up-regulation with transfected pcDNA3.1-GAEC1 | pSilencer-ve control |
| 221040_at | Calpain 10 | 15.3033010 | 2.1643467 | 1.0476209 |
| 1561417_x_at | Not assigned | 12.1628650 | 2.0130675 | 1.1347373 |
| 1562828_at | Not assigned | 9.9816000 | 2.6808436 | 1.1661105 |
| 229929_at | splA/ryanodine receptor domain and SOCS box containing 4 | 8.3420770 | 2.2365010 | 1.1751518 |
| 235209_at | Chromosome 8 open reading frame 84 | 7.6294910 | 2.1356385 | 1.1450081 |
| 220090_at | Cornulin | 7.6125007 | 2.3401918 | 1.1664450 |
| 242713_at | Not assigned | 7.3507795 | 2.0578532 | 1.1959343 |
| 224499_s_at | Activation-induced cytidine deaminase | 5.8917794 | 3.3146940 | 1.1664389 |
| 229543_at | Not assigned | 5.3493247 | 2.0206234 | 1.0065930 |
| 242064_at | Sidekick homolog 2 (chicken) | 5.0322995 | 2.8469403 | 1.0170712 |
| 1561041_at | Trinucleotide repeat containing 6C | 7.5979643 | 2.1234870 | 1.0152589 |
| 216787_at | Not assigned | 5.3369575 | 2.2657390 | 1.1658608 |
| 206725_x_at | Bone morphogenetic protein 1 | 4.8828310 | 2.7046654 | 1.1015952 |
| 206276_at | Lymphocyte antigen 6 complex, locus D | 4.7652740 | 2.7379642 | 1.0686288 |
| 1560482_at | Not assigned | 4.2348604 | 3.0122058 | 1.1875614 |
| 211362_s_at | Serpin peptidase inhibitor, clade B (ovalbumin), member 13 | 4.0584164 | 2.4208739 | 1.0186443 |
| 216491_x_at | Immunoglobulin heavy constant mu | 3.4819565 | 2.8060850 | 1.0186309 |
| 238415_at | Not assigned | 3.2034543 | 2.9523630 | 1.0138865 |
| 241028_at | RPGRIP1-like | 3.0331728 | 2.3752263 | 1.0001514 |
All the listed genes were selected based on more than 2-fold of the corresponding up-regulation when GAEC1 was overexpressed with transfected pcDNA3.1-GAEC1 vector and no significant fold change with transfected pSilencer-negative control vector compared with parental cells.
The expression of CAPN10 and TNRC6C proteins in TMA sections sampling 132 paired tumor and non-tumor tissues from ESCC specimens was investigated using immunohistochemistry. Fourteen out of 132 tumors (10.61%) were found to belong to the high expression group of CAPN10 expression. However, TNRC6C did not show any significant expression signals in all the ESCC cases analyzed except eight non-tumor esophageal tissues which also served as the positive controls. Representative examples of immunohistochemical staining of CAPN10 and TNRC6C are shown in Figure 4. Correlation between expression level of CAPN10 and clinicopathological features are summarized in Table 2. There was no significant correlation of any clinicopathological features with the expression level of CAPN10. The median survival of patients with high expression level of CAPN10 was 38 mo whereas that of low expression level was 13 mo, and the survival range is from 0.72 to 65.15 mo. The difference was significant on both univariant and multi-variant analysis (P = 0.032 and 0.035 respectively; Figure 5).
Figure 4.
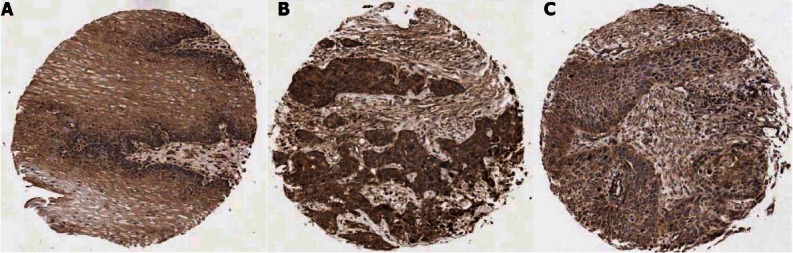
Immunohistochemical staining of CAPN10. A: In normal esophageal epithelial tissue showing weak CAPN10 staining; B: esophageal squamous cell carcinoma (ESCC) tissue showing strong CAPN10 staining; C: ESCC tissue showing weak CAPN10 staining in tumor. CAPN10 was mainly localized in the cytoplasm of the cancer cells (original magnification, ×100).
Table 2.
Relationship between CAPN10 expression and clinicopathological features
| Characteristics | Patients | Low expression | High expression | P value |
| Age, yr (mean ± SD) | 132 | 65.64 ± 10.55 | 63.50 ± 13.39 | 0.572 |
| Gender | 0.086 | |||
| Male | 102 | 94 | 8 | |
| Female | 30 | 24 | 6 | |
| TNM stage | 0.762 | |||
| 0/I/II | 18 | 15 | 9 | |
| III/IV | 83 | 72 | 11 | |
| Tumor depth | 0.729 | |||
| T1-3 | 79 | 67 | 12 | |
| T4 | 22 | 20 | 2 | |
| Lymph node metastasis | 0.363 | |||
| N0 | 32 | 26 | 6 | |
| N1 | 69 | 61 | 8 | |
| Distant metastasis | 1 | |||
| M0 | 66 | 57 | 9 | |
| M1 | 35 | 30 | 5 | |
| Differentiation | 0.459 | |||
| Well | 16 | 15 | 1 | |
| Moderate | 59 | 51 | 8 | |
| Poor | 26 | 21 | 5 | |
Figure 5.
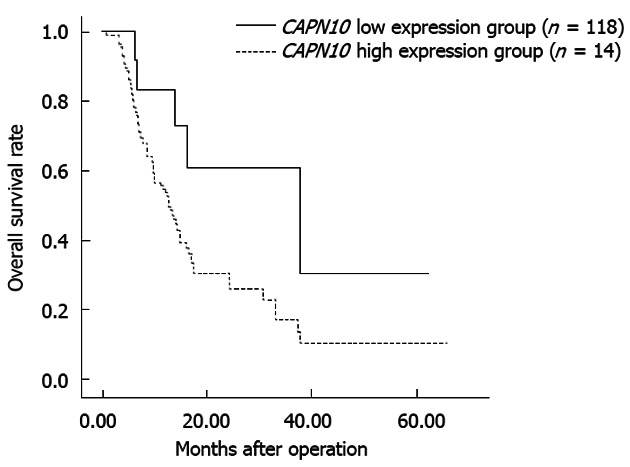
Overall 5-year survival rates as determined by the expression level of CAPN10 in esophageal squamous cell carcinoma patients. Low expression group of CAPN10 in ESCC patients showed a significantly lower 5-year survival rate than those of high expression group.
DISCUSSION
Our previous study reported the oncogenic role of GAEC1 in esophageal carcinogenesis and high expression level of GAEC1 caused malignant transformation of normal cells[15]. High DNA copy number of GAEC1 was also observed in colorectal adenocarcinoma and significant difference was reported in cancer sub-sites and tumor types[16]. From our previous study[15], however, no significant correlation was observed between GAEC1 amplification and clinicopathological parameters and prognosis in ESCC tumors, and thus the DNA amplification study of GAEC1 is not included in this report. In the present study, an attempt was made to investigate the downstream- regulated genes when GAEC1 expression was suppressed in an ESCC cell line KYSE150. Our group also investigated the ESCC cell lines which showed overexpression of GAEC1 as we reported previously[15]. KYSE150 showed the more stable and consistent overexpression with time compared among the ESCC cell lines. Reduction of proliferation rate and increase in apoptotic population were observed in association with reduced GAEC1 expression in ESCC cells. Thus our study is the first report to demonstrate the significance of suppressing GAEC1 as a target of reducing the malignant properties of ESCC. In order to assess whether the tumors are more proliferative, the use of other histological markers for assessing proliferation, such as Ki-67[20] and AgNOR[21], in parallel to CAPN10 is suggested in future studies to determine whether the CAPN10 level is associated with progression of the disease. Similar targeting approach against potential oncogenes is now being explored intensively in the direction of gene therapy for various types of cancers[22]. Examples include the suppression of MTA1 in esophageal carcinoma[23], alpha-actinin-4 in oral carcinoma[24], osteopontin in colon carcinoma[25] and EGFR in hepatocellular carcinoma[26]. The application of RNAi approach has been recognized as having high potential for the clinical application of targeted cancer therapy[27]. To date, clinical trials at different stages were reported and they targeted against various oncogenic components in various cancers, including metastatic melanoma, liver cancer, chronic myelogenous leukemia, pancreatic cancer and colon cancer[22]. Moreover, the RNAi approach for targeting specifically on transforming growth factor-β has been developed as a “cancer vaccine” against ovarian cancer[22]. Thus our present study offers a new direction for exploring the application of RNAi-based method for suppressing the oncogenic target GAEC1 as a novel gene therapy approach in our future investigations.
Calpain 10 (CAPN10) is a member of the mitochondrial calpain system[28]. Mitochondrial calpain system has been shown to promote caspase-independent programmed cell death via the apoptotic inducing factor-mediated mechanism[28] and its expression has been correlated to insulin-stimulated glucose uptake[29] and type 2 diabetes[30]. However, the correlation and functional roles of CAPN10 in tumorigenesis are still not fully understood, although CAPN10 has been linked to laryngeal[31], colorectal[32] and pancreatic cancers[33]. In the present study, the RNAi-based suppression of GAEC1 in KYSE150 resulted in the suppression of CAPN10 expression (approximately 15-fold compared with the parental cells). For those ESCC tumors belonging to the CAPN10 low expression group, the 5-year survival rate is significantly lower than those belonging to the CAPN10 high expression group. From the study of Moreno-Luna et al[31], CAPN10 genotype 12 was reported to be related with a worse prognosis in laryngeal cancer, which is similar to our present study which is newly described in ESCC. Our observation from the low CAPN10 expression group implied the possibility that the oncogene GAEC1 overexpression within this group might involve more prominently at the initial stage of molecular carcinogenesis, so that the expression level of CAPN10 was lower in ESCC at the time of operation. Similar pattern of oncogenic expression happening at the earlier stage of carcinogenesis was also observed from fibroblast growth factor-2 in melanoma[34] and KLF4 in cutaneous squamous epithelial neoplasia[35]. The verification of this hypothesis can be followed up with the future development of GAEC1-specific antibody, which is still unavailable in market, for the future analysis of GAEC1 expression in various stages in ESCC. This important finding also paves the path for the further investigation for the roles of CAPN10 in the molecular pathogenesis of esophageal carcinoma. Moreover, in the present study, no significant correlation of any other clinicopathological features with the expression level of CAPN10 was found, but CAPN10 predicted the poor survival of ESCC patients. Similar results were also reported previously in which the overexpression of a chemokine CXCL12 in ovarian cancer[36] and a protein Rad51 for homologous recombination in ESCC[37] also showed a correlation to the survival of patients, but no correlation to other clinicopathological features was found. The level of CAPN10 is also not associated with local lymph node and distant metastasis in the ESCC cases, implying the possibility that GAEC1 expression may not be relevant to the control of metastasis in ESCC.
TNRC6C has been reported to be the miRNA regulation-related genes and their mutation was correlated to cancer development through deregulating the miRNA regulation[38]. TNRC6C was also shown in the present study to undergo up-regulation with the suppression of GAEC1 expression by the RNAi approach, but there was no significant expression of TNRC6C in the ESCC cases studied. This may be due to the down-regulation of TNRC6C in ESCC by other unknown mechanisms which are subjected to further investigation. Future study of TNRC6C mutation in ESCC is required to investigate the possible roles of TNRC6C in carcinogenesis. Moreover, among the down-regulated genes identified by cDNA microarray with suppressed GAEC1 expression, activation-induced cytidine deaminase (AID) was reported to show overexpression in Barrett’s esophagus and Barrett’s adenocarcinoma[39], but there was no investigation on the roles of AID in molecular pathogenesis in ESCC. AID has been shown to induce somatic mutations in host genes and implicated in the carcinogenesis of lung[40], colorectal[41] and gastric cancers[42]. Therefore, our findings provide a new evidence for prompting future study on the role of AID in the development of ESCC.
In conclusion, the suppression of GAEC1 expression resulted in reduced tumor cell proliferation, increased apoptotic population in ESCC cells and also regulated CAPN10 and TNRC6C expression. The low expression of CAPN10 predicted the poor survival of ESCC patients.
ACKNOWLEDGMENTS
Special thanks are given to Professor George SW Tsao of the Department of Anatomy of The University of Hong Kong for giving us the cell line NE1 as the control. Tang JCO would also like to thank the Griffith University of Australia for awarding the Visiting Research Fellowship (2012).
COMMENTS
Background
Esophageal squamous cell carcinoma (ESCC) has a multifactorial etiology which involves environmental and/or genetic factors. More investigations at the molecular level are essential for a better understanding the molecular pathogenesis of this disease. Authors subsequently identified and characterized the role of a novel oncogene GAEC1 which is located at 7q22 region. In this report, GAEC1 was further characterized by identifying the downstream partners. The prominently downstream-regulated genes were then studied by immunohistochemistry on ESCC tissues to determine their clinicopathological significance.
Research frontiers
The anti-proliferative effect of knocking-down the expression of GAEC1 in ESCC cells was studied. The research hotspot is to find out the target genes which are most regulated by GAEC1 and to determine their clinicopathological significance in ESCC.
Innovations and breakthroughs
The RNA interference (RNAi) approach showed effective suppression of GAEC1 expression in ESCC cells to inhibit cell proliferation and increase apoptosis. cDNA microarray analysis for differentially expressed genes identified the greatest levels of downregulation of calpain 10 (CAPN10) and upregulation of trinucleotide repeat containing 6C when GAEC1 expression was suppressed. High level expression of calpain 10 was significantly associated with longer patient survival. This is the first study to explore the regulatory roles of GAEC1 on the downstream targets and to report the association of CAPN10 to the survival of ESCC patients.
Applications
This study suggested that the potential use of CAPN10 as a prognostic marker to predict the survival of ESCC patients after operation. The findings of the present study pave the path for the future related studies in other human cancers.
Terminology
Squamous cell carcinoma: It is a cancer of a kind of epithelial cell called squamous cell. Squamous cells also occur in the lining of the digestive tract, such as the esophagus; Oncogene: An oncogene is a gene that has the potential to cause cancer. In tumor cells, they are often mutated or expressed at high levels.
Peer review
This is a good study in which the authors employed GAEC1 RNAi to knockdown the expression of GAEC1, investigated its effects on GAEC1-overexpressed esophageal carcinoma cell line KYSE150, and then explored the possible mechanisms. The study design is reasonable, statistical methods are appropriate.
Footnotes
Supported by The General Research Fund, offered by Research Grant Council of Hong Kong to Tang JCO and Lam AKY, PolyU 5627/08M; Griffith Health Institute Project Grant
P- Reviewers Ghigna C, Guo YM, Takeno S, Ding MX, Guerra C, Lin CH S- Editor Gou SX L- Editor A E- Editor Li JY
References
- 1.Wang JM, Xu B, Rao JY, Shen HB, Xue HC, Jiang QW. Diet habits, alcohol drinking, tobacco smoking, green tea drinking, and the risk of esophageal squamous cell carcinoma in the Chinese population. Eur J Gastroenterol Hepatol. 2007;19:171–176. doi: 10.1097/MEG.0b013e32800ff77a. [DOI] [PubMed] [Google Scholar]
- 2.Lam AK. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol. 2000;33:71–90. doi: 10.1016/s1040-8428(99)00054-2. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Ghadimi MR, Rasouli M, Mahmoodi M, Mohammad K. Prognostic factors for the survival of patients with esophageal cancer in Northern Iran. J Res Med Sci. 2011;16:1261–1272. [PMC free article] [PubMed] [Google Scholar]
- 5.Bellini MF, Silva AE, Varella-Garcia M. Genomic imbalances in esophageal squamous cell carcinoma identified by molecular cytogenetic techniques. Genet Mol Biol. 2010;33:205–213. doi: 10.1590/S1415-47572010005000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuwano H, Kato H, Miyazaki T, Fukuchi M, Masuda N, Nakajima M, Fukai Y, Sohda M, Kimura H, Faried A. Genetic alterations in esophageal cancer. Surg Today. 2005;35:7–18. doi: 10.1007/s00595-004-2885-3. [DOI] [PubMed] [Google Scholar]
- 7.Tang JC, Lam KY, Law S, Wong J, Srivastava G. Detection of genetic alterations in esophageal squamous cell carcinomas and adjacent normal epithelia by comparative DNA fingerprinting using inter-simple sequence repeat PCR. Clin Cancer Res. 2001;7:1539–1545. [PubMed] [Google Scholar]
- 8.Touchman JW, Bouffard GG, Weintraub LA, Idol JR, Wang L, Robbins CM, Nussbaum JC, Lovett M, Green ED. 2006 expressed-sequence tags derived from human chromosome 7-enriched cDNA libraries. Genome Res. 1997;7:281–292. doi: 10.1101/gr.7.3.281. [DOI] [PubMed] [Google Scholar]
- 9.Glöckner G, Scherer S, Schattevoy R, Boright A, Weber J, Tsui LC, Rosenthal A. Large-scale sequencing of two regions in human chromosome 7q22: analysis of 650 kb of genomic sequence around the EPO and CUTL1 loci reveals 17 genes. Genome Res. 1998;8:1060–1073. doi: 10.1101/gr.8.10.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaefer IM, Enders C, Polten A, Haller F, Frölich AM, Cameron S, Schüler P, Schweiger P, Gunawan B, Beham A, et al. Common genomic aberrations in basaloid squamous cell carcinoma and carcinosarcoma of the esophagus detected by CGH and array CGH. Am J Clin Pathol. 2011;135:579–586. doi: 10.1309/AJCPZ1O7UUUISPNR. [DOI] [PubMed] [Google Scholar]
- 11.Pinto AE, Roque L, Rodrigues R, André S, Soares J. Frequent 7q gains in flow cytometric multiploid/hypertetraploid breast carcinomas: a study of chromosome imbalances by comparative genomic hybridisation. J Clin Pathol. 2006;59:367–372. doi: 10.1136/jcp.2005.027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang MC, Chang YT, Tien YW, Sun CT, Wu MS, Lin JT. Distinct chromosomal aberrations of ampulla of Vater and pancreatic head cancers detected by laser capture microdissection and comparative genomic hybridization. Oncol Rep. 2005;14:867–872. [PubMed] [Google Scholar]
- 13.Baudis M. Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data. BMC Cancer. 2007;7:226. doi: 10.1186/1471-2407-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyata T, Yonekura K, Utsunomiya A, Kanekura T, Nakamura S, Seto M. Cutaneous type adult T-cell leukemia/lymphoma is a characteristic subtype and includes erythema/papule and nodule/tumor subgroups. Int J Cancer. 2010;126:1521–1528. doi: 10.1002/ijc.24874. [DOI] [PubMed] [Google Scholar]
- 15.Law FB, Chen YW, Wong KY, Ying J, Tao Q, Langford C, Lee PY, Law S, Cheung RW, Chui CH, et al. Identification of a novel tumor transforming gene GAEC1 at 7q22 which encodes a nuclear protein and is frequently amplified and overexpressed in esophageal squamous cell carcinoma. Oncogene. 2007;26:5877–5888. doi: 10.1038/sj.onc.1210390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopalan V, Smith RA, Nassiri MR, Yasuda K, Salajegheh A, Kim SY, Ho YH, Weinstein S, Tang JC, Lam AK. GAEC1 and colorectal cancer: a study of the relationships between a novel oncogene and clinicopathologic features. Hum Pathol. 2010;41:1009–1015. doi: 10.1016/j.humpath.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 18.Deng W, Tsao SW, Guan XY, Lucas JN, Si HX, Leung CS, Mak P, Wang LD, Cheung AL. Distinct profiles of critically short telomeres are a key determinant of different chromosome aberrations in immortalized human cells: whole-genome evidence from multiple cell lines. Oncogene. 2004;23:9090–9101. doi: 10.1038/sj.onc.1208119. [DOI] [PubMed] [Google Scholar]
- 19.Yuen HF, Chan YP, Chan KK, Chu YY, Wong ML, Law SY, Srivastava G, Wong YC, Wang X, Chan KW. Id-1 and Id-2 are markers for metastasis and prognosis in oesophageal squamous cell carcinoma. Br J Cancer. 2007;97:1409–1415. doi: 10.1038/sj.bjc.6604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown DC, Gatter KC. Ki67 protein: the immaculate deception? Histopathology. 2002;40:2–11. doi: 10.1046/j.1365-2559.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- 21.Derenzini M. The AgNORs. Micron. 2000;31:117–120. doi: 10.1016/s0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- 22.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian H, Lu N, Xue L, Liang X, Zhang X, Fu M, Xie Y, Zhan Q, Liu Z, Lin C. Reduced MTA1 expression by RNAi inhibits in vitro invasion and migration of esophageal squamous cell carcinoma cell line. Clin Exp Metastasis. 2005;22:653–662. doi: 10.1007/s10585-006-9005-2. [DOI] [PubMed] [Google Scholar]
- 24.Yamada S, Yanamoto S, Yoshida H, Yoshitomi I, Kawasaki G, Mizuno A, Nemoto TK. RNAi-mediated down-regulation of alpha-actinin-4 decreases invasion potential in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2010;39:61–67. doi: 10.1016/j.ijom.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Likui W, Hong W, Shuwen Z, Yuangang Y, Yan W. The potential of osteopontin as a therapeutic target for human colorectal cancer. J Gastrointest Surg. 2011;15:652–659. doi: 10.1007/s11605-011-1445-6. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, Shen Y, Ji B, Wang L, Zhang Z, Zhang Y. Combinational RNAi gene therapy of hepatocellular carcinoma by targeting human EGFR and TERT. Eur J Pharm Sci. 2011;42:387–391. doi: 10.1016/j.ejps.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Vázquez-Vega S, Contreras-Paredes A, Lizano-Soberón M, Amador-Molina A, García-Carrancá A, Sánchez-Suárez LP, Benítez-Bribiesca L. [RNA interference (RNAi) and its therapeutic potential in cancer] Rev Invest Clin. 2010;62:81–90. [PubMed] [Google Scholar]
- 28.Kar P, Samanta K, Shaikh S, Chowdhury A, Chakraborti T, Chakraborti S. Mitochondrial calpain system: an overview. Arch Biochem Biophys. 2010;495:1–7. doi: 10.1016/j.abb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Brown AE, Yeaman SJ, Walker M. Targeted suppression of calpain-10 expression impairs insulin-stimulated glucose uptake in cultured primary human skeletal muscle cells. Mol Genet Metab. 2007;91:318–324. doi: 10.1016/j.ymgme.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Horikawa Y. Calpain-10 (NIDDM1) as a Susceptibility Gene for Common Type 2 Diabetes. Endocr J. 2006;53:567–576. doi: 10.1507/endocrj.kr-70. [DOI] [PubMed] [Google Scholar]
- 31.Moreno-Luna R, Abrante A, Esteban F, González-Moles MA, Delgado-Rodríguez M, Sáez ME, González-Pérez A, Ramírez-Lorca R, Real LM, Ruiz A. Calpain 10 gene and laryngeal cancer: a survival analysis. Head Neck. 2011;33:72–76. doi: 10.1002/hed.21404. [DOI] [PubMed] [Google Scholar]
- 32.Frances CP, Conde MC, Saez ME, Diez SF, Rey CM, Ramírez-Armengol JA, Pascual MH, Gonzalez-Perez A, Torres PP, Real LM, et al. Identification of a protective haplogenotype within CAPN10 gene influencing colorectal cancer susceptibility. J Gastroenterol Hepatol. 2007;22:2298–2302. doi: 10.1111/j.1440-1746.2007.04843.x. [DOI] [PubMed] [Google Scholar]
- 33.Fong PY, Fesinmeyer MD, White E, Farin FM, Srinouanprachanh S, Afsharinejad Z, Mandelson MT, Brentnall TA, Barnett MJ, Goodman GE, et al. Association of diabetes susceptibility gene calpain-10 with pancreatic cancer among smokers. J Gastrointest Cancer. 2010;41:203–208. doi: 10.1007/s12029-010-9130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsunoda S, Nakamura T, Sakurai H, Saiki I. Fibroblast growth factor-2-induced host stroma reaction during initial tumor growth promotes progression of mouse melanoma via vascular endothelial growth factor A-dependent neovascularization. Cancer Sci. 2007;98:541–548. doi: 10.1111/j.1349-7006.2007.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang CC, Liu Z, Li X, Bailey SK, Nail CD, Foster KW, Frost AR, Ruppert JM, Lobo-Ruppert SM. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Biol Ther. 2005;4:1401–1408. doi: 10.4161/cbt.4.12.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popple A, Durrant LG, Spendlove I, Rolland P, Scott IV, Deen S, Ramage JM. The chemokine, CXCL12, is an independent predictor of poor survival in ovarian cancer. Br J Cancer. 2012;106:1306–1313. doi: 10.1038/bjc.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Yu H, Luo RZ, Zhang Y, Zhang MF, Wang X, Jia WH. Elevated expression of Rad51 is correlated with decreased survival in resectable esophageal squamous cell carcinoma. J Surg Oncol. 2011;104:617–622. doi: 10.1002/jso.22018. [DOI] [PubMed] [Google Scholar]
- 38.Kim MS, Oh JE, Kim YR, Park SW, Kang MR, Kim SS, Ahn CH, Yoo NJ, Lee SH. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J Pathol. 2010;221:139–146. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 39.Morita S, Matsumoto Y, Okuyama S, Ono K, Kitamura Y, Tomori A, Oyama T, Amano Y, Kinoshita Y, Chiba T, et al. Bile acid-induced expression of activation-induced cytidine deaminase during the development of Barrett’s oesophageal adenocarcinoma. Carcinogenesis. 2011;32:1706–1712. doi: 10.1093/carcin/bgr194. [DOI] [PubMed] [Google Scholar]
- 40.Shinmura K, Igarashi H, Goto M, Tao H, Yamada H, Matsuura S, Tajima M, Matsuda T, Yamane A, Funai K, et al. Aberrant expression and mutation-inducing activity of AID in human lung cancer. Ann Surg Oncol. 2011;18:2084–2092. doi: 10.1245/s10434-011-1568-8. [DOI] [PubMed] [Google Scholar]
- 41.Endo Y, Marusawa H, Chiba T. Involvement of activation-induced cytidine deaminase in the development of colitis-associated colorectal cancers. J Gastroenterol. 2011;46(Suppl 1):6–10. doi: 10.1007/s00535-010-0326-1. [DOI] [PubMed] [Google Scholar]
- 42.Goto A, Hirahashi M, Osada M, Nakamura K, Yao T, Tsuneyoshi M, Takayanagi R, Oda Y. Aberrant activation-induced cytidine deaminase expression is associated with mucosal intestinalization in the early stage of gastric cancer. Virchows Arch. 2011;458:717–724. doi: 10.1007/s00428-011-1086-x. [DOI] [PubMed] [Google Scholar]


