Abstract
Fragile X syndrome (FXS) is a trinucleotide repeat disorder caused by a CGG repeat expansion in FMR1, and loss of its protein product FMRP. Recent studies have provided increased support for the role of FMRP in translational repression via ribosomal stalling and the microRNA pathway. In neurons, particular focus has been placed on identifying the signaling pathways such as PI3K and mTOR downstream of group 1 metabotropic glutamate receptors (mGluR1/5) that regulate FMRP. New evidence also suggests that loss of FMRP causes presynaptic dysfunction and abnormal adult neurogenesis. In addition, studies on FXS stem cells especially induced pluripotent stem (iPS) cells and new sequencing efforts hold out promise for deeper understanding of the silencing process and mutation spectrum of FMR1.
Keywords: fragile X syndrome, intellectual disability (ID), translational regulation, ribosomal stalling, microRNAs, long term depression (LTD), group 1 metabotropic glutamate receptors, presynapse, adult neurogenesis, human embryonic stem cell (hESC), induced pluripotent stem cell (iPS), missense mutation
Introduction
Fragile X syndrome (FXS) is the most common cause of inherited intellectual disability (ID) and the leading monogenic cause of autism spectrum disorders [1]. In almost all known cases of FXS, the causative mutation is a trinucleotide CGG expansion in the 5'-untranslated region of the fragile X mental retardation gene, FMR1. In humans, the number of CGG repeats is highly polymorphic. Normal individuals have between 6–54 repeats, with 29 or 30 repeats being the most common allele. When the number of repeats expands to between 60–200, it is referred to as a premutation allele. When the repeat number reaches over 200, it is known as a full mutation and leads to hypermethylation and epigenetic silencing of FMR1, resulting in the loss of its protein product, fragile X mental retardation protein, FMRP, which in turn causes FXS [2, 3]. FMRP is a selective RNA-binding protein found to have a major role in negatively regulating the translation of bound mRNAs, especially at synapses in neurons. Loss of FMRP impairs normal synaptic plasticity, which is believed to be the molecular basis for ID in FXS patients [4]. Recent studies are continuing to uncover new aspects of FMRP function in translational regulation and neural function. Particular focus has also been placed on identifying the signaling pathways that regulate FMRP in hopes of revealing new therapeutic targets. Furthermore, research on induced pluripotent stem (iPS) cells and next-generation sequencing efforts hold out promise for deeper understanding of the silencing and mutation spectrum of the FMR1 gene.
Mechanisms of FMRP-Mediated Translational Regulation
Loss of FMRP results in increased translation of FMRP-bound transcripts, implying that FMRP normally acts as a translational repressor. The exact mechanisms of translational regulation by FMRP are not entirely clear, although mounting evidence suggests that FMRP inhibits translation of its target mRNAs by stalling ribosomes and via association with microRNAs (miRNAs). Recent findings are providing further support that these mechanisms are critical for the translational function of FMRP.
The majority of cytoplasmic FMRP is associated with polyribosomes and evidence that FMRP causes ribosome stalling was observed by treating cells with the translational inhibitors sodium azide or puromycin, which cause actively translocating ribosomes to be released or “run-off” the transcript [5–7]. Interestingly, following treatment, some FMRP was still associated with polyribosomes, suggesting that sodium azide and puromycin-resistant ribosomes were “stalled” in an inactive state. By using an in vitro translation system with endogenous brain polyribosomes, Darnell et al.[8] recently uncovered more evidence of ribosome stalling. They found that the presence of FMRP increased the number of ribosomes associated with FMRP-target transcripts following puromycin run-off, but did not affect the number of ribosomes on non-target mRNAs. The increased number of ribosomes stalled on FMRP-target transcripts implies that FMRP associated with stalled polyribosomes is a major mechanism of translational control.
Notably, high throughput sequencing of RNA transcripts co-immunoprecipitated with crosslinked FMRP revealed that FMRP is closely associated with transcripts throughout both the coding and non-coding regions and does not appear limited to known RNA binding motifs such as G-quadruplexes [8]. It is unclear how FMRP mediates such broad binding patterns, since RNA binding domains (KH domains and an RGG box) of FMRP are believed to only recognize RNA secondary structures such as “kissing complexes” and G-quadruplexes. It is possible that FMRP binds specific recognition sequences in its targets and then spreads along the transcript by associating with ribosomes as they are loaded on the transcript. Or perhaps, FMRP is merely part of a complex whose specificity is determined by one or more of the other components. Creating mutations in known G-quadruplex motifs and seeing how they might affect these results would be instructive. Despite the unanswered questions, the study by Darnell et al. provides strong support that FMRP regulates translation by stalling ribosomal translocation (Figure 1a).
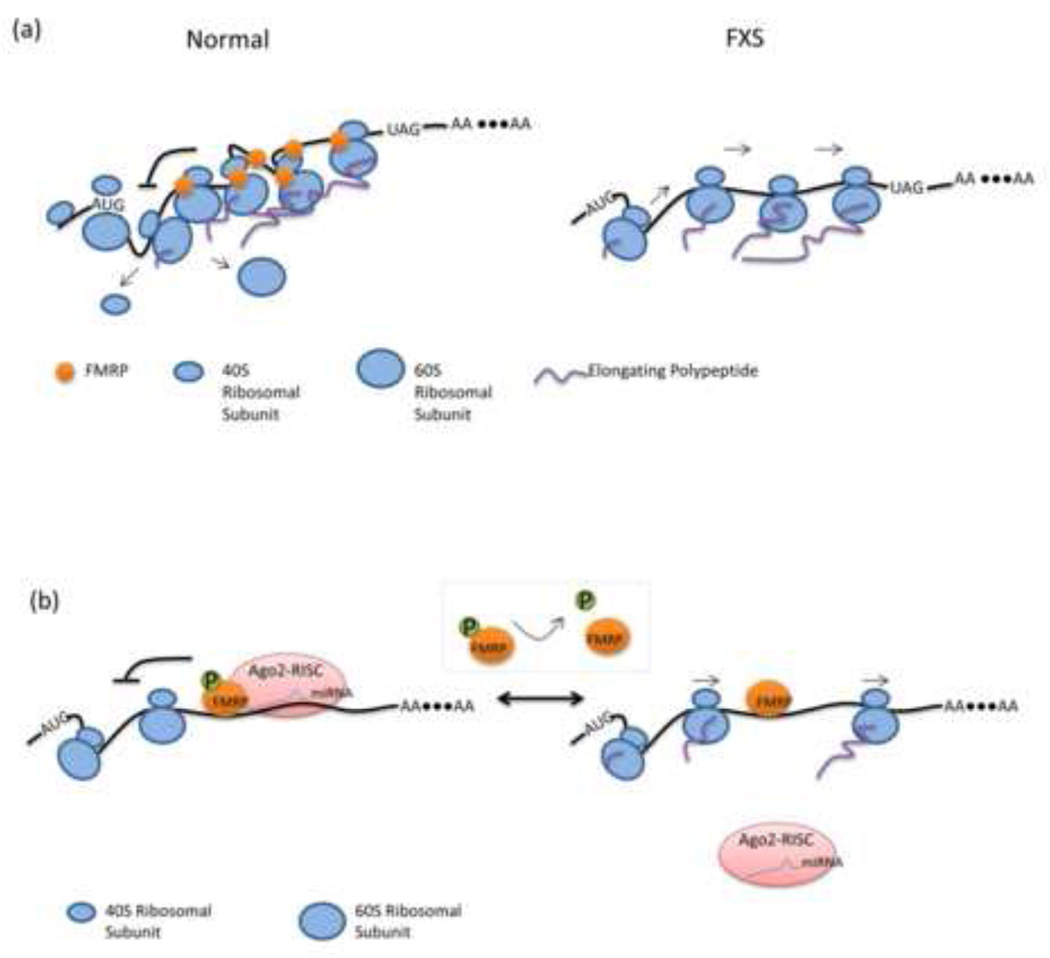
Figure 1.
Mechanisms of FMRP mediated translational regulation. (a) FMRP inhibits translation of target mRNAs by ribosome stalling. In normal cells, increased number of ribosomes are stalled on FMRP-target transcripts, which results in reduced translation. Darnell et al.[8] shows that a predominance of tags among FMRP target transcripts are distributed within the coding sequence by HITS-CLIP experiment. The stalling event occurs not only on target mRNAs bearing known secondary structures such as kissing complex and G-quadruplexes, but also on some mRNAs without these structures. In FXS cells, translational repression by FMRP via ribosome stalling is absent. (b) MiRNA-mediated translational repression by FMRP. FMRP is found to interact with miRNAs and members of the RISC complex. MiRNAs such as miR-125a, miR-125b and miR-132 are selectively associated with the FMRP-RISC RNP complex. These miRNAs in turn facilitate the selection and repression of target mRNAs by FMRP. Phosphorylation of FMRP acts as a switch for this mechanism. Dephosphorylation of serine 499 causes the dissociation of the RISC complex from target mRNAs and relieves the translational repression. However, dephosphorylated FMRP remains associated with target mRNAs.
In Drosophila and mammalian cells, FMRP is also found to interact with miRNAs and members of the RISC complex, including Dicer and Argonaute 2/eIF2C2 [9, 10]. FMRP has no miRNA/siRNA-binding domain, which indicates that miRNAs are likely associated with FMRP via interaction with other members of the RISC complex. In mice, this interaction is also evident in the postsynapse underscoring its relevance for synaptic translational control. Several miRNAs, such as miR-125b and miR-132, are selectively associated with the FMRP-RISC RNP complex in the mouse brain. Interestingly, regulation of a known FMRP target, NR2A, was recently shown to depend in part on miR-125b [11]. This study provides direct evidence that specific miRNAs facilitate the selection and repression of target mRNAs by FMRP. In another recent study, the FMRP-miR-125a complex was shown to bind the 3’UTR of PSD-95 mRNA, inhibiting translation of the PSD-95 protein, a key component of postsynaptic organization. The authors further revealed that the translation inhibition of PSD-95 by FMRP-miR-125a is dependent on the phosphorylation of FMRP at serine 499 (human serine 500). Stimulation of mGluR, which leads to dephosphorylation of FMRP, or substitution of an unphosphorylatable mutant FMRP, S499A, relieved the translational repression and caused the dissociation of the RISC complex from PSD-95 mRNA [12]. Interestingly, FMRP does not disassociate from the message following the loss of the RISC complex suggesting that FMRP may not itself suppress ranslation but rather utilizes this function of the RISC complex (Figure 1b). Phosphorylation of FMRP was also reported to inhibit its association with Dicer, while increasing its affinity for pre-miRNA complexes [13]. Together, these studies suggest that microRNAs function as a critical component to modulate FMRP-mediated translation and that phosphorylation of FMRP acts as a switch in this pathway.
How ribosome stalling and miRNA-directed translational repression are temporally and spatially coordinated with each other remains to be determined. It seems reasonable to speculate that these mechanisms can occur separately in spatially distinct locations or at different times of the mRNA life-cycle, for example, prior to transport to the synapse or after synaptic stimulation. In other situations, the mechanisms may be coordinated on a single transcript, such that miRNAs facilitate the ability of FMRP to stall ribosomes. In recent findings with miRNA-mediated repression and in early studies of ribosome stalling, phosphorylation of FMRP appears to be the primary trigger for releasing the translation repression of FMRP-bound transcripts.
Besides the well-established ribosome stalling and miRNA-directed translational repression model, Napoli et al. suggest that FMRP can also suppress translation via inhibition of translation initiation [14, 15]. Recently, FMRP was also found to behave as a translational activator of the Sod1 mRNA, with the absence of FMRP resulting in decreased expression of Sod1 [16]. In addition, FMRP is shown to be involved in regulating mRNA stability [17, 18]. All these studies demonstrate that much remains to be learned about the role of FMRP in translational regulation.
Neuronal Dysfunction in FXS
Many FMRP target transcripts are localized in neuronal dendrites and play important roles in synaptic structure and function. The current working model is that FMRP accompanies specific target mRNAs to dendritic spines, where it regulates their translation in response to synaptic stimuli. In FXS, loss of FMRP leads to misregulation of activity-dependent local protein synthesis, which is evidenced by impaired synaptic plasticity. Unraveling the neuronal signaling pathways that are regulated by FMRP is a main focus for developing treatments to rescue FXS cognitive phenotypes. In wild-type neurons, activation of group I mGluR receptors rapidly increases protein synthesis of synaptic transcripts, including FMRP-bound transcripts, via mTOR and ERK-dependent pathways. Both pathways converge to increase eIF4E activity and initiate the assembly of the initiation complex 4F (eIF4F), the first step in the initiation of mRNA cap-dependent translation [19–21]. This group I mGluR-dependent protein synthesis induces long-term depression (LTD), a molecular basis of learning and memory, which is impaired in FXS [22, 23]. Recently, different observations on how the loss of FMRP affects the relative levels of mTOR and ERK signaling molecules have emerged. In one set of studies, increased activities of PI3K, Akt, and mTOR have been detected in cortical synaptoneurosomes and hippocampal lysates from Fmr1 KO mice [19, 21]. Additionally, the inhibition of PI3K, but not inhibition of ERK, specifically rescued excess translation and subsequent AMPA receptor endocytosis seen in the KO [19]. However, another study failed to observe any increased levels of mTOR pathway components in cultured brain slices from Fmr1 KO mice and additionally showed that inhibition of ERK, but not mTOR, could rescue excess protein synthesis in the KO slices [24]. Differences in experimental procedures may cause such discrepancies; therefore, it remains to be determined how those results explain the in vivo status of the mGluR downstream signals in the absence of FMRP. Nevertheless, these studies suggest that FMRP modulates translation of its mRNA targets in an activity-dependent manner such as in response to mGluR stimulation.
Amygdala dysfunction is also a hallmark characteristic in FXS. It has been implicated that alterations in the GABA system, including dramatic changes in levels of expression of GABA receptors and the defects in GABAergic neurotransmission could contribute to circuit dysfunction in FXS [25, 26]. Initial findings of exaggerated LTD in FXS mouse models have largely focused on the postsynaptic function of FMRP. However, several studies now report that the loss of FMRP causes morphological and functional presynaptic abnormalities. Quantitative proteomic analysis shows that many presynaptic proteins involved in presynaptic specialization, vesicle recycling, excitability and neurotransmitter release are affected when FMRP is absent [27, 28]. High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) results also reveal that FMRP directly binds mRNAs encoding nearly one-third of the presynaptic proteome [8]. In addition, the loss of FMRP leads to altered short-term plasticity in excitatory synapses and excessive calcium influx in the presynaptic neurons during spike trains. Furthermore, faster vesicle recycling and enlarged vesicle pools are also observed in the absence of FMRP, which leads to reduced short-term depression (STD) [27]. Additionally, FMRP also directly interacts with the membrane protein Slack-B, a sodium-activated potassium channel, which is presumably expressed presynaptically. Further data also support the idea that FMRP functions as a potent activator of Slack-B to modulate neuronal firing patterns [29]. All these studies point to presynaptic dysfunction as a potential contributor to neurologic impairments in FXS.
Recent studies also reveal an unexpected regulatory role for FMRP in adult neurogenesis. Adult neurogenesis occurs mainly at the subgranular zone of the dentate gyrus in hippocampus and the subventricular zone of the lateral ventricles, both of which contain adult neural stem cells/progenitor cells (aNSCs), generating new neurons and glia. Although the precise role of adult neurogenesis in learning and memory is unclear, mounting evidence suggests that it plays a role in adult neuroplasticity and hippocampus-dependent learning [30, 31]. Deletion of FMRP from adult neural stem cells in mice leads to impaired performance in hippocampus-dependent learning tasks and these defects can be rescued by restoring FMRP expression [32]. FMRP-deficient aNSCs also display increased proliferation, decreased neuronal differentiation, and increased glial differentiation, which in turn alters the fate specification of aNSCs. The altered aNSC function is partially dependent on CDK4 and GSK3β signaling, which are both known FMRP targets of translational repression [33].
Silencing of the FMR1 full mutation
Understanding how and when the expanded CGG repeat is methylated and silenced in FXS is critical for a broader understanding of the disease etiology and potential development of therapies aimed at rescuing FMR1 expression. The identification of high-functioning fragile X males who have near normal intelligence and carry unmethylated full mutation alleles holds out additional promise that preventing or rescuing FMR1 silencing is a viable pursuit [34, 35].
Studies investigating the developmental timing of FMR1 silencing, using human embryonic stem cells (hESCs) and chorionic villi (CV) samples from FXS pregnancies, have found that, at early embryogenesis, when extra embryonic tissue separates from the embryo proper, the FXS full mutation alleles are still active. Thus, in early development, FMR1 remains unsilenced, presumably producing transcripts with long CGG tracts [36, 37]. The mechanisms behind this unique epigenetic event in response to DNA sequence variation remains unclear. Use of hESCs may be useful, although the continuous generation of hESCs presents an ethical challenge. Moreover, incorporation of the CGG repeat expansion in mice fails to recapitulate the human hypermethylation seen in FXS. One emerging solution may lie in continuing advances to reprogram somatic cells into iPS cells. Generating iPS cells bypasses the ethical problems with creating hESCs and offers a great opportunity to dissect changes in methylation during development and differentiation. Recent studies reported the derivation of the first FXS-iPSC lines from FXS patients and surprisingly found that the CGG repeat region remained hypermethylated, which differs from findings in FXS hESC [38, 39]. A possible explanation for the discrepancy may be that FMRP-dependent signaling pathways, dysfunctioning in FXS cells, are required for FMR1 reactivation. In this scenario, FMRP may directly regulate the silencing of its own transcript or indirectly modulate its methylation via translational control of target mRNAs. Another, perhaps more plausible, explanation for the difference in FMR1 methylation between iPSCs and hESCs could be that human iPS cells represent a later stage of development in which silencing of the full mutation has already occurred; thus, FXS-iPS cells may not have all the full characteristics of early pluripotency. FXS-iPS cells show very similar pluripotent characteristics with hESCs [36, 38], nevertheless, studies have found that different stages of pluripotency are critical for certain epigenetic events, such as X-inactivation. Investigators have since discovered how to create iPSCs that represent an earlier developmental stage, called the ground state/naïve pluripotent stage, which shares more similar characteristic of mouse ES cells, such as both active X chromosomes in females [40, 41]. The generation of iPS cells to a state before X-inactivation is encouraging because CV sampling from FMR1 full mutation carriers suggests that X-inactivation occurs prior to inactivation of the full mutation allele. In these samples, X-inactivation was evident by 10 weeks gestational age but FMRP silencing did not occur until 10–12.5 weeks [37]. It is worth noting that in humans, the epigenetic regulation in extra-embryonic tissues such as CV is different compared with the embryo, thus, the exact time for full mutation silencing in embryos remains unclear. Nevertheless, the successful generation of naïve FXS-iPS cells should allow new investigations into the epigenetic status of the full mutation in this naïve pluripotent stage.
Successful generation of naïve/ground state FXS-iPS cells with reactivated FMR1 will allow the exploration of many hypotheses including one intriguing mechanism of transcriptional silencing. RNA-directed transcriptional gene silencing (RDTS), which occurs in both plants and animals, uses small 22–26nt RNA fragments processed by the RNAi machinery to induce methylation of the target transcript [42]. This is interesting because FMRP is known to bind its own FMR1 transcript and also associates with Argonaute and Dicer in the RISC complex. In addition, transcripts with long CGG tracts are known to be cut by Dicer in vitro [43]. Thus, FMRP might direct binding of the RISC complex on the FMR1 transcript and lead to production of 22–26nt CGG fragments, which then facilitate FMR1 methylation and silencing by directing histone modifying proteins to the locus. This is just one example of how studying FXS-iPS cells may give new insight into understanding these processes.
FMR1 mutations in FXS
Currently, clinical testing of the CGG repeat size is the standard of care for the diagnosis of FXS. However, identification of point mutations, insertions, or deletions at the FMR1 locus, could increase the overall diagnostic yield and help account for a portion of undiagnosed ID. Indeed, several cases have been reported for these non-conventional mutations in FXS patients. Many examples of FMR1 deletion have been reported in patients with FXS like phenotypes [44, 45]. Also, a missense mutation of I304N in the second FMRP KH-type RNA-binding domain which alters the biological function of FMRP was found in a patient with severe ID [46–48].
To find additional pathogenic sequence variants, a recent study systematically screened 963 males who were clinically referred for FXS testing but showed normal CGG-repeat length. This study discovered several additional mutations in FMR1. One of the mutations, R138Q, occurs in a highly conserved residue within the nuclear localization signal of FMRP. Three additional mutations were found in the promoter region, all of which were shown to reduce reporter transcription in vitro. The authors postulate that the frequency of FMR1 sequence variants causing developmental delay would be up to 0.8%. However, thorough functional testing will be needed to investigate the causality of these variants [49]. Regardless, it remains surprising that in the 20 years since the cloning of FMR1 only two missense mutations have been uncovered while other X-linked ID loci, such as MECP2, have revealed over a hundred missense mutations [50]. While this paradox maybe partly due to the exclusive testing of the CGG repeat length, rather than DNA sequences, the study described above still suggests a deficiency of FMR1 missense mutations. Such mutations must exist in the population and, indeed, the NHLBI Exome Variant Project has found seven additional missense mutations of FMR1 (by Oct, 2011) in a heterogeneous group of samples drawn largely from adult onset common disease [51]. Therefore it is reasonable to conclude that missense mutations in FMR1 may either have less impact on the protein function than missense mutations in many other proteins or that the correct population has not been sampled.
Conclusion
There has been great progress in our understanding of the role of FMRP in neurogenesis, presynaptic signaling, and translational regulation, as well as the FMR1 mutation spectrum. In addition, identifying pathways up- and downstream of FMRP activation have revealed several possible targets for drug intervention. This avenue of research has been the impetus for several current clinical trials aimed at down-regulating the exaggerated mGluR activity seen in FXS by using mGluR antagonists or GABA agonists. Other downstream signals, such as PI3K, may also be potential targets. An alternative area of therapeutic potential is to identify the mechanisms for rescuing the silencing of FMR1. However, the translational repression caused by expanded CGG repeats needs to be overcome. Pluripotent stem cells derived from FXS patients may provide invaluable model systems for studying FMR1 epigenetic silencing mechanism, as well as drug screening and creating in vitro neuronal model. With new discoveries will undoubtedly also come new complexities and the study of FXS has been replete with both.
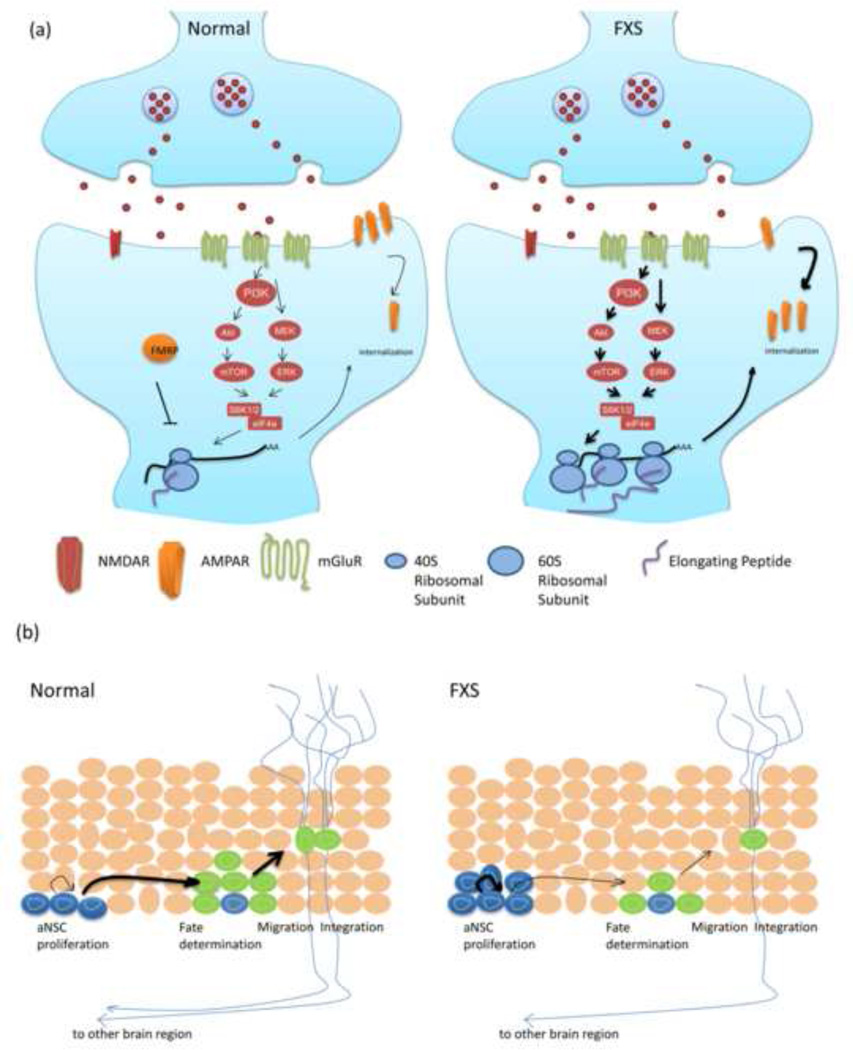
Figure 2.
(a) FMRP is a negative regulator in Group I mGluR-dependent protein synthesis. In wild-type synpase, activation of Group I mGluR receptors increases protein synthesis via the mTOR and ERK signaling pathways. Both pathways converge to increase eIF4E activity and initiate the assembly of eIF4F, the first step in the initiation of mRNA cap-dependent translation. This group I mGluR-dependent protein synthesis induces long-term depression (LTD), a molecular basis of learning and memory, which is impaired in FXS. In FXS, stimulation of Group I mGluR receptors causes excessive protein synthesis via increased mTOR and ERK signaling pathways, which leads to abnormal synaptic plasticity such as increased AMPAR internalization. (b) The role of FMRP in adult neurogenesis. The panel shows the cells from subgranular zone of the dentate gyrus in hippocampus in mice. Compared with wild-type cells, Fmrp-deficient aNSCs display increased proliferation, decreased neuronal differentiation, and increased glial differentiation, which in turn alter the fate specification of aNSCs. Subsequently, the abnormal adult neurogenesis leads to impaired hippocampus-dependent learning.
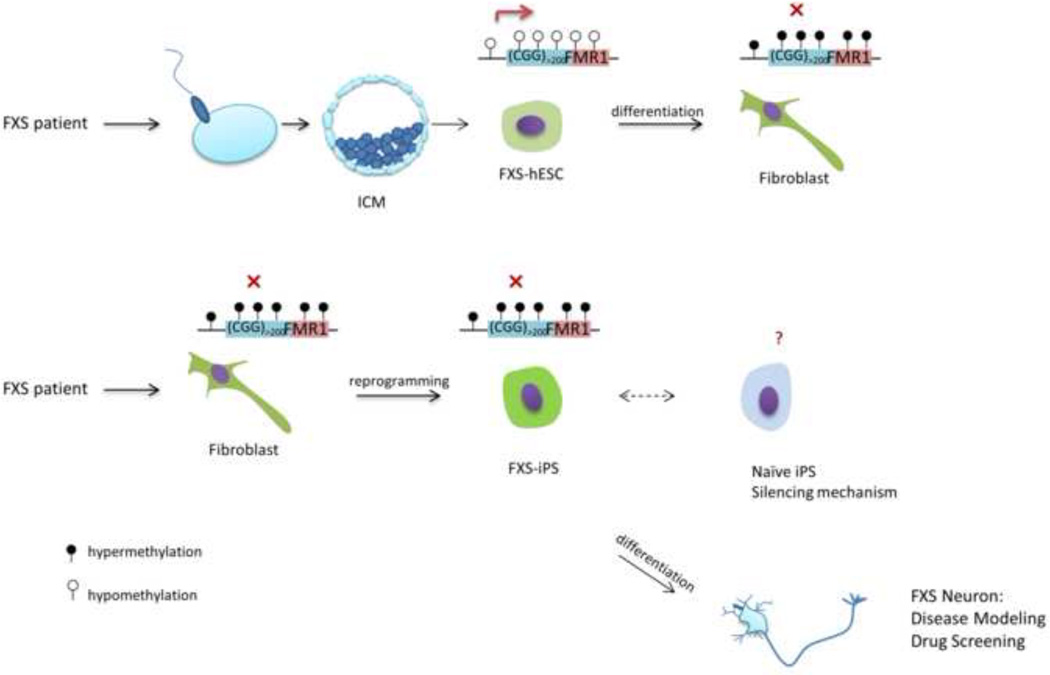
Figure 3.
The paradox of FMR1 full mutation epigenetic status between FX-iPSC and FX-hESC. In FXS, during early embryonic development, the FMR1 full mutation allele remains unsilenced. Analysis from one FXS-hESC line shows that the full mutation allele is still active, while during in vitro differentiation, FMR1 undergoes epigenetic silencing. In contrast, the full mutation allele remains methylated in iPS cells reprogrammed from FXS patients’ fibroblasts. The successful generation of naïve/ground state FXS-iPS cells will allow new investigations into the epigenetic status of the full mutation. These pluripotent stem cells derived from FXS patients provide invaluable model systems for studying FMR1 epigenetic regulation, drug screening and in vitro neuronal modeling.
Acknowledgements
We are grateful to Dr. Joshua Suhl, Dr. Mika Kinoshita, and Michael Santoro from Warren Lab, Cheryl Strauss, Dr. Peng Jin, and Dr. Brad Coffee from the Department of Human Genetics, Dr. Yue Feng from Department of Pharmacology at Emory University for their critical review of the manuscript. We apologize to colleagues whose recent work we could not cite or discuss thoroughly due to space constraints. This work is supported by the NIH grant HD020521 and HD024064.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Boyle L, Kaufmann WE. The behavioral phenotype of FMR1 mutations. Am J Med Genet C Semin Med Genet. 2010;154C:469–476. doi: 10.1002/ajmg.c.30277. [DOI] [PubMed] [Google Scholar]
- 2.Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 4.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 6.Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Dictenberg JB, Ku L, Li W, Bassell GJ, Feng Y. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell. 2008;19:105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. •• This study uses high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) to identify FMRP bounding mRNAs associated with polyribosomes. It shows that FMRP stalls ribosomes specifically on its target mRNAs, one of the key mechanisms for FMRP mediated translational repression.
- 9.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Jin P. Macro role(s) of microRNAs in fragile X syndrome? Neuromolecular Med. 2009;11:200–207. doi: 10.1007/s12017-009-8081-2. [DOI] [PubMed] [Google Scholar]
- 11. Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR- 125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. • This study provides direct evidence that specific miRNAs such as miR-125b and miR-132 facilitate the selection and repression of target mRNAs by FMRP.
- 12. Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. • This study provides an example for FMRP regulated translational repression via miRNA pathway: FRMP inhibits PSD-95 mRNA translation via miR-125a regulated by gp1 mGluR signaling. More importantly, it shows that phosphorylation of FMRP acts as a switch in this pathway.
- 13.Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. RNA. 2009;15:362–366. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rubeis S, Bagni C. Regulation of molecular pathways in the Fragile X Syndrome: insights into Autism Spectrum Disorders. J Neurodev Disord. 2011;3:257–269. doi: 10.1007/s11689-011-9087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 16.Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Wang Q, Huang Y. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc Natl Acad Sci U S A. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross C, Nakamoto M, Yao X, Chan CB, Yim SY, Ye K, Warren ST, Bassell GJ. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamoto M, Nalavadi V, Epstein MP, Narayanan U, Bassell GJ, Warren ST. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc Natl Acad Sci U S A. 2007;104:15537–15542. doi: 10.1073/pnas.0707484104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olmos-Serrano JL, Paluszkiewicz SM, Martin BS, Kaufmann WE, Corbin JG, Huntsman MM. Defective GABAergic neurotransmission and pharmacological rescue of neuronal hyperexcitability in the amygdala in a mouse model of fragile X syndrome. J Neurosci. 2010;30:9929–9938. doi: 10.1523/JNEUROSCI.1714-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levenga J, de Vrij FM, Oostra BA, Willemsen R. Potential therapeutic interventions for fragile X syndrome. Trends Mol Med. 2010;16:516–527. doi: 10.1016/j.molmed.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng PY, Sojka D, Klyachko VA. Abnormal presynaptic short-term plasticity and information processing in a mouse model of fragile X syndrome. J Neurosci. 2011;31:10971–10982. doi: 10.1523/JNEUROSCI.2021-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klemmer P, Meredith RM, Holmgren CD, Klychnikov OI, Stahl-Zeng J, Loos M, van der Schors RC, Wortel J, de Wit H, Spijker S, et al. Proteomics, ultrastructure, and physiology of hippocampal synapses in a fragile X syndrome mouse model reveal presynaptic phenotype. J Biol Chem. 2011;286:25495–25504. doi: 10.1074/jbc.M110.210260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown MR, Kronengold J, Gazula VR, Chen Y, Strumbos JG, Sigworth FJ, Navaratnam D, Kaczmarek LK. Fragile X mental retardation protein controls gating of the sodium-activated potassium channel Slack. Nat Neurosci. 2010;13:819–821. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci. 2010;11:339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo W, Allan AM, Zong R, Zhang L, Johnson EB, Schaller EG, Murthy AC, Goggin SL, Eisch AJ, Oostra BA, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17:559–565. doi: 10.1038/nm.2336. • The authors demonstrate that removal of Fmrp from adult neural stem cells will lead to hippocampusdependent learning deficits, which suggests that besides synaptic plasticity, altered adult neurogenesis could also contribute to the cognitive deficits associated with FXS.
- 33.Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, Barkho BZ, et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han XD, Powell BR, Phalin JL, Chehab FF. Mosaicism for a full mutation, premutation, and deletion of the CGG repeats results in 22% FMRP and elevated FMR1 mRNA levels in a high-functioning fragile X male. Am J Med Genet A. 2006;140:1463–1471. doi: 10.1002/ajmg.a.31291. [DOI] [PubMed] [Google Scholar]
- 35.Pietrobono R, Tabolacci E, Zalfa F, Zito I, Terracciano A, Moscato U, Bagni C, Oostra B, Chiurazzi P, Neri G. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum Mol Genet. 2005;14:267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- 36.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Willemsen R, Bontekoe CJ, Severijnen LA, Oostra BA. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110:601–605. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- 38. Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. •• This study presents the first iPS cells derived from FXS fibroblasts, showing that full mutation of FMR1 including the CGG repeat region remains hypermethylated, which differs from findings in FXS hESC.
- 39.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile x syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am J Med Genet A. 2008;146A:1358–1367. doi: 10.1002/ajmg.a.32261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meijer H, de Graaff E, Merckx DM, Jongbloed RJ, de Die-Smulders CE, Engelen JJ, Fryns JP, Curfs PM, Oostra BA. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet. 1994;3:615–620. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- 46.De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 47.Valverde R, Pozdnyakova I, Kajander T, Venkatraman J, Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 48.Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, Zwick ME, Warren ST. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 2010;152A:2512–2520. doi: 10.1002/ajmg.a.33626. • This study reveals several non-conventional mutations in FMR1, such as R138Q, which may cause FXS.
- 50.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Exome Variant Server. Seattle, WA: NHLBI Exome Sequencing Project (ESP); (URL: http://evs.gs.washington.edu/EVS/) [Oct, 2011 accessed] [Google Scholar]