Abstract
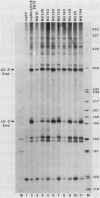
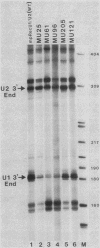
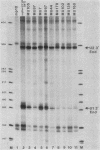
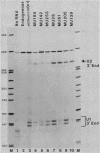
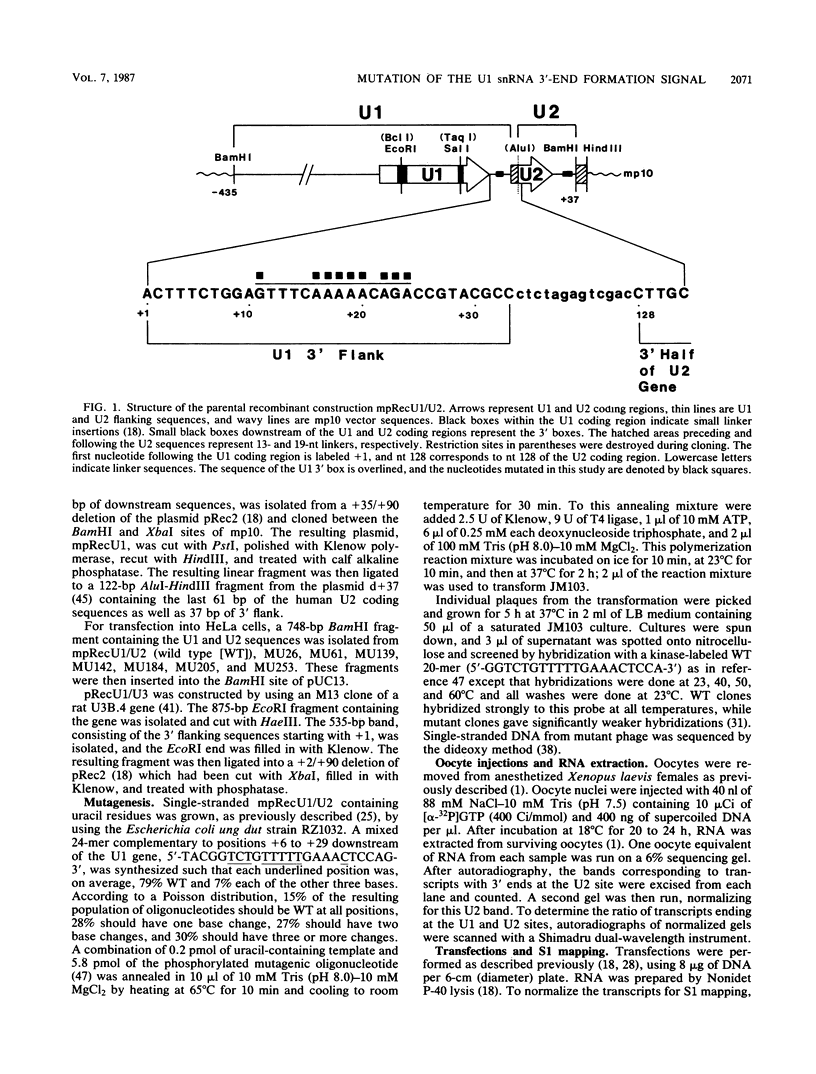
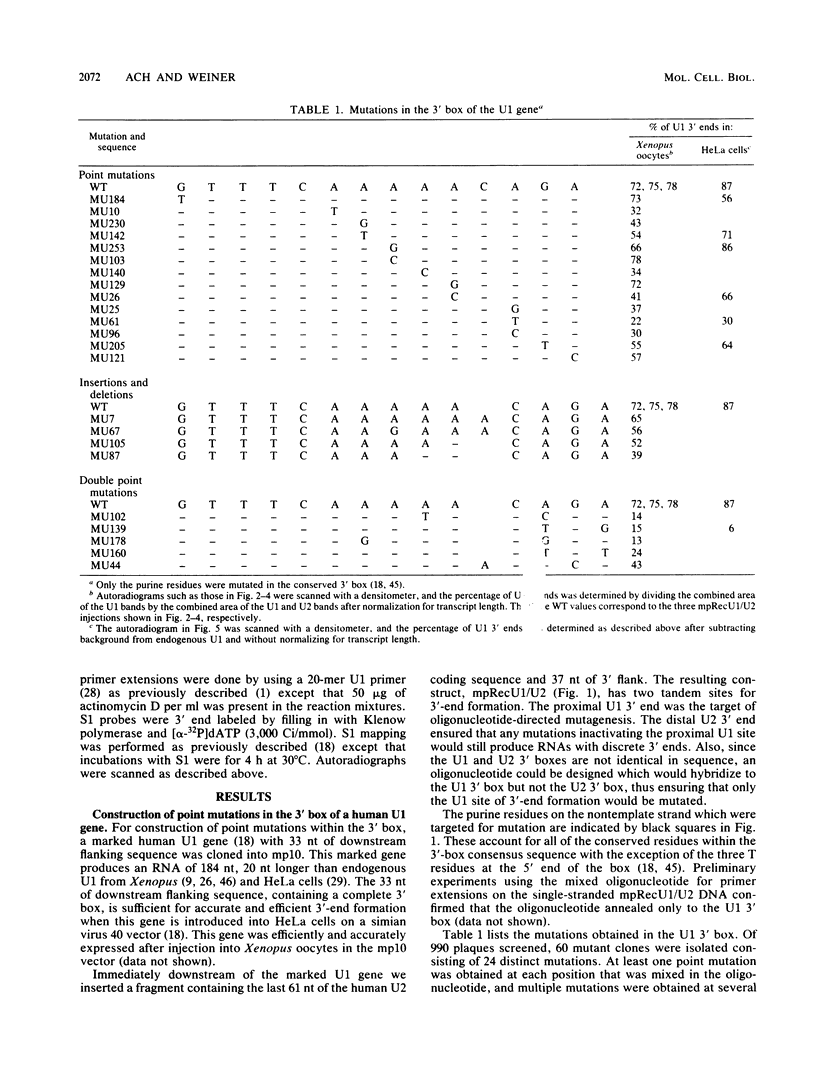
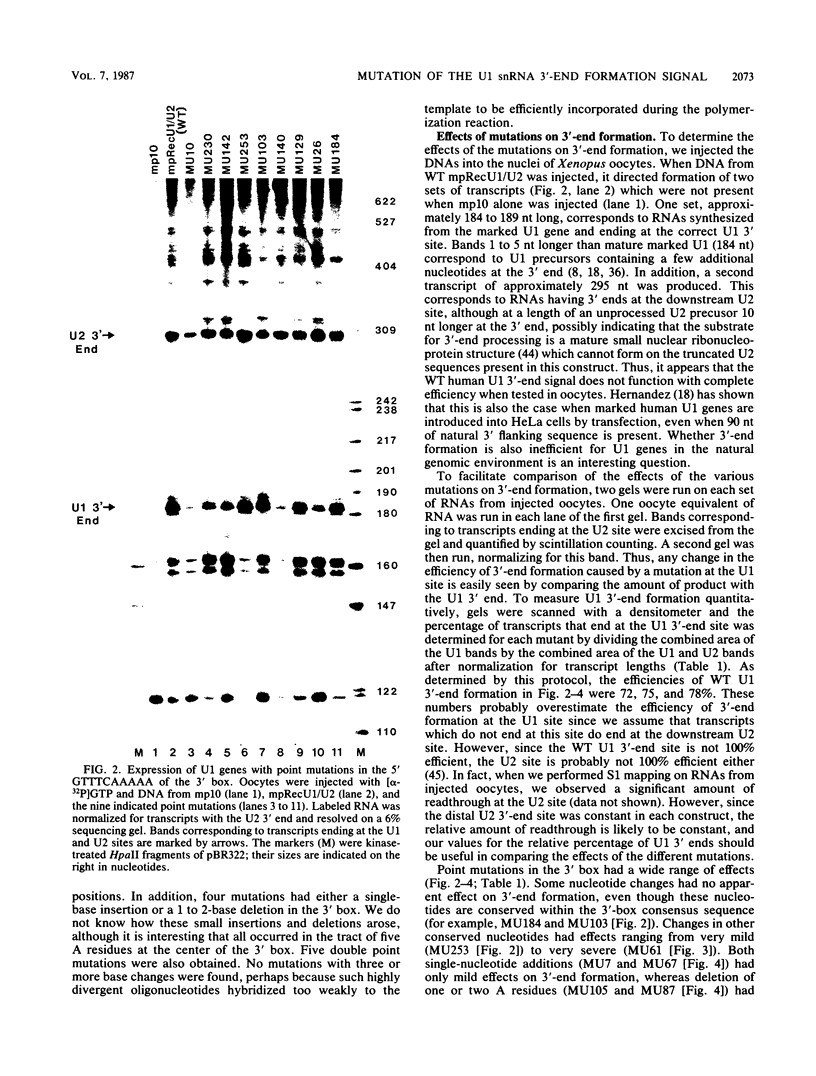
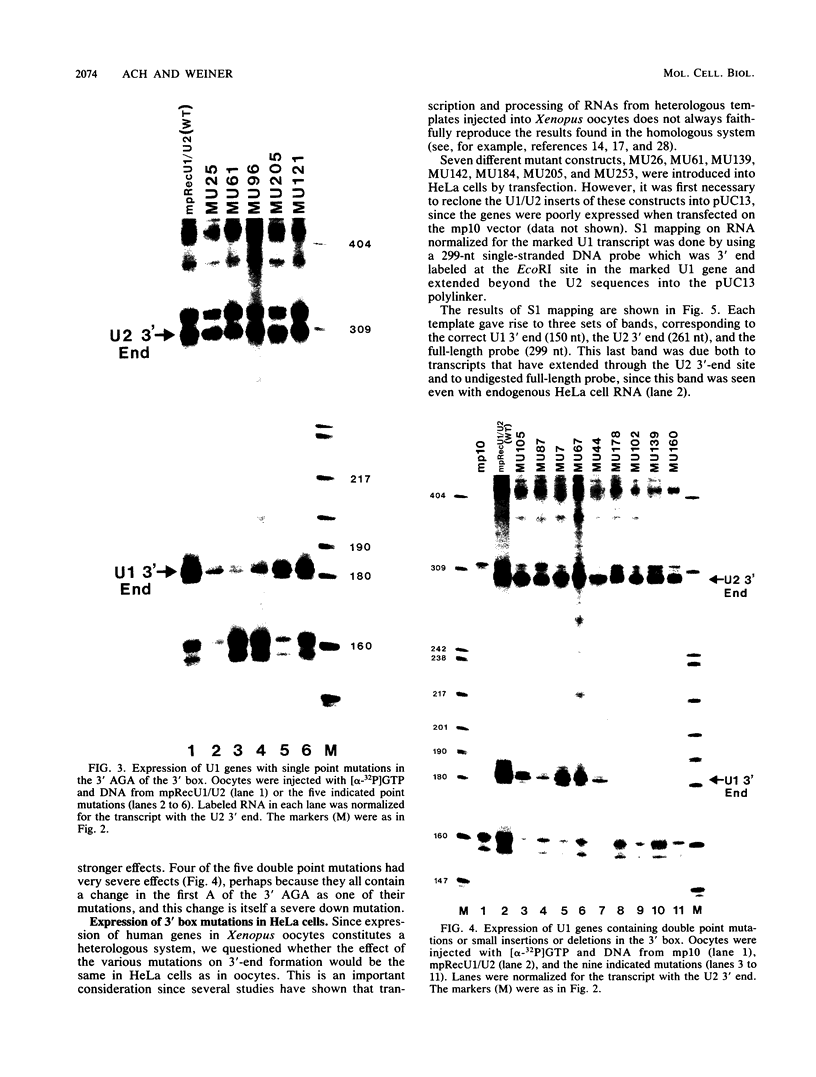
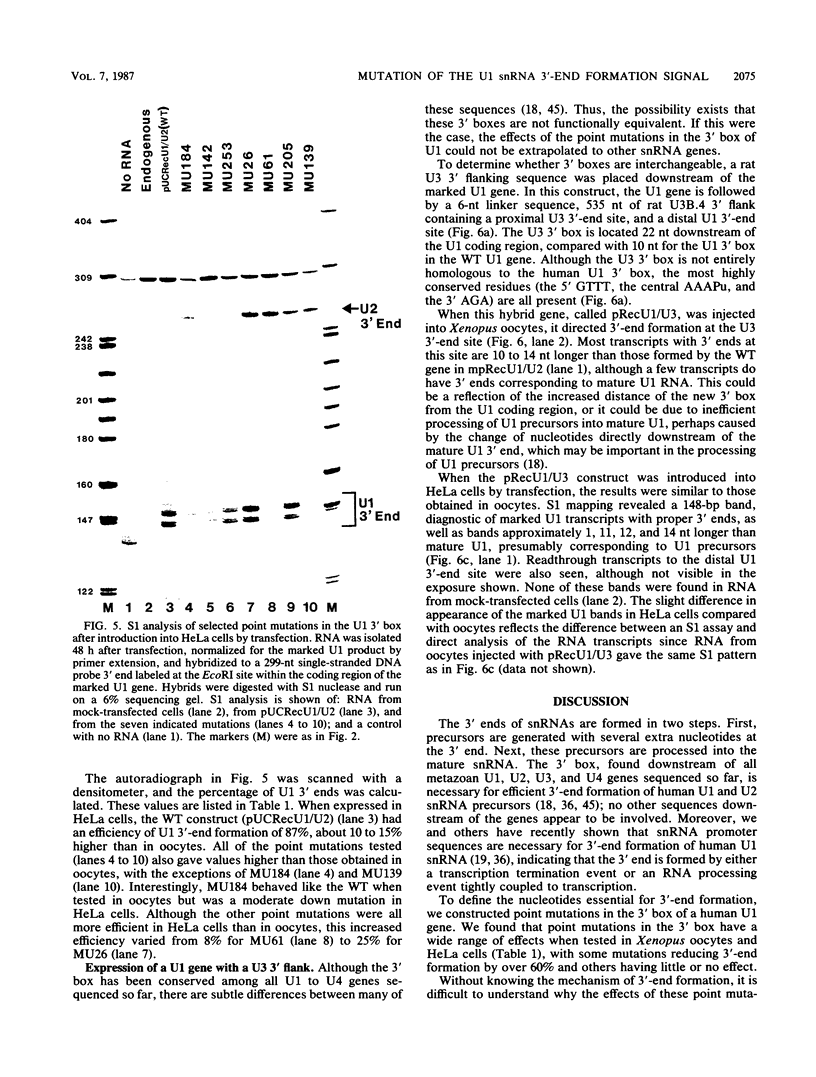
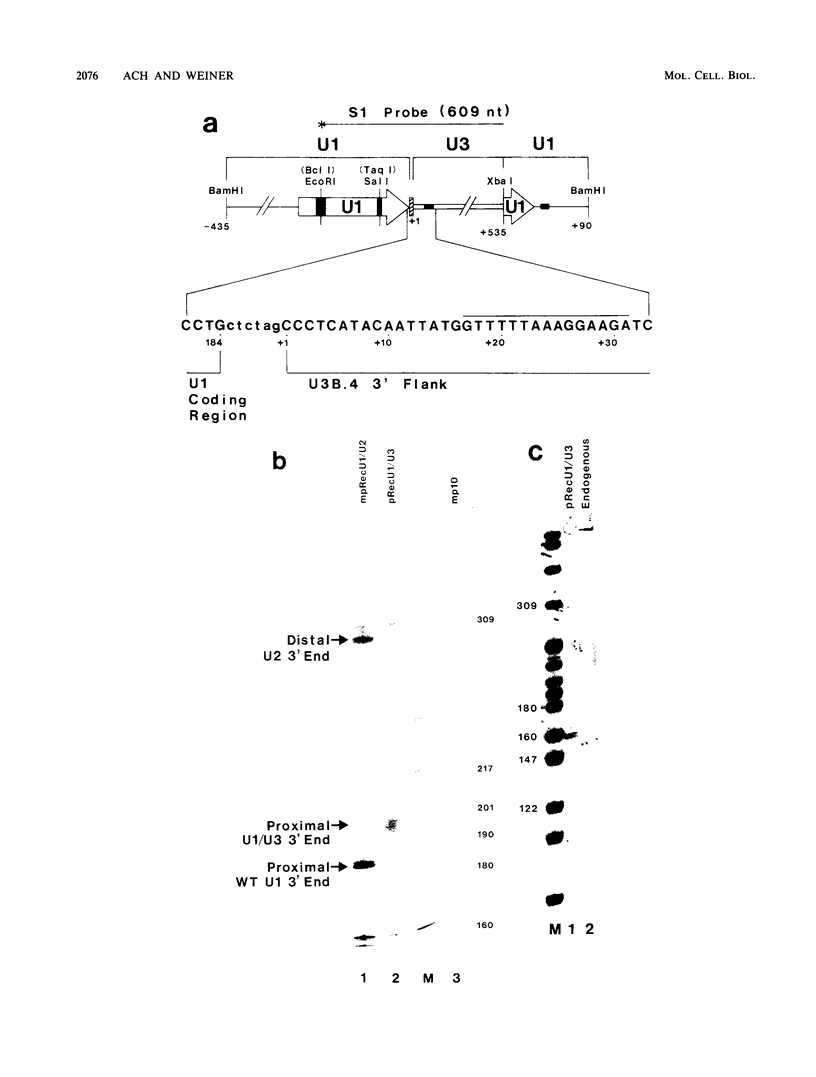
Formation of the 3' end of U1 and U2 small nuclear RNA (snRNA) precursors is directed by a conserved sequence called the 3' box located 9 to 28 nucleotides downstream of all metazoan U1 to U4 snRNA genes sequenced so far. Deletion of part or all of the 3' box from human U1 and U2 genes drastically reduces 3'-end formation. To define the essential nucleotides within this box that direct 3'-end formation, we constructed a set of point mutations in the conserved residues of the human U1 3' box. The ability of the various mutations to direct 3'-end formation was tested by microinjection into Xenopus oocytes and transfection into HeLa cells. We found that the point mutations had diverse effects on 3'-end formation, ranging from no effect at all to severe inhibition; however, no single or double point mutation we tested completely eliminated 3'-end formation. We also showed that a rat U3 3' flank can effectively substitute for the human U1 3' flank, indicating that the 3' boxes of the different U snRNA genes are functionally equivalent.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr, Mangin M., Weiner A. M. Orientation-dependent transcriptional activator upstream of a human U2 snRNA gene. Mol Cell Biol. 1985 Jul;5(7):1560–1570. doi: 10.1128/mcb.5.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Chandrasekharappa S. C., Smith J. H., Eliceiri G. L. Biosynthesis of small nuclear RNAs in human cells. J Cell Physiol. 1983 Nov;117(2):169–174. doi: 10.1002/jcp.1041170206. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Buckland R., Cortese R., Philipson L. Transcription signals in embryonic Xenopus laevis U1 RNA genes. EMBO J. 1985 Jun;4(6):1537–1543. doi: 10.1002/j.1460-2075.1985.tb03814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Dathan N., Frank R., Philipson L., Mattaj I. W. Formation of the 3' end on U snRNAs requires at least three sequence elements. EMBO J. 1986 Nov;5(11):2931–2937. doi: 10.1002/j.1460-2075.1986.tb04589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley J. M., 3rd, Roebuck K. A., Stumph W. E. Three linked chicken U1 RNA genes have limited flanking DNA sequence homologies that reveal potential regulatory signals. Nucleic Acids Res. 1984 Oct 11;12(19):7411–7421. doi: 10.1093/nar/12.19.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri G. L. Formation of low molecular weight RNA species in HeLa cells. J Cell Physiol. 1980 Feb;102(2):199–207. doi: 10.1002/jcp.1041020211. [DOI] [PubMed] [Google Scholar]
- Eliceiri G. L., Sayavedra M. S. Small RNAs in the nucleus and cytoplasm of HeLa cells. Biochem Biophys Res Commun. 1976 Sep 20;72(2):507–512. doi: 10.1016/s0006-291x(76)80070-8. [DOI] [PubMed] [Google Scholar]
- Forbes D. J., Kirschner M. W., Caput D., Dahlberg J. E., Lund E. Differential expression of multiple U1 small nuclear RNAs in oocytes and embryos of Xenopus laevis. Cell. 1984 Oct;38(3):681–689. doi: 10.1016/0092-8674(84)90263-0. [DOI] [PubMed] [Google Scholar]
- Frederiksen S., Hellung-Larsen P., Gram Jensen E. The differential inhibitory effect of alpha-amanitin on the synthesis of low molecular weight RNA components in BHK cells. FEBS Lett. 1978 Mar 15;87(2):227–231. doi: 10.1016/0014-5793(78)80338-x. [DOI] [PubMed] [Google Scholar]
- Georgiev O., Birnstiel M. L. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3' termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985 Feb;4(2):481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Eisenman R. N., McKnight S. L. Delineation of transcriptional control signals within the Moloney murine sarcoma virus long terminal repeat. Mol Cell Biol. 1985 Aug;5(8):1948–1958. doi: 10.1128/mcb.5.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Ali H., Nevins J. R. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol Cell Biol. 1985 Nov;5(11):2975–2983. doi: 10.1128/mcb.5.11.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. A small nuclear ribonucleoprotein associates with the AAUAAA polyadenylation signal in vitro. Cell. 1986 May 23;45(4):581–591. doi: 10.1016/0092-8674(86)90290-4. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Probst E., Birnstiel M. L. Transcriptional fidelity of histone genes injected into Xenopus oocyte nuclei. Nature. 1980 Nov 6;288(5786):100–102. doi: 10.1038/288100a0. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hoffman M. L., Korf G. M., McNamara K. J., Stumph W. E. Structural and functional analysis of chicken U4 small nuclear RNA genes. Mol Cell Biol. 1986 Nov;6(11):3910–3919. doi: 10.1128/mcb.6.11.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf G. M., Stumph W. E. Chicken U2 and U1 RNA genes are found in very different genomic environments but have similar promoter structures. Biochemistry. 1986 Apr 22;25(8):2041–2047. doi: 10.1021/bi00356a031. [DOI] [PubMed] [Google Scholar]
- Krol A., Lund E., Dahlberg J. E. The two embryonic U1 RNA genes of Xenopus laevis have both common and gene-specific transcription signals. EMBO J. 1985 Jun;4(6):1529–1535. doi: 10.1002/j.1460-2075.1985.tb03813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Pederson T. Transcription boundaries of U1 small nuclear RNA. Mol Cell Biol. 1985 Sep;5(9):2332–2340. doi: 10.1128/mcb.5.9.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E., Forbes D. J. The two embryonic U1 small nuclear RNAs of Xenopus laevis are encoded by a major family of tandemly repeated genes. Mol Cell Biol. 1984 Dec;4(12):2580–2586. doi: 10.1128/mcb.4.12.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madore S. J., Wieben E. D., Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol. 1984 Jan;98(1):188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin M., Ares M., Jr, Weiner A. M. Human U2 small nuclear RNA genes contain an upstream enhancer. EMBO J. 1986 May;5(5):987–995. doi: 10.1002/j.1460-2075.1986.tb04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser T., Gesteland R. F. Human U1 loci: genes for human U1 RNA have dramatically similar genomic environments. Cell. 1982 May;29(1):257–264. doi: 10.1016/0092-8674(82)90110-6. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Zeller R. Xenopus laevis U2 snRNA genes: tandemly repeated transcription units sharing 5' and 3' flanking homology with other RNA polymerase II transcribed genes. EMBO J. 1983;2(11):1883–1891. doi: 10.1002/j.1460-2075.1983.tb01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteucci M. D., Heyneker H. L. Targeted random mutagenesis: the use of ambiguously synthesized oligonucleotides to mutagenize sequences immediately 5' of an ATG initiation codon. Nucleic Acids Res. 1983 May 25;11(10):3113–3121. doi: 10.1093/nar/11.10.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985 Jul;41(3):845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984 Mar;36(3):581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Murphy J. T., Burgess R. R., Dahlberg J. E., Lund E. Transcription of a gene for human U1 small nuclear RNA. Cell. 1982 May;29(1):265–274. doi: 10.1016/0092-8674(82)90111-8. [DOI] [PubMed] [Google Scholar]
- Saba J. A., Busch H., Wright D., Reddy R. Isolation and characterization of two putative full-length Drosophila U4 small nuclear RNA genes. J Biol Chem. 1986 Jul 5;261(19):8750–8753. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F., Gilmartin G. M., Bannwarth W., Birnstiel M. L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3' end of H3 messenger RNA. 1986 Oct 30-Nov 5Nature. 323(6091):777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- Skuzeski J. M., Lund E., Murphy J. T., Steinberg T. H., Burgess R. R., Dahlberg J. E. Synthesis of human U1 RNA. II. Identification of two regions of the promoter essential for transcription initiation at position +1. J Biol Chem. 1984 Jul 10;259(13):8345–8352. [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. Genes and pseudogenes for rat U3A and U3B small nuclear RNA. J Mol Biol. 1985 Jul 20;184(2):183–193. doi: 10.1016/0022-2836(85)90372-9. [DOI] [PubMed] [Google Scholar]
- Van Arsdell S. W., Weiner A. M. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol. 1984 Mar;4(3):492–499. doi: 10.1128/mcb.4.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Lund E., Murphy J. T., Pettersson U., Dahlberg J. E. Human U2 and U1 RNA genes use similar transcription signals. EMBO J. 1984 Dec 20;3(13):3295–3301. doi: 10.1002/j.1460-2075.1984.tb02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo C. Y., Ares M., Jr, Weiner A. M. Sequences required for 3' end formation of human U2 small nuclear RNA. Cell. 1985 Aug;42(1):193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- Zeller R., Carri M. T., Mattaj I. W., De Robertis E. M. Xenopus laevis U1 snRNA genes: characterisation of transcriptionally active genes reveals major and minor repeated gene families. EMBO J. 1984 May;3(5):1075–1081. doi: 10.1002/j.1460-2075.1984.tb01931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]