Summary
WNT signaling controls many biological processes including cell differentiation in metazoans. However, how WNT reprograms cell identity is not well understood. We have investigated the potential role of cellular metabolism in WNT-induced osteoblast differentiation. WNT3A induces aerobic glycolysis known as Warburg effect by increasing the level of key glycolytic enzymes. The metabolic regulation requires LRP5 but not β-catenin, and is mediated by mTORC2-AKT signaling downstream of RAC1. Suppressing WNT3A-induced metabolic enzymes impairs osteoblast differentiation in vitro. Deletion of Lrp5 in the mouse, which decreases postnatal bone mass, reduces mTORC2 activity and glycolytic enzymes in bone cells and lowers serum lactate levels. Conversely, mice expressing a mutant Lrp5 that causes high bone mass exhibit increased glycolysis in bone. Thus, WNT-LRP5 signaling promotes bone formation in part through direct reprogramming of glucose metabolism. Moreover, regulation of cellular metabolism may represent a general mechanism contributing to the wide-ranging functions of WNT proteins.
Introduction
WNT signaling controls cell proliferation, fate decision, polarity and migration throughout the evolution of metazoans (Croce and McClay, 2008). WNT proteins, by engaging various receptors and coreceptors at the cell membrane, activate an intracellular signaling network highly dependent on the cellular context, to induce diverse biological responses (van Amerongen and Nusse, 2009). WNT signaling through β-catenin has been most extensively studied. In this mechanism, binding of WNT to a Frizzled (Fz) receptor and a LRP5/6 co-receptor leads to stabilization of β-catenin, which subsequently translocates to the nucleus where it interacts with members of the TCF/LEF transcription factors to activate transcription of downstream target genes (Clevers, 2006). In addition, WNT proteins can activate the Rho family of small GTPases (Habas et al., 2003; Habas et al., 2001; Wu et al., 2008), the Ca2+ pathway (Kuhl et al., 2000) and PKCδ (Kinoshita et al., 2003; Tu et al., 2007). WNT has also been shown to activate mTORC1 (mammalian target of rapamycin complex 1), one of the two complexes formed by mTOR (Inoki et al., 2006). Whereas mTORC1 uniquely contains raptor and is the main target of rapamycin, mTORC2 contains rictor and is relatively insensitive to the drug (Laplante and Sabatini, 2012; Wullschleger et al., 2006). Among many other functions, mTORC1 is best known to control protein synthesis through phosphorylation of the translational regulators 4E-BP1 and S6K1, the latter of which in turn phosphorylates the ribosomal protein S6 and other substrates (Ma and Blenis, 2009). mTORC2 is known to activate Akt through phosphorylation at Ser473, which is necessary for its activity towards some but not all substrates (Guertin et al., 2006; Hresko and Mueckler, 2005; Jacinto et al., 2006; Sarbassov et al., 2005). Other targets of mTORC2 include PKCα, FOXO3 and SGK1 (Garcia-Martinez and Alessi, 2008; Guertin et al., 2006). Earlier studies with siRNA have implicated mTORC2 signaling in regulating the actin cytoskeleton (Jacinto et al., 2004; Sarbassov et al., 2004), but this function was not confirmed in embryonic fibroblasts derived from knockout mice (Guertin et al., 2006). On the other hand, mTORC2 has been shown to regulate whole-body glucose and lipid metabolism through its action in the liver and the adipose tissue (Cybulski et al., 2009; Hagiwara et al., 2012; Kumar et al., 2010; Lamming et al., 2012). Recent studies have shown that the small GTPase RAC1 localizes mTOR to specific membranes and mediates the activation of both mTORC1 and mTORC2 in response to growth factors (Saci et al., 2011). In addition, ribosomes activate mTORC2 through physical association (Zinzalla et al., 2011). Whether or not WNT signaling activates mTORC2 has not been explored.
WNT signaling has emerged as an important mechanism regulating bone formation in mammals (Long, 2012). In the mouse embryo, deletion of β-catenin, or both LRP5 and LRP6, in the skeletogenic progenitors abolishes osteoblast differentiation, indicating that WNT signaling through β-catenin is critical for embryonic osteoblastogenesis (Day et al., 2005; Hill et al., 2005; Hu et al., 2005; Joeng et al., 2011; Rodda and McMahon, 2006). Postanally, loss- and gain-of-function mutations in LRP5 cause low and high bone mass syndromes, respectively, in humans (Boyden et al., 2002; Gong et al., 2001; Little et al., 2002). Moreover, deficiency in SOST, a secreted inhibitor that prevents the binding of WNT to LRP5 or LRP6, results in high bone mass in human patients (Balemans et al., 2001; Balemans et al., 2002). In the mouse, deletion of LRP5 causes osteopenia (Cui et al., 2011; Kato et al., 2002), whereas loss of SOST increases bone mass (Li et al., 2008). The mechanism through which WNT signaling stimulates osteoblast differentiation, however, remains to be elucidated.
Emerging evidence has implicated WNT signaling in the regulation of cellular metabolism. A missense mutation in LRP6 has been linked with abnormal whole-body metabolism in humans (Mani et al., 2007). Genomic polymorphism of TCF7L2, a transcriptional effector of WNT/β-catenin signaling is associated with Type II diabetes (Grant et al., 2006). In cell culture models, prolonged WNT treatment induced mitochondria biogenesis in a β-catenin-dependent manner (Yoon et al., 2010). In the mouse, hepatic manipulation of β-catenin was shown to regulate glucose and glutamine metabolism (Cadoret et al., 2002; Chafey et al., 2009; Liu et al., 2011). However, whether WNT regulates cellular metabolism via β-catenin-independent mechanisms has not been examined. Moreover, it is not known whether metabolic regulation by WNT contributes to cell differentiation.
Here we investigate the potential regulation of glucose metabolism by WNT during osteoblast differentiation. We report that multiple WNT proteins acutely stimulate aerobic glycolysis to control osteoblast differentiation. Distinct from the previous findings, the metabolic regulation described here is independent of β-catenin signaling, but requires mTORC2 activation. Importantly, mouse genetic models demonstrate that WNT-LRP5 signaling concurrently increases glycolysis and bone formation in vivo. Thus, WNT signaling reprograms glucose metabolism through a novel mechanism, and WNT-induced metabolic reprogramming contributes to osteoblast differentiation.
Results
WNT induces aerobic glycolysis independent of β-catenin
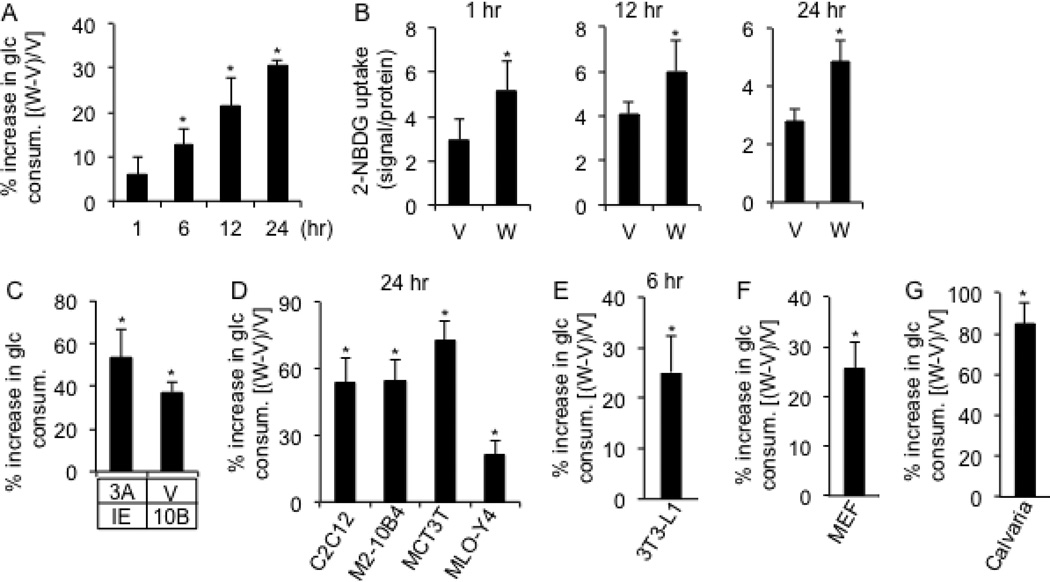
To investigate a potential link between WNT signaling, cellular metabolism and osteoblast differentiation, we examined the effect of WNT proteins on glucose metabolism in ST2 cells, a mouse bone marrow stromal cell line known to undergo osteoblast differentiation in response to WNT (Tu et al., 2007). Because we were interested in direct regulation by WNT instead of adaptive effects secondary to the differentiated state, we focused on the response within the first 24 hours of treatment. Purified WNT3A progressively increased glucose consumption over the control, reaching statistical significance at 6 hrs and exhibiting a marked increase at 12 and 24 hrs (Fig. 1A). Importantly, during this time period, WNT3A did not increase the number of cells, or alter the cell cycle distribution (Fig. S1A, B). Compared to WNT3A, insulin at high concentrations (1 or 2 µg/ml) was less effective in stimulating glucose consumption in ST2 cells (Fig. S1C). Moreover, WNT3A stimulated glucose consumption in the absence of serum even though overall glucose consumption was lower at both basal and stimulated conditions (Fig. S1D). Consistent with the increased glucose consumption, glucose uptake, as assayed by fluorescently labeled 2-deoxyglucose, was enhanced following 1, 12 or 24 hours of WNT3A treatment (Fig. 1B). Thus, WNT3A acutely induces glucose consumption in ST2 cells.
Figure 1. WNT stimulates glucose consumption in cell culture.
(A) Increase in glucose consumption by ST2 cells treated with WNT3A (W) over vehicle (V) for indicated times. (B) Glucose uptake assay following WNT3A (W) or vehicle (V) treatment for indicated times. (C) Induced glucose consumption after 24 hours by purified WNT3A (3A) or virally expressed WNT10B (10B). Increases as % over control cells incubated with vehicle (V) and a GFP-producing virus (IE). (D–G) Induced glucose consumption by WNT3A in indicated cell lines, MEFs and mouse calvarial cells. Increases as % over vehicle-treated cells. * denotes significant difference over respective controls, n=3, p<0.05. Error bars: STDEV.
We then examined whether the regulation was limited to WNT3A and undifferentiated ST2 cells. WNT10B has been shown to induce osteoblast differentiation in ST2 cells (Kang et al., 2007). We found that virally expressed WNT10B increased glucose consumption to a comparable extent as purified WNT3A (Fig. 1C). In contrast, recombinant WNT5A did not have a similar effect (Fig. S1E). BMP2, a known inducer of osteoblast differentiation in ST2 cells, did not stimulate glucose consumption after 24 hours of treatment (Fig. S1F). To determine the effect of WNT3A on differentiating ST2 cells, we stimulated them with an established osteogenic media containing dexamethasone, β-glycerophosphate and ascorbate for up to 15 days, and then assessed their response to WNT3A for 24 hours at each differentiation stage. WNT3A stimulated glucose consumption in ST2 cells at all stages (Fig. S1G, H). We then tested the effect of WNT3A on other cell lines as well as primary cell cultures. We found that WNT3A stimulated glucose consumption in C2C12 (myoblast), M2-10B4 (bone marrow stromal cell), MC3T3 (preosteoblast), MLO-Y4 cells (osteocyte), 3T3-L1 cells (preadipocytes), as well as primary cultures of mouse embryonic fibroblasts (MEFs) and osteoblast-lineage cells from the mouse calvaria (Fig. 1D–G). Thus, multiple osteogenic WNT ligands increase glucose consumption, but all osteogenic signals do not exhibit the same regulation. Furthermore, WNT3A stimulates glucose consumption in a variety of cell types.
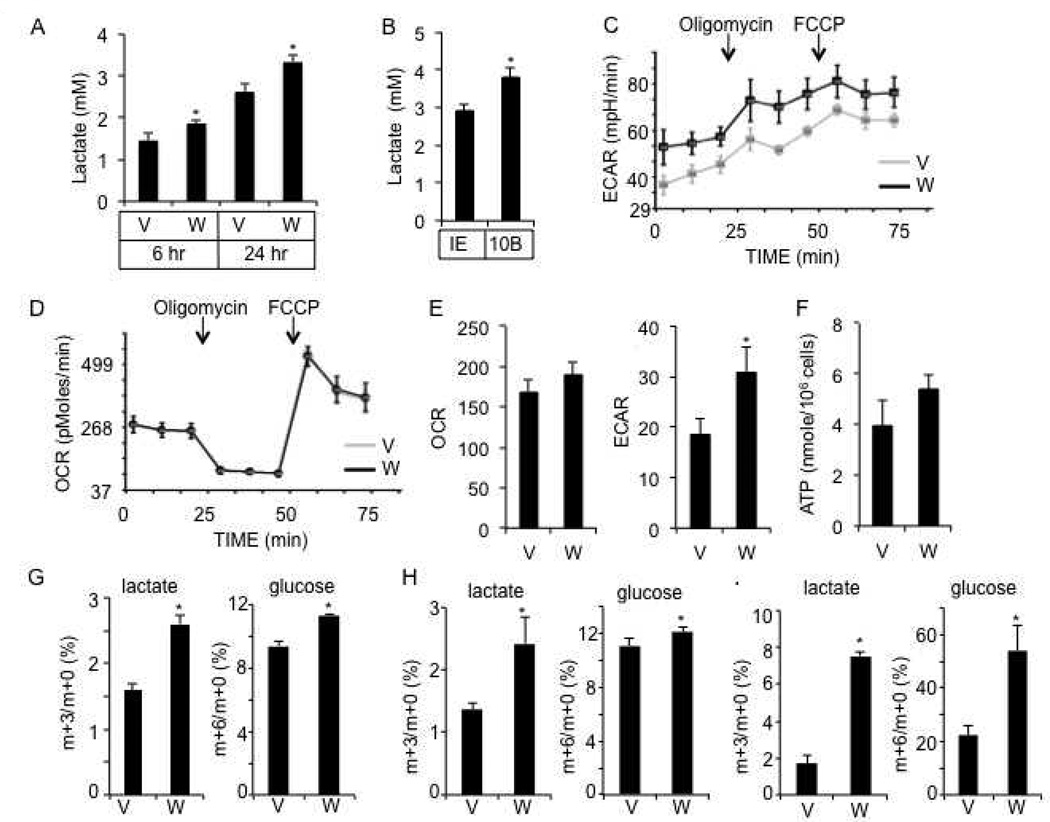
We next examined potential metabolic changes in ST2 cells. WNT3A markedly increased the concentration of lactate in the culture media at both 6 and 24 hours of treatment (Fig. 2A). Similarly, WNT10B increased lactate levels at 24 hours (Fig. 2B). WNT5A, on the other hand, did not affect lactate levels (Fig. S1I), even though it induced phosphorylation of MARKS in ST2 cells (Fig. S1J). We next measured extracellular acidification rate (ECAR) as an indicator for lactate production rate, and the oxygen consumption rate (OCR) with the Extracellular Flux Analyzer after 6 hours of WNT stimulation. WNT3A notably increased ECAR both at the basal state, and during mitochondria stress tests with oligomycin or FCCP, but had no effect on the oxygen consumption rate (OCR) under all conditions (Fig. 2C, D). Similarly, after 24 hours, WNT3A increased ECAR but not OCR (Fig. 2E). Moreover, WNT3A did not alter the intracellular ATP levels after 24 hours of treatment (Fig. 2F). Thus, WNT signaling stimulated lactate production but not oxidative phosphorylation.
Figure 2. WNT3A stimulates aerobic glycolysis in ST2 cells.
(A) Media lactate levels following vehicle (V) or WNT3A (W) treatments. (B) Media lactate levels following viral expression of GFP (IE) or WNT10B for 24 hours. (C–D) Extracellular acidification rate (ECAR) (C) and oxygen consumption rate (OCR) (D) after 6 hours of WNT3A (W) or vehicle (V) treatment. (E) Measurements of ECAR and OCR after 24 hours of treatment. (F) Intracellular ATP levels after 24 hours of treatment. (G–I) Isotopomer enrichment of [U-13C3]-lactate and [U-13C6]-glucose in cell lysates after 6 (G), 12 (H) or 24 (I) hours of WNT3A (W) or vehicle (V) treatment. *: n=3, p<0.05. Error bars: STDEV.
To demonstrate that WNT3A stimulates lactate production directly from glucose, we tracked the fate of glucose through isotopomer distribution analyses of stable isotopically labeled substrates through GC/MS. Briefly, cells were first stimulated with WNT3A for 6, 12 or 24 hours and then incubated with 13C-labeled glucose ([U-13C6]-glucose, m+6 isotopomer tracer) for 1 hour, and its contribution to lactate through glycolysis was determined by measuring the abundance of the labeled [U-13C3]-lactate (m+3 isotopomer) relative to the unlabeled (m+0) pool. After 6, 12 or 24 hours of stimulation, WNT3A markedly increased the relative abundance of m+3 lactate in the cell lysate, indicating a greater portion of lactate derived through glycolysis (Fig. 2G–I). Moreover, at all time points, the WNT3A-induced enrichment of labeled lactate was greater than that of the intracellular labeled glucose (m+6/m+0), indicating that a greater portion of the intracellular glucose underwent glycolysis in response to WNT3A (Fig. 2G–I). Thus, WNT signaling stimulates glycolysis despite the abundance of oxygen, a phenomenon known as the Warburg effect.
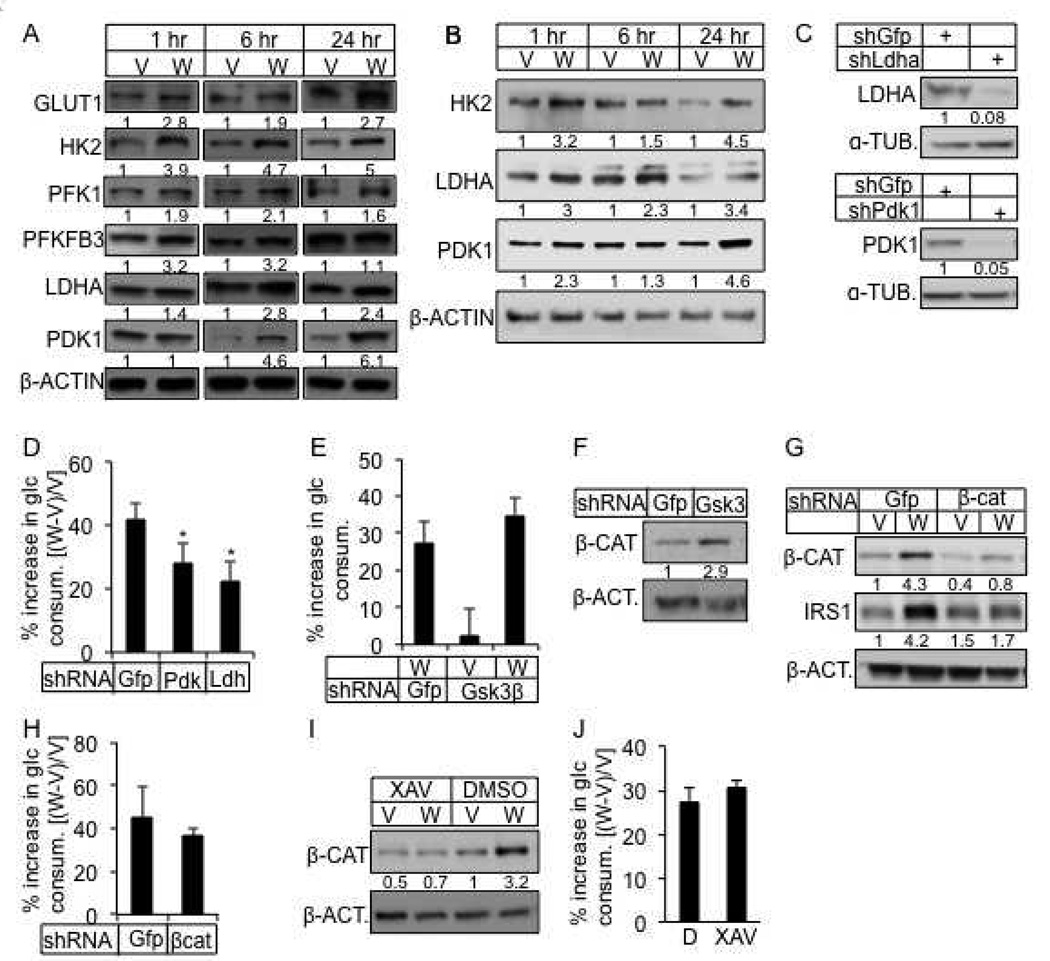
We next examined the molecular basis for the increased glycolysis. GLUT1, a main glucose transporter, and hexokinase II (HK2) that catalyzes the first rate-limiting step of glucose catabolism, were both induced by WNT3A at 1 hour, and remained high at 6, 12 and 24 hours of treatment (Fig. 3A, and data not shown). Phosphofructokinase 1 (PFK1), a key regulatory enzyme for the “committed step” of glycolysis, and 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 3 (PFKFB3), which controls the concentration of fructose 2,6-bisphosphate, a potent allosteric activator of PFK1, were both induced by WNT3A, although PFKFB3 returned to control levels by 24 hours of treatment (Fig. 3A). Finally, lactate dehydrogenase A (LDHA), which catalyzes the conversion of pyruvate to lactate, and pyruvate dehydrogenase kinase 1 (PDK1) that inactivates the pyruvate dehydrogenase complex to suppress pyruvate from entering the TCA cycle, were both induced by 6 hours of WNT3A stimulation, and remained high at the later time points (Fig. 3A). Quantification of Western blots from multiple independent experiments confirmed that the glycolytic regulators was consistently induced by WNT3A after 1, 6 and 24 hours of treatment (Fig. S2A–C). Interestingly, when ST2 cells were first starved for serum, WNT3A induced not only HK2 but also LDHA and PDK1 after 1 hour of stimulation (Fig. 3B). Further experiments revealed that these enzymes were in fact upregulated by WNT3A within 5 minutes of stimulation in the serum-starved cells (Fig. S2D). The quick induction of the enzymes is unlikely due to transcriptional regulation, as their mRNA levels stayed relatively unchanged even after 6 hours of WNT3A treatment (Fig. S2E). After 24 hours, only Ldha and Pdk1 mRNA but not the others were increased over the control (Fig. S2F). Knockdown of either LDHA or PDK1 partially suppressed WNT3A-induced glucose consumption (Fig. 3C, D). Thus, WNT3A acutely increases the protein levels of a number of key glycolytic regulators to stimulate glycolysis.
Figure 3. WNT3A induces glycolysis independent of GSK3β and β-catenin.
(A, B) Representative Western blots of glycolytic regulators in ST2 cells treated with WNT3A or vehicle in the presence (A) or absence of serum (B) for indicated times. Cells in B starved for serum for 12 hours before treatment. Protein abundance normalized to β-ACTIN. (C) Knockdown of LDHA or PDK1. shGfp as negative control. Protein abundance normalized to α-TUBULIN. (D) Effect of LDHA or PDK1 knockdown on WNT3A-induced glucose consumption. (E) Effect of GSK3β knockdown on WNT3A-induced glucose consumption. Increase in glucose consumption as % over cells treated with shGfp and vehicle. (F) Effect of GSK3β knockdown on β-catenin. (G) Knockdown of β-catenin and its effect on IRS1. (H) Effect of β-catenin knockdown on WNT3A-induced glucose consumption. (I, J) Effect of XAV-939 on β-catenin (I) and WNT3A-induced glucose consumption (J). All glucose consumption measured after 24 hours of WNT3A or vehicle treatment. D: DMSO; W: WNT3A; V: vehicle. *: n=3, p<0.05. Error bars: STDEV.
We next investigated the signal transduction mechanism through which WNT3A induces glycolysis. Because WNT3A inhibits GSK3β activity, we first investigated the potential importance of GSK3β inhibition. Inhibition of GSK3β activity by either genetic knockdown or LiCl did not increase glucose consumption by itself, nor did it affect WNT3A-induced glucose consumption (Fig. 3E, and data not shown), even though it increased β-catenin levels as expected (Fig. 3F, and data not shown). Similarly, knockdown of β-catenin did not alter WNT3A-induced glucose consumption, although it suppressed the induction of IRS1, known to be induced transcriptionally by β-catenin (Yoon et al., 2010) (Fig. 3G, H). Knockdown of β-catenin with a second shRNA also did not suppress WNT3A-induced glucose consumption (Fig. S3A). Finally, stabilization of AXIN1/2 with the tankyrase inhibitor XAV939 inhibited β-catenin stabilization by WNT3A, but did not impair the induction of glucose consumption (Huang et al., 2009) (Fig. 3I, J). Thus, regulation of AXIN, GSK3 or β-catenin is not the principle mechanism for WNT3A to induce glycolysis.
WNT-LRP5 signaling activates mTORC2 via RAC1 to induce glycolysis
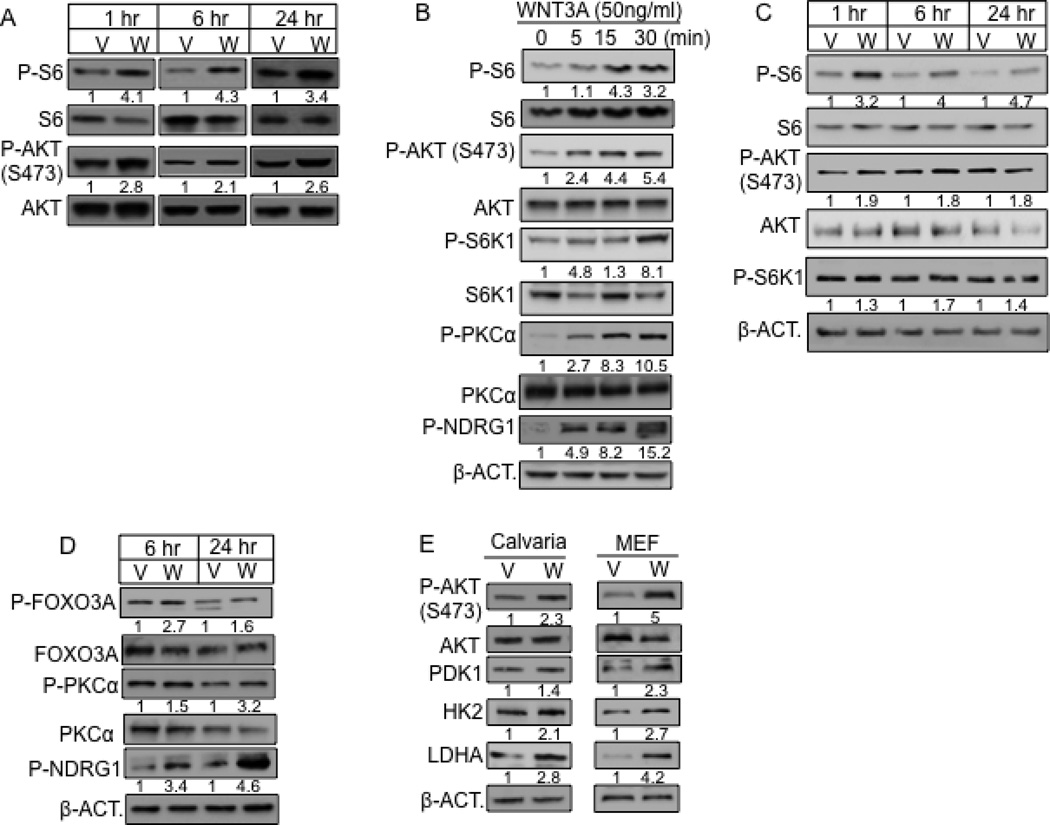
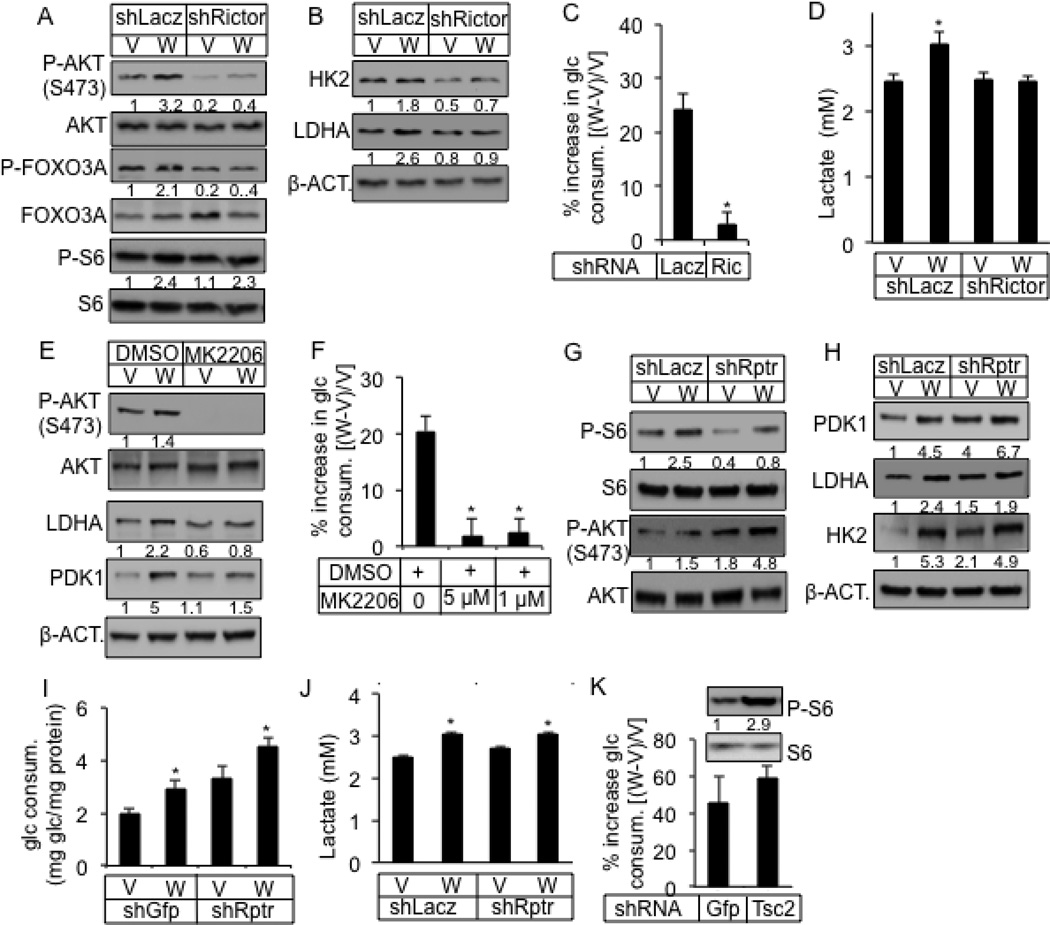
We next investigated the potential role of mTOR signaling in WNT-induced glycolysis. WNT3A acutely activated mTORC1 as indicated by increased phosphorylation of the ribosomal protein S6, which was evident at 1 hour of stimulation and maintained after 24 hours (Fig. 4A). In addition, WNT3A activated mTORC2 as Ser473-phosphorylation of AKT was elevated at these time points (Fig. 4A). Quantification of Western blots from multiple independent experiments confirmed these findings (Fig. S2A–C). We further explored the temporal regulation of mTOR signaling under serum-starved condition. We found that WNT3A activated both mTOR complexes in serum-starved ST2 cells within 5 minutes of stimulation, and throughout 24 hours of treatment (Fig. 4B, C). In contrast, WNT5A, which did not induce glycolysis, did not stimulate mTORC2 (Fig. S1J). Confirming the activation of mTORC2 signaling by WNT3A, phosphorylation of PKCα at S657, FOXO3A at T32 and NDRG1 at T346, all previously shown to require mTORC2 activity (Garcia-Martinez and Alessi, 2008; Guertin et al., 2006), were also induced (Fig. 4B, D). Furthermore, like in ST2 cells, WNT3A activated mTORC2 and induced the glycolytic enzymes in MEFs, and primary osteoblast-lineage cells from the mouse calvaria (Fig. 4E). Knockdown of RICTOR, an mTORC2-specific component, greatly reduced both basal and induced mTORC2 signaling, but not mTORC1 (Fig. 5A, S4A). RICTOR knockdown also abolished the upregulation of HK2 and LDHA by WNT3A, as well as WNT3A-induced glucose consumption, lactate production and media acidification (Fig. 5B–D, S4B). The effects of RICTOR knockdown on WNT3A-induced mTOR signaling, glycolytic enzymes and glucose consumption were all confirmed with a second shRNA (Fig. S4C–E). Similarly, PP242 and Torin 1, inhibitors of both mTORC1 and mTORC2, greatly suppressed WNT3A-induced glucose consumption (Fig. S4F, G). MK-2206, an allosteric AKT inhibitor preventing S473 phosphorylation, completely abolished WNT3A-induced glucose consumption, as well as LDHA and PDK1 upregulation (Fig. 5E, F). Use of a lower concentration (0.1 µM) of MK2206 suppressed WNT3A-induced glucose consumption to a lessor degree but an even lower concentration (0.01 µM) did not have an effect (Fig. S4H). MK-2206 at the effective dosages did not affect cell numbers (Fig. S4I). In contrast to RICTOR, knockdown of RAPTOR, an mTORC1-specific component, suppressed mTORC1, but not mTORC2 activation or the induction of PDK1, LDHA and HK2 by WNT3A (Fig. 5G, H, S4J). Interestingly, RAPTOR knockdown activated basal mTORC2, and increased the basal levels of the glycolytic enzymes without WNT stimulation (Fig. 5G, H), which was likely responsible for the increased basal glucose consumption (Fig. 5I). Nonetheless, RAPTOR knockdown did not prevent further stimulation of glucose consumption or lactate production by WNT3A (Fig. 5I, J). Conversely, activation of mTORC1 by knockdown of TSC2 did not increase glucose consumption by itself, nor did it affect WNT3A-induced glucose consumption (Fig. 5K, data not shown). Thus, WNT3A stimulates glycolysis predominantly through mTORC2 activation.
Figure 4. WNT3A activates mTORC2.
(A–D) Representative Western blots of mTOR targets in ST2 cells treated with WNT3A (W) or vehicle (V) with (A) or without serum (B–D) for indicated time. Cells in B–D serum-starved for 24 (B) or 12 (C, D) hours before treatment. “0 min” samples in B treated with vehicle for 30 minutes. Phospho-protein normalized to respective total protein, except for P-NDRG1 and P-S6K1 in C normalized to β-ACTIN. (E) Effect of WNT3A on mouse calvarial cells or MEFs. Calvarial cells treated with WNT3A or vehicle for 1 hour after 12-hr serum starvation. MEFs treated with WNT3A or vehicle for 6 hours in the presence of serum.
Figure 5. WNT3A induces glucose consumption via mTORC2 activation.
(A–D) Effect of RICTOR knockdown (shRictor) on mTORC1 versus mTORC2 (A), glycolytic enzymes (B), glucose consumption (C) and lactate production (D) after 24 hours of WNT3A (W) or vehicle (V) treatment. (E, F) Effect of AKT inhibitor MK2206 on glycolytic enzymes (E) and glucose consumption (F) after 24 hours of WNT3A (W) or vehicle (V) treatment. (G–J) Effect of RAPTOR knockdown (ShRptr) on mTORC1 versus mTORC2 (G), glycolytic enzymes (H), glucose consumption (I) and lactate production (J) after 24 hours of WNT3A (W) or vehicle (V) treatment. (K) Effect of TSC2 knockdown (ShTSC2) on mTORC1 and glucose consumption after 24 hours of treatment. shGFP or shLacz as negative control. *: n=3, p<0.05. Error bars: STDEV.
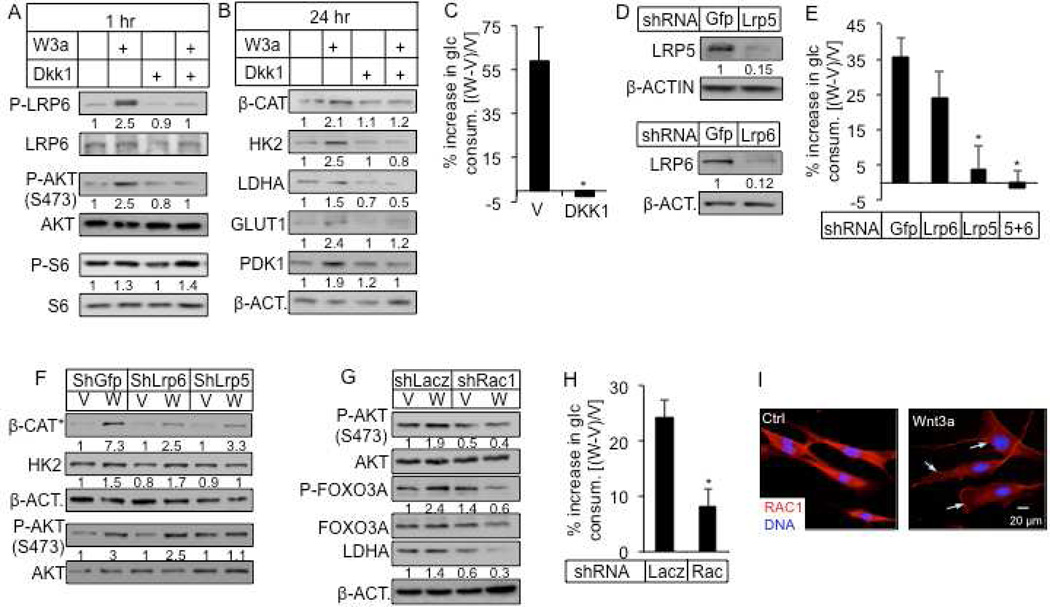
How does WNT signaling activate mTORC2? DKK1, which prevents WNT from binding to LRP5 or LRP6, abolished LRP6 phosphorylation, mTORC2, but not mTORC1 activation by WNT3A at 1 hour (Fig. 6A). Moreover, DKK1 abrogated the induction of β-catenin, HK2, LDHA, GLUT1 and PDK1 after 24 hours of WNT3A treatment (Fig. 6B). Importantly, DKK1 abolished the increase in glucose consumption in response to WNT3A (Fig. 6C). To distinguish the relative contribution of LRP5 versus LRP6 in this regulation, we performed knockdown experiments. Remarkably, knockdown of LRP5 alone essentially recapitulated the effect of either DKK1, or double knockdown of LRP5 and LRP6, in abolishing WNT3A-induced glucose consumption, whereas knockdown of LRP6 had a relatively minor effect (Fig. 6D, E). The differential effect of LRP5 versus LRP6 knockdown was confirmed with a second shRNA for each molecule (Fig. S3A, B). We further examined potential compensation between LRP5 and LRP6 when either molecule was knocked down. Interestingly, LRP6 knockdown doubled Lrp5 mRNA but did not increase its protein level, whereas LRP5 knockdown did not affect either mRNA or protein of LRP6 (data not shown). Confirming the importance of mTORC2, knockdown of LRP5 but not LRP6 suppressed mTORC2 activation, even though either knockdown similarly suppressed the accumulation of the stabilized form of β-catenin in response to WNT3A (Fig. 6F). Moreover, knockdown of LRP5 but not LRP6 eliminated the induction of HK2 by WNT3A (Fig. 6F). Thus, LRP5 appears to be the principle mediator for WNT3A to stimulate glycolysis.
Figure 6. WNT3A stimulates glucose consumption via LRP5 and RAC1.
(A–B) Effects of DKK1 on WNT3A-induced phosphorylation and glycolytic enzymes. P-LRP6, P-AKT, P-S6 normalized to respective total protein. Other proteins normalized to β-ACTIN. (C) Effect of DKK1 on WNT3A-induced glucose consumption. (D–F) Effect of LRP5 or LRP6 knockdown (D) on WNT3A-induced glucose consumption (E), and on signaling events after 1 hour of treatment (F). β-CAT* denotes β-CATENIN unphosphorylated at N-terminus. (G) Effect of RAC1 knockdown on mTORC2 and LDHA induction by 24-hour WNT3A treatment. (H) Effect of RAC1 knockdown on WNT3A-induced glucose consumption. (I) Representative confocal images of RAC1 immunofluorescence. ST2 cells were serum-starved overnight before being treated for 2 hours with vehicle (Ctrl) or Wnt3a. Arrows denote RAC1 membrane localization. All glucose consumption measured after 24 hours of WNT3A or vehicle treatment. V: vehicle; W: WNT3A. *: n=3, p<0.05. Error bars: STDEV.
We next investigated how WNT-LRP5 signaling activates mTORC2. Because we have previously shown that WNT-LRP5/6 signaling activates the Rho-family small GTPase RAC1 (Wu et al., 2008), and others have reported that RAC1 mediates both mTORC1 and mTORC2 activation (Saci et al., 2011), we examined the relevance of RAC1 in WNT3A-induced mTORC2 signaling. Knockdown of RAC1 suppressed the induction of P-AKT, P-FOXO3A and LDHA by WNT3A (Fig. 6G, S5A), as well as WNT3A-induced glucose consumption (Fig. 6H). A second shRNA against RAC1 confirmed its role in mTORC2 activation and glycolysis stimulation by WNT3A (Fig. S5B–D). Because the previous study demonstrated that RAC1 membrane translocation but not its GFP-bound form mediates mTOR activation (Saci et al., 2011), we examined the effect of WNT on RAC1 subcellular localization by confocal microscopy. Indeed, WNT3A induced accumulation of RAC1 at the plasma membrane (Fig. 6I). Specifically, out of 85 cells counted, 23 in the control but 61 in the WNT3A-treated sample showed membrane localization of RAC1. Similarly, virally expressed WNT10B also induced RAC1 accumulation at the plasma membrane (Fig. S5E). Thus, WNT activates mTORC2 through LRP5 and RAC1 to stimulate glycolysis.
Metabolic regulation contributes to WNT-induced osteoblast differentiation
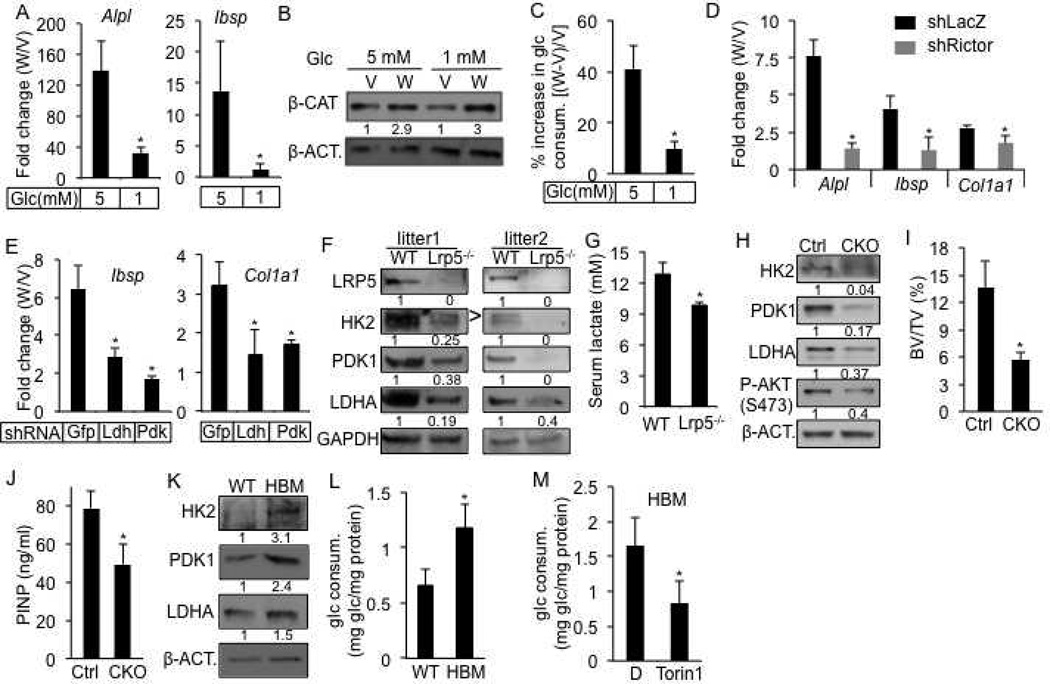
We then tested whether the metabolic regulation plays a role in WNT-induced osteoblast differentiation in ST2 cells. Because WNT induced glucose consumption, we hypothesized that glucose concentrations may impact osteoblast differentiation. Indeed, reducing glucose concentration from the normal 5mM to 1mM greatly impaired osteoblast differentiation in response to WNT3A, as indicated by the decreased expression of Alpl and Ibsp (Fig. 7A), even though β-catenin was similarly stabilized by WNT3A under both conditions (Fig. 7B). The lower glucose concentration did not affect cell numbers, but markedly reduced the extent of induction in glucose consumption by WNT3A (Fig. 7C, S6A). The cells also produced much less lactate with or without WNT3A (Fig. S6B). The inhibitory effect of low glucose was specific to WNT3A, as BMP2 induced osteoblast differentiation similarly with either 5mM or 1mM glucose (Fig. S6C). Next, we tested the role of mTORC2 in WNT3A-induced osteoblast differentiation. RICTOR knockdown suppressed osteoblast differentiation (Fig. 7D). Similarly, Torin 1, which inhibits both mTORC1 and mTORC2, greatly diminished the expression of Alpl and Ibsp in response to WNT3A (Fig. S6D). Finally, we examined the roles of glycolytic enzymes. Knockdown of either LDHA or PDK1, both normally induced by WNT3A, greatly reduced the induction of osteoblast marker genes Col1a1 and Ibsp by WNT3A (Fig. 7E). In addition, either knockdown suppressed the level of Alpl expression in response to WNT3A, whereas PDK1 knockdown also reduced the basal level (Fig. S6E). In contrast, neither knockdown impaired osteoblast differentiation in response to BMP2 (Fig. S6F, and data not shown). Thus, reprogramming of glucose metabolism specifically contributes to WNT-induced osteoblast differentiation.
Figure 7. Metabolic reprogramming by WNT-LRP5 signaling in osteoblast lineage.
(A) Effect of glucose concentration on WNT3A-induced osteoblast differentiation in ST2 cells after 4 days of stimulation. Expression of osteoblast markers determined by qPCR. Fold changes between WNT3A- and vehicle-treated cells calculated after normalization to 18S RNA. (B–C) Effect of glucose concentration on β-CATENIN stabilization (B) and WNT3A-induced glucose consumption (C) after 24 hours of treatment. (D–E) Effect of RICTOR (D), LDHA or PDK1 (E) knockdown on WNT3A-induced osteoblast differentiation in ST2 cells after 4 days of treatment. Expression of osteoblast markers determined by qPCR. Fold changes between WNT3A- and vehicle-treated cells calculated after normalization to 18S RNA. (F) Western blots of glycolytic enzymes in bone protein extracts from Lrp5−/− versus wild-type littermates. > denotes correct band for HK2. (G) Serum lactate levels from Lrp5−/− versus wild-type littermates. (H) Western blots with bone protein extracts from Lrp5CKO versus Osx-Cre littermates (Ctrl). (I, J) Bone phenotype analyses of Lrp5CKO versus Osx-Cre littermates (Ctrl) by µCT (I) and serum P1NP assays (J). (K) Western blots of glycolytic enzymes in bone protein extracts from homozygous LRP5 HBM mice versus wild-type littermates. (L) Glucose consumption after 48 hours of culture by BMSC from LRP5 HBM (heterozygous or homozygous) mice versus wild-type littermates. (M) Effect of Torin1 on 48-hour glucose consumption by BMSC from homozygous LRP5 HBM mice. D: DMSO. *: n≥3, p<0.05. Error bars: STDEV.
WNT-LRP5 signaling increases glycolysis in vivo
Lastly, we tested whether WNT-LRP5 signaling reprograms glucose metabolism in vivo. We first examined the Lrp5−/− mice that are known to be defective in bone formation (Holmen et al., 2004; Kato et al., 2002). These mutants at six weeks of age contained a much lower level of HK2, LDHA and PDK1 in their bones when compared to the littermate controls (Fig. 7F). Moreover, the serum lactate levels in one-month-old Lrp5−/− mice were significantly lower than those in their littermate controls (Fig. 7G). To rule out the possibility that the metabolic changes in bone were secondary to the effects on other tissues, we generated Osx-Cre;Lrp5f/f mice (CKO) containing bone-specific deletion of Lrp5. We found a notable decrease in the levels of HK2, PDK1 and LDHA in the bones of CKO mice at ten weeks of age, coupled with reduced mTORC2 activity (Fig. 7H). Analyses with µCT techniques revealed obvious osteopenia in the CKO mice when compared to the littermate control (Fig. 7I, S7A, B). Consistent with reduced bone formation, the serum P1NP (procollagen type I N-terminal propeptide) level was lower in the CKO mice than the control (Fig. 7J). To further establish the link between LRP5 and metabolic regulation in bone, we studied the high-bone-mass (HBM) mice harboring the point mutation of A214V in LRP5 (Cui et al., 2011). We confirmed by µCT analyses that mice either heterozygous or homozygous for the mutant allele exhibited markedly higher bone mass at two months of age (data not shown). Importantly, bones from the HBM mice expressed higher levels of HK2, PDK1 and LDHA (Fig. 7K). The bone marrow stromal cells isolated from the HBM mice consumed more glucose than their control counterpart when cultured in vitro, and the increase in glucose consumption was suppressed by the mTOR inhibitor Torin 1 (Fig. 7L, M). Thus, LRP5 signaling modulates glucose metabolism in bone cells in the mouse.
Discussion
We have provided evidence that WNT signaling directly regulates glucose metabolism independent of β-catenin signaling. Specifically, WNT3A signals through LRP5 and RAC1 to activate mTORC2 and AKT, resulting in upregulation of key glycolytic enzymes. Functionally, the metabolic regulation contributes to WNT-induced osteoblast differentiation in vitro, and correlates with the bone forming activity of LRP5 signaling in vivo. This study not only uncovers a novel mechanism through which WNT signaling regulates cellular metabolism, but also demonstrates that metabolic regulation contributes to WNT-induced cell differentiation.
The present study further expands the repertoire of signaling cascades activated by WNT3A. In addition to β-catenin stabilization, we have previously shown that WNT3A signals through heterotrimeric G-proteins to activate both PLCβ-PKCδ and PI3K-RAC1 signaling in ST2 cells (Tu et al., 2007; Wu et al., 2008). Here we show that WNT3A activates mTORC2 downstream of RAC1. Activation of the different pathways by WNT3A may require distinct cell-surface receptor complexes, as β-catenin stabilization and RAC1 activation are inhibited by DKK1, but PKCδ activation is not. Taken together, these studies support the notion that WNT proteins activate multiple intracellular signaling cascades highly dependent on the cellular context, and do not possess intrinsic “canonical” or “noncanonical” signaling properties (van Amerongen et al., 2008).
The mechanism through which mTORC2 induces glycolytic enzymes remains to be further elucidated. The fact that the induction occurs abruptly following WNT3A treatment with no change in mRNA levels indicates a transcription-independent mechanism at work. We show that AKT, a direct target of mTORC2, is critical for the induction of glycolytic enzymes and glycolysis in response to WNT3A. Future studies are necessary to determine whether and how mTORC2-AKT signaling affects protein stability or translation of the glycolytic enzymes.
Our data identifies LRP5 as a major co-receptor for WNT3A to induce glycolysis. Although RNA-seq experiments revealed that ST2 cells expressed three times as much Lrp6 mRNA as Lrp5 (data not shown), knockdown of LRP6 did not have a major effect on WNT3A-induced glucose consumption. In light of the finding that LRP6 has a more potent function in mediating β-catenin signaling (MacDonald et al., 2011), the two homologous co-receptors may have evolved to preferentially execute different WNT signaling cascades. However, because we have analyzed glucose metabolism only within the first 24 hours of WNT3A treatment, LRP6 may regulate cell metabolism at later time points through β-catenin signaling. Indeed, we found that both Ldha and Pdk1 mRNA were induced by WNT3A at 24 hours (Fig. S2F), and that knockdown of β-catenin partially suppressed the induction of LDHA and PDK1 proteins at this time point (data not shown). Furthermore, induction of IRS1 by β-catenin signaling may contribute to glucose metabolism in response to insulin (Yoon et al., 2010) (this study). Thus, WNT signaling may control glucose metabolism both through the fast-acting, β-catenin-independent and the slow-acting, β-catenin–dependent mechanisms, which may be preferentially mediated by LRP5 and LRP6, respectively. This conclusion is in agreement with the previous reports that implicated LRP5, LRP6 and β-catenin in the regulation of whole-body metabolism (Fujino et al., 2003; Liu et al., 2011; Mani et al., 2007).
The finding that LRP5 mediates WNT-induced metabolic reprogramming may have important implications for understanding the pathogenesis of bone disorders caused by LRP5 mutations. Although the role of LRP5 in regulating both osteoblast number and function in postnatal mice is well established, the mechanism underlying LRP5 function has been a matter of debate (Cui et al., 2011; Yadav et al., 2008). We have recently reported that β-catenin is necessary for normal osteoblast life span and activity in postnatal mice, lending support to the notion that β-catenin may mediate some aspects of LRP5 signaling in postnatal bones (Chen and Long, 2012). The current study provides an additional mechanism through which LRP5 may regulate osteoblast differentiation and function independent of β-catenin. Future studies are necessary to determine the relative contributions of the different mechanisms to LRP5 function in vivo.
Beyond cells of the osteoblast lineage, WNT signaling may be a general paracrine mechanism that modulates cellular metabolism in the body. Besides cell differentiation, changes in cellular metabolism are likely to influence other aspects of cell physiology, as well as whole body metabolism. Furthermore, because insulin is an endocrine signal that controls glucose metabolism, we expect that WNT may intersect with insulin signaling to coordinate cellular metabolism. Indeed, a recent report has shown physical interaction between LRP5 and insulin receptor and interdependence between WNT and insulin signaling (Palsgaard et al., 2012). We observed greater potency for WNT3A than insulin in inducing glucose consumption in ST2 cells, and that WNT3A exerted a similar effect in the absence of serum (hence no insulin). Thus, the WNT can operate independent of insulin signaling in our setting. Future studies are necessary to elucidate the interaction between WNT and insulin signaling in regulating glucose metabolism.
Warburg effect originally describes the phenomenon that cancer cells often utilize glucose through glycolysis over oxidative phosphorylation despite the abundance of oxygen (Warburg, 1956). The phenomenon is now known to be common to proliferating cells in culture. The reason for Warburg effect continues to be an area of active research, but it has been proposed that glycolysis produces the necessary intermediate metabolites for fueling cell proliferation (Vander Heiden et al., 2009). In our experiments, WNT induced cell differentiation without an obvious effect on proliferation. How increased glycolysis promotes differentiation remained to be investigated in the future, but it may alter the levels of key intermediate metabolites that regulate gene expression. Overall, our study has identified metabolic regulation as a new mechanism for WNT proteins to induce cell differentiation.
Experimental procedures
Mouse strains
Osx-Cre, Lrp5f/f, Lrp5−/−, Lrp5HBM mice are as previously described (Cui et al., 2011; Holmen et al., 2004; Joeng et al., 2011; Rodda and McMahon, 2006). The Animal Studies Committee at Washington University has reviewed and approved all mouse procedures used in this study.
Antibodies
Antibodies for p-Lrp6 (cat#2568), p-Akt(S473)(cat#9271), Akt (cat#9272), p-S6 (cat#2215), S6 (cat#2217), Raptor (cat#2280), Rictor (cat#2140), Lrp5 (cat#5731), β-actin (cat#4970), FoxO3a (cat#2497S), p-FoxO3a–T32 (cat#9464S), p-NDRG1-Thr346 (cat#3217S), P-S6K (cat#9205) are from Cell Signaling Technologies. Hk2 (sc-6521), Ldha (sc-27230), Pfk1 (sc-31712), Lrp6 (sc-25317), α-tubulin (sc-8035), p-PKCα-Ser 657 (sc-12356) total PKCα (sc-208) antibodies are from Santa Cruz Biotechnology. Pfkfp3 (ab96699) antibody is from Abcam. Antibody for unphosphorylated β-catenin (05–665) is from Millipore. Pdk1 (KAP-pk112) antibody is from Assay Designs. RAC1 antibody (610650) is from BD Biosciences. Glut1 polyclonal antibody F350 is as previously described and kindly provided by Dr. Michael Mueckler (Washington University School of Medicine) (Haney et al., 1991). HRP-conjugated anti-rabbit secondary antibody is from GE healthcare (NA934V), HRP-conjugated anti-mouse (sc-2005) and anti-goat (sc-2352) secondary antibodies are from Santa Cruz Biotechnology.
Western blot
Protein extracts from cells or bone were prepared in RIPA buffer containing phosphatase and proteinase inhibitors. Membranes were imaged with Molecular Imager ChemiDoc™ XRS+ System (Bio-Rad). Quantification of western blots was performed with ImageLab software or Photoshop CS3. Detailed procedure is provided in Supplemental Materials and Methods.
Quantitative PCR
RNA was isolated from whole cells with QIAGEN RNeasy kit (#74104) and was transcribed into cDNA using iScript cDNA synthesis kit (Bio-rad). Fast-start SYBR green (Bio-rad) and 0.05 µM primers were used in each reaction. 18S RNA was used for normalization. The primer sequences and additional information are provided in Supplemental Materials and Methods.
Cell culture
For routine cultures, ST2 cells were grown in α-MEM (Gibco, cat#12561), MC3T3 subclone 4 cells were grown in α-MEM (GIBCO cat# A1049001), HEK293T and 3T3-L1 cells were grown in DMEM (Gibco cat#11965), M2-10B4 cells (ATCC CRL-1972) were grown in RPMI (Gibco, cat#11875), all supplemented with 10% heat inactivated FBS (Gibco) and Pen Strep (Gibco, cat#14140). MLO-Y4 cells were grown on pre-collagen coated plates in α-MEM containing nucleosides (Gibco, cat#12571-063) supplemented with heat inactivated FBS (2.5 %), calf serum (2.5%) and Pen Strep.
For all experiments unless indicated otherwise, cells were cultured in glucose- and glutamine-free α-MEM (Gibco, custom-made from cat#12561) containing 10% FBS and Pen Strep, and freshly supplemented with 5.5 mM glucose plus 2mM glutamine. Recombinant mouse Wnt3a (R&D systems) was used at 50 ng/ml unless indicated otherwise. As a vehicle control for Wnt3a, PBS with 0.5% CHAPS and 0.1 mM EDTA was used. Recombinant human BMP2 (R&D systems) was used at 300 ng/ml. Recombinant mouse Dkk1 (R&D systems) was used at 500 ng/ml and cells were pretreated with Dkk1 for 30 minutes before the addition of Wnt3a. Torin1 (Tocris biosciences), PP242 (sigma), rapamycin (sigma), all dissolved in DMSO was used at 20, 10 and 100 nM, respectively. Insulin (sigma) was used at 1 or 2 µg/ml.
Bone marrow stromal cells (BMSCs), calvarial cells and mouse embryonic fibroblasts (MEFs) were isolated from mouse adult long bones, newborn calvaria and E13.5 embryos, respectively, according to established protocols. Detailed methods are provided in Supplemental Materials and Methods.
Glucose consumption and uptake assays, lactate and ATP measurements
For glucose consumption measurements, aliquots of the media and glucose standards were assayed with Glucose (HK) Assay Kit (Sigma cat# GAHK20) and read at 340 OD using a plate reader (BioTek model SAMLFTA, Gen5 software). For glucose uptake assays, cells were incubated with 100 µM 2-NBDG for 30 minutes, and then prepared for fluorescence reading following the manufacturer’s instructions (Glucose Uptake Cell-based Assay Kit, Cayman Chemical). Fluorescence intensity measured at 485/535 nm (excitation/emission) using a plate reader (BioTek model SAMLFTA, Gen5 software) was normalized to the protein content in each well. For lactate measurements, L-lactate assay kit from Eton biosciences (cat# 1200011002) was used. To measure lactate levels in the serum, mice were fasted for 8 hours before blood was collected from the peri-orbital venous sinus under anesthesia. Intracellular ATP was measured based on a method previously described (Chi et al., 2002). Additional information is provided in Supplemental Materials and Methods.
OCR and ECAR measurements with Seahorse Cellular Flux assays
ST2 cells were plated in XF96 plates at 20,000 cells/well after coating the plates with cell-tak (BD Biosciences). The next day, the cells were treated with 100 ng/ml Wnt3a for 6 hours, then switched to XF Assay Medium Modified DMEM (Seahorse cat#101022-100) supplemented with 5.5 mM glucose, and further incubated in CO2-free incubator for 1 hour. Oligomycin and FCCP (Seahorse Stress Kit) were prepared in XF assay medium with final concentration of 5 µM and 1 µM, respectively and were injected during the measurements. At the end of the assays, protein concentrations were measured for normalization.
shRNA knockdown and retroviral infection
Lentiviral shRNA targeting vectors were purchased from Genome Center at Washington University. Targeted sequences and virus production procedure are provided in Supplemental Materials and Methods. Retroviruses expressing WNT10B or GFP were generated according to a procedure as previously described (Hu et al., 2005).
Analyses of postnatal mouse bones
Micro-CT analyses were performed with Scanco µCT 40 (Scanco Medical AG) according to ASBMR guidelines (Bouxsein et al., 2010). Quantification of the trabecular bone in the proximal tibia was performed with 100 µCT slices (1.6 mm total) immediately below the growth plate. To measure P1NP in the serum, serum was collected from mice after 6 hours of fasting, and analyzed with Rat/Mouse P1NP EIA kit (Immunodiagnostic Systems, Ltd., cat# DS-AC33F1).
Bone protein extracts were prepared from femurs and tibias of postnatal mice with RIPA buffer. After removal of both epiphyses of each bone, bone marrow cells were removed by brief centrifugation. The remaining bone shafts were rinsed twice in the cold PBS, flash-frozen in liquid nitrogen, and then manually ground into a fine power with a mortar/pestle. The bone power was incubated on ice for 30 minutes, with 200µl RIPA buffer containing phosphatase inhibitors (Roche, Cat#04906845001) and proteinase inhibitors (Roche, Cat#11 836 170 001). The protein extracts were then collected after centrifugation for 10 minutes.
GC/MS analyses
At the end of the Wnt3a treatment, [U-13C6] glucose was added to the medium at a final concentration of 0.55 mM and incubated for 1 hour. At the end of the [U-13C6] incubation, an aliquot of medium from each sample was collected for analyses. The cells were then washed with cold PBS and extracted three times with −80 C methanol on dry ice. Extracts were dried in a centrifugal concentrator (Savant SpeedVac, Thermo Scientific, Millford, MA) and derivatized at 70°C, 30 min with either 1:1 acetonitrile:N-methyl-N(tert-butyldimethylsilyl)trifluoroacetamide (MTBSTFA, to make tert-butyldimethylsilyl derivatives (tBDMS) of lactate) or 10% heptafluorobutyric anhydride in ethyl acetate (to make heptafluorobutyryl derivative (HFB) of glucose).
GC-MS/EI analyses were performed with Hewlett-Packard 6890 series gas chromatograph interfaced to an Agilent 5973N mass spectrometer. Helium was the carrier gas at constant flow rate of 1.0 mL/min. The injector and the transfer line temperatures were set at 250 °C and 280 °C respectively. GC analysis was performed with a DB-5MS column (30 m × 0.25 mm × 0.25 mm; Agilent). The initial temp of the oven was 90 °C held for 0 min and then ramped at 20°C /min to 230 °C, and next ramped at 70°C /min to 275 °C held for 3.5 min for a total run of 11 min. 2.0 µL of sample in heptane was injected with a 7683 autosampler (split mode; split ratio of 5:1). The electron energy was set at 70 eV and the ion source temperature kept at 230 °C.
Isotopomer distributions were measured by electron impact ionization for tBDMS derivative of lactate (m/z 261–264) and for the HFB derivative of glucose (m/z 519–525). All isotopomer distributions were corrected for natural abundance and for spectral overlap (Wolfe and Chinkes, 2004).
Statistical analyses
All quantitative data are presented as mean ± STDEV with a minimum of three independent samples. Statistical significance is determined by Student’s t-test.
Supplementary Material
Highlights.
WNT3A stimulates aerobic glycolysis independent of GSK3 and β-catenin
WNT3A stimulates aerobic glycolysis through LRP5-RAC1-mTORC2-AKT signaling
Metabolic reprogramming is necessary for WNT3A-induced osteoblast differentiation
LRP5 promote glycolysis and bone formation in vivo
Acknowledgements
This work is supported by NIH grants AR060456, AR055923 (FL) and NIH P30 DK056341 (Washington University Nutrition Obesity Research Center). We thank Dr. Michael Mueckler (Washington University School of Medicine) for providing the GLUT1 antibody, Dr. Lynda Bonewald (University of Missouri-Kansas City) for MLO-Y4 cells, Dr. Roberto Civitelli (Washington University School of Medicine) for MC3T3 cells and Dr. Clay Semenkovich (Washington University School of Medicine) for 3T3-L1 cells. We thank Maggie Chi and Dr. Kelle Moley (Washington University School of Medicine) for help with ATP measurements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum Mol Genet. 2001;10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39:91–97. doi: 10.1136/jmg.39.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Cadoret A, Ovejero C, Terris B, Souil E, Levy L, Lamers WH, Kitajewski J, Kahn A, Perret C. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- Chafey P, Finzi L, Boisgard R, Cauzac M, Clary G, Broussard C, Pegorier JP, Guillonneau F, Mayeux P, Camoin L, et al. Proteomic analysis of beta-catenin activation in mouse liver by DIGE analysis identifies glucose metabolism as a new target of the Wnt pathway. Proteomics. 2009;9:3889–3900. doi: 10.1002/pmic.200800609. [DOI] [PubMed] [Google Scholar]
- Chen J, Long F. beta-catenin promotes bone formation and suppresses bone resorption in postnatal growing mice. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2012 doi: 10.1002/jbmr.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi MM, Hoehn A, Moley KH. Metabolic changes in the glucose-induced apoptotic blastocyst suggest alterations in mitochondrial physiology. American journal of physiology Endocrinology and metabolism. 2002;283:E226–E232. doi: 10.1152/ajpendo.00046.2002. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Croce JC, McClay DR. Evolution of the Wnt pathways. Methods in molecular biology. 2008;469:3–18. doi: 10.1007/978-1-60327-469-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Niziolek PJ, Macdonald BT, Zylstra CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon RA, et al. Lrp5 functions in bone to regulate bone mass. Nature medicine. 2011;17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9902–9907. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Fujino T, Asaba H, Kang MJ, Ikeda Y, Sone H, Takada S, Kim DH, Ioka RX, Ono M, Tomoyori H, et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) The Biochemical journal. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature genetics. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Developmental cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Habas R, Dawid IB, He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell metabolism. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Haney PM, Slot JW, Piper RC, James DE, Mueckler M. Intracellular targeting of the insulin-regulatable glucose transporter (GLUT4) is isoform specific and independent of cell type. The Journal of cell biology. 1991;114:689–699. doi: 10.1083/jcb.114.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF, Glatt V, Bouxsein ML, Ai M, Warman ML, et al. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2004;19:2033–2040. doi: 10.1359/JBMR.040907. [DOI] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. The Journal of biological chemistry. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature cell biology. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Joeng KS, Long F. The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development. 2009;136:4177–4185. doi: 10.1242/dev.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeng KS, Schumacher CA, Zylstra-Diegel CR, Long F, Williams BO. Lrp5 and Lrp6 redundantly control skeletal development in the mouse embryo. Developmental biology. 2011;359:222–229. doi: 10.1016/j.ydbio.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. The Journal of biological chemistry. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Iioka H, Miyakoshi A, Ueno N. PKC delta is essential for Dishevelled function in a noncanonical Wnt pathway that regulates Xenopus convergent extension movements. Genes Dev. 2003;17:1663–1676. doi: 10.1101/gad.1101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl M, Sheldahl LC, Park M, Miller JR, Moon RT. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16:279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC, Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59:1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ominsky MS, Niu QT, Sun N, Daugherty B, D’Agostin D, Kurahara C, Gao Y, Cao J, Gong J, et al. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2008;23:860–869. doi: 10.1359/jbmr.080216. [DOI] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Wu JJ, Rovira II, Liu J, Gavrilova O, Lu T, Bao J, Han D, Sack MN. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4:ra6. doi: 10.1126/scisignal.2001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, et al. Building strong bones: molecular regulation of the osteoblast lineage. Nature reviews. Molecular cell biology. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews. Molecular cell biology. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Semenov MV, Huang H, He X. Dissecting molecular differences between Wnt coreceptors LRP5 and LRP6. PloS one. 2011;6:e23537. doi: 10.1371/journal.pone.0023537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsgaard J, Emanuelli B, Winnay JN, Sumara G, Karsenty G, Kahn CR. Cross-talk between Insulin and Wnt Signaling in Preadipocytes: ROLE OF WNT CORECEPTOR LOW DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN-5 (LRP5) The Journal of biological chemistry. 2012;287:12016–12026. doi: 10.1074/jbc.M111.337048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Molecular cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology : CB. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Tu X, Joeng KS, Nakayama KI, Nakayama K, Rajagopal J, Carroll TJ, McMahon AP, Long F. Noncanonical Wnt Signaling through G Protein-Linked PKCdelta Activation Promotes Bone Formation. Dev Cell. 2007;12:113–127. doi: 10.1016/j.devcel.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R. Alternative wnt signaling is initiated by distinct receptors. Sci Signal. 2008;1:re9. doi: 10.1126/scisignal.135re9. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wolfe RR, Chinkes DL. Determination of isotope enrichment In Isotope Tracers in Metabolic Research: Principles and Practice of Kinetics Analysis (John Wiley & Sons) 2004 [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yadav VK, Ryu JH, Suda N, Tanaka KF, Gingrich JA, Schutz G, Glorieux FH, Chiang CY, Zajac JD, Insogna KL, et al. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell. 2008;135:825–837. doi: 10.1016/j.cell.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes & development. 2010;24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.