Abstract
The high level of 5-hydroxymethylcytosine (5hmC) present in neuronal genomes suggests that mechanisms interpreting 5hmC in the central nervous system (CNS) may differ from those present in embryonic stem cells. Here we present quantitative, genome-wide analysis of 5hmC, 5-methylcytosine (5mC) and gene expression in differentiated CNS cell types in vivo. We report that 5hmC is enriched in active genes, and that surprisingly strong depletion of 5mC is observed over these regions. The contribution of these epigenetic marks to gene expression depends critically on cell type. We identify methyl-CpG binding protein 2 (MeCP2) as the major 5hmC binding protein in the brain, and demonstrate that MeCP2 binds 5hmC and 5mC containing DNA with similar high affinities. The Rett Syndromecausing mutation R133C preferentially inhibits 5hmC binding. These findings support a model in which 5hmC and MeCP2 constitute a cell specific epigenetic mechanism for regulation of chromatin structure and gene expression.
Introduction
The appearance of the nucleus and the architecture of chromatin vary substantially in terminally differentiated CNS cell types (Palay, Chan-Palay, 1974). The recent discovery of 5hmC in the mammalian genome and the demonstration that it is approximately ten fold more abundant in neurons than in some peripheral tissues or embryonic stem (ES) cells (Kriaucionis and Heintz, 2009; Munzel et al., 2010; Szulwach et al., 2011) suggests that 5hmC is a stable epigenetic mark that engages cell specific mechanisms to carry out its functions in the brain. Consistent with this view, recent studies mapping the genomic distribution of 5hmC in the hippocampus and cerebellum have established that the distribution of 5hmC varies between brain areas, and that the location of 5hmC in the genome differs significantly between the brain and ES cells (Szulwach et al., 2011). For example, 5hmC is present preferentially in specific classes of promoter and enhancer elements that regulate the pluripotent state in ES cells (Pastor et al., 2011; Yu et al., 2012; Booth at al., 2012), whereas in the brain it is enriched in gene bodies and depleted from TSS (Szulwach et al., 2011). To understand the importance of these distinctions, and to determine whether the mechanisms that decode cytosine methylation status are cell and tissue specific, we have pursued two strategies in parallel: quantitative, genome wide, cell specific measurements of gene expression and genomic cytosine modification in defined CNS cell types; and biochemical analysis of proteins involved in 5hmC binding in the nervous system.
We have chosen for these studies two classically defined neuronal cell types, Purkinje cells (PC) and granule cells (GC), and the terminally differentiated and specialized Bergmann glial (BG) cell population that is co-resident with them in the cerebellum. PCs are among the largest cells in the brain. PC nuclei are large and pale, and the majority of heterochromatin detectable at the ultra-structural level is present surrounding the large, centrally located nucleolus (Palay, Chan-Palay, 1974). GCs are the smallest and most numerous neurons of the cerebellum, present at several hundred times the abundance of PCs (Palay, Chan-Palay, 1974; Lange, 1975). GC nuclei are small, compact, and contain large blocks of condensed heterochromatin localized in nuclear periphery. BGs, originally referred to as Golgi epithelial cells (Palay, Chan-Palay, 1974) have a radial morphology that distinguishes them from the much more abundant, typical protoplasmic astrocytes. BG nuclei are intermediate in size and structure between PC and GC nuclei, and contain a few small, dense clumps of heterochromatin. The distinctive structures of these three neural cell types suggests that their analysis can provide important insights into cell specific relationships between genomic cytosine modification and nuclear function.
We demonstrate here that the relationship between the genomic distribution of 5hmC, 5mC and gene expression is cell specific. We identify MeCP2 as the major 5hmC binding protein in the brain, and show that the R133C mutation present in some RTT patients preferentially impacts 5hmC binding. Loss of MeCP2 does not alter the genomic distribution of 5hmC, although the preferential nuclease sensitivity of 5hmC containing chromatin is no longer present in the absence of MeCP2. Our data support a model in which MeCP2 binding to 5hmC can facilitate transcription in neural cell types while at the same time acting in repression when bound to 5mC containing DNA. Deciphering the relationships between these functions and how they are used in specific cell types will be essential for understanding the pathophysiology of RTT.
Results
TRAP-Seq profiling of gene expression in PCs, GCs and BG
We have previously employed Translating Ribosome Affinity Purification (TRAP) method to determine that each of these cell types expresses a unique complement of cell specific gene products (Doyle et al., 2008; Heiman et al., 2008). Although these microarray data might suffice for comparative analysis of gene expression and the cytosine modification status, we sought to improve our analysis by collecting gene expression data from these cell types using the more comprehensive and quantitative high throughput sequencing (HTS) of TRAP isolated mRNA (referred to hereafter as TRAP-Seq). Datasets for PCs, GCs and BGs and their input tissue (whole cerebellum) were generated in four biological replicates for each cell type, resulting in an average 76.5x10−6 reads per sample (Fig. S1A). In total we obtained ~1.36x10−9 mapped reads, enabling deep analysis of the expressed genes in each of these neural cell types (Fig. S1). Principal component and hierarchical clustering algorithms demonstrated tight relation of TRAP-Seq data between the replicas and gender (Fig. S1C, D). The quality of the TRAP-Seq datasets collected from PCs, GCs and BG is further supported by the fact that the correlation coefficients between datasets obtained from a single cell type were between 0.94–0.99 (Fig. S1B).
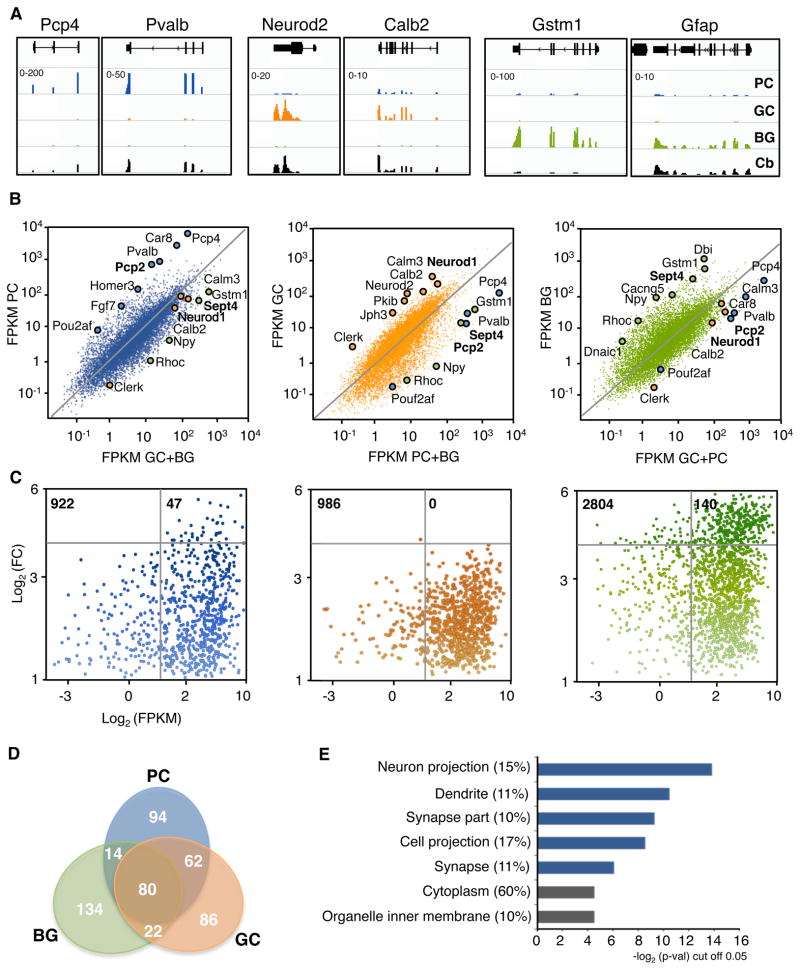
Inspection of TRAP-Seq data from individual, well known genes illustrates the importance of cell type specific analysis for the evaluation of gene expression (Table S1A). Alignment of RNA-Seq data collected from the whole cerebellum (Fig. 1A, bottom, black traces) demonstrates that each of the six genes represented is expressed at detectable levels in the cerebellum, and that differences in their expression levels are evident even by visual inspection of the aligned data. However, it is apparent from the top three traces that display the levels of expression of these genes in PCs (blue), GCs (orange) and BGs (green) that cell-specific expression levels cannot be evaluated in whole tissue RNA-Seq data. For example, both Pcp4 and Gstm1 mRNAs are present at quite low levels in cerebellar mRNA preparations, yet they are amongst the most abundantly expressed genes in cells in which they are specifically expressed. In contrast, the relationship between the GC specific mRNAs in total cerebellar samples and GCs TRAP-Seq data is much more robust because of their abundancein the cerebellum.
Figure 1. Cell type specific gene expression in cerebellar cell types by TRAP-Seq.
(A) Visualization of FPKM values of TRAP-Seq data of various examples in PC, GC and BG. Pcp4 and Pvalb are enriched in PC (blue), Neurod2 and Calb2 are enriched in GC (orange) and Gstm1 and Gfap are enriched in BG (green). Values of RNA sequencing for these genes from total cerebellum (black) are also shown for comparison. Scale (minimum-maximum) is indicated on the top left corner of PC line for each gene. Windows show the following locations: Pcp4 chr16:96,683,159–96,757,502; Pvalb chr15:78,019,548–78,036,586 ; Neurod2 chr11:98,186,324–98,191,364; Calb2 chr8:112,663,312–112,696,669; Gstm1 chr3:107,814,027–107,821,968; Gfap chr11:102,746,534–102,760,963.
(B) Scatter plots comparing FPKM of TRAP-Seq of an individual cell type [y-axis; blue (left) PC, orange (center) GC, green (right) BG] vs the averaged values of the other two (x-axis). Remarked dots in each panel represent genes enriched in each cell type (PC blue, GC orange and BG green) as previously described by microarray analysis in the literature. Genes in bold indicate the reporter gene of each cell type (n=4 per cell type)
(C) Gene distribution showing enrichment (y-axis; Log2 FC: fold change) and expression values (Log2 FPKM) of enriched genes (> 2 FC) of each cell type compared individually with the other two, and averaged. The number of genes represented are indicated on the top left corner. Intensity of the color represents the level of enrichment. Grey bars delimit genes that are highly enriched (horizontal, cut off: 15 fold) and medium to highly expressed (vertical, cut off: 6 FPKM).
(D) Venn diagram of the 250 most expressed genes of each cell type.
(E) GO analysis of the 94 most expressed genes in PC that are not highly expressed in GC or BG as shown in panel D, p-val cut off 0.05. In parenthesis, percentage of genes that are included in each term. GO terms that explain the main biological features of PCs are highlighted in blue.
To identify those genes that are differentially expressed between PCs, GCs, and BG, TRAP-Seq data collected from each individual cell type was compared to the summed TRAP-Seq data from the remaining two cell types (Fig. 1B, Table S2A). These data were consistent with our previously collected TRAP microarray data (Doyle et al, 2008), as shown by the enrichment of positive control genes from each cell type in the corresponding TRAP samples. However, the quantitative nature of HTS relative to microarrays is readily apparent from the improved linear range of the TRAP-Seq analysis, as illustrated by the significant enrichment of cell specific mRNAs throughout the range of expression, including those that occur at low levels (for example Pou2af and Fgf7 in PC) and those in very high abundance (for example Pcp4 and Car8 in PC) (Fig. 1B). Given the quantitative nature of HTS, and the fact that TRAP-Seq data measures those mRNAs that are engaged by the ribosome and actively involved in protein synthesis, we were interested in determining the biosynthetic signatures of these very different cell types. Analysis of the TRAP-Seq data generally confirms the conclusion reached previously that each of these cell types is characterized by the enriched expression of a large set of genes (Fig. 1C) (Doyle et al., 2008; Heiman et al., 2008). However, comparison of the cell specific enrichment of these products and their absolute values of expression levels leads to an additional interesting insight. In each of these cell types, significant fractions of the most actively translated mRNAs are cell type specific (Fig. 1C, D). Furthermore, analysis of the Gene Ontology (GO) terms for these highly expressed and cell specific transcripts yields a biochemical signature of each cell type. For example, in PCs 94 of the 250 mostly highly expressed mRNAs are not expressed in either GCs or BG (Fig 1D). The GOs covered by these transcripts clearly reflect the fact that PCs have a very large dendritic arbor and make hundreds of thousands of synapses with GCs (Fig 1E). GO categories revealed in this analysis for GCs and BG also reflect their main functions: axonal maintenance and neuronal support, respectively (Fig S1E).
5hmC is enriched in euchromatin and 5mC in heterochromatin in cerebellar cell types
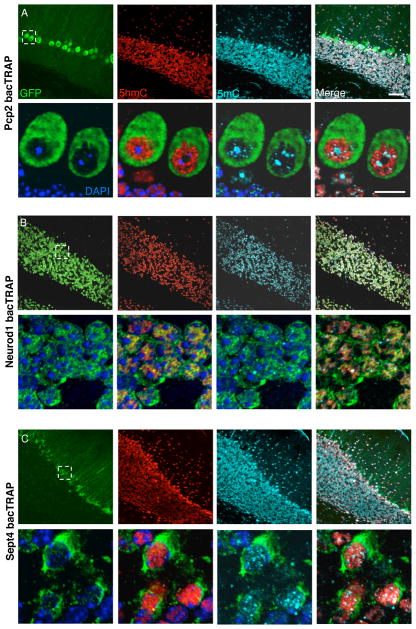
To gain an initial appreciation of the relative distribution of 5hmC, 5mC and chromatin domains in the distinctive nuclei of PCs, GCs, and BG, immunofluorescence studies of eGFP/L10a bacTRAP transgenic mice were conducted (Fig. 2). As previously reported (Doyle et al., 2008; Kriaucionis and Heintz, 2009), each of these cell types is readily visualized by the high levels of fluorescence present on translating polysomes in the cell soma. 5hmC is distributed throughout the nucleoplasm of all three cell types, and its distribution is clearly different from either 5mC or DAPI. 5hmC staining is evident in a dappled pattern that nearly fills the PC nucleus, yet is excluded from both the nucleolus and the adjacent, DAPI bright heterochromatic caps (Fig 2A, lower panels). Although the fraction of chromatin stained with 5mC and/or DAPI in both GCs (Fig 2B lower panels) and BG (Fig 2C lower panels) is evidently increased, the exclusion of 5hmC from nuclear domains with very high 5mC content or bright DAPI staining is maintained.
Figure 2. 5hmC is enriched in euchromatin in mature cerebellar cells.
(A) (B) (C) Inmunohistochemistry in cerebellar sections showing the enriched distribution of 5hmC and 5mC. Upper panels: Confocal microscopy image of the granule cell layer of the cerebellum from a Pcp2 bacTRAP (+/−) (A), Neurod1 bacTRAP (+/−) (B) and Sept4 bacTRAP (+/−) (C) expressed in PC, GC and BG respectively. GFP expression (green), and inmunostaining for 5hmC (red) and 5mC (cyan) are shown. Scale bar, 50 μm. Lower panels: Magnification of these areas delimited by dotted lines in A, B and C respectively. In the 3 first panels also DAPI staining in blue is shown. Scale bar, 10 μm.
Cell type specific Utilization of 5hmC and 5mC in the nervous system
Given the distinctive structure of chromatin in PCs, GCs, and BG, and the large number of gene products that are characteristic of each of these cell types, we were next interested in determining whether the relationships between cytosine modification status, and the rate and specificity of gene expression are cell type specific. Accordingly, genomic DNA for each cell type was obtained by fluorescence activated cell sorting (FACS) of EGFP/L10a labeled nuclei from bacTRAP transgenic lines as previously described (Kriaucionis and Heintz, 2009). Genome wide enrichment of 5hmC containing DNA fragments was done using a selective chemical labeling strategy (Song et al., 2010) ; 5mC was enriched using methylated DNA immunoprecipitation (MeDIP) (Jin et al., 2010; Weber et al., 2005) followed by sequencing using Illumina platform. Two biological replicas were done for each cell type and DNA modification, resulting the total of 198x10−6 uniquely mapped reads. 33 x10−6 reads per condition provided enough coverage to achieve correlation of 0.90 between two half’s of the sample, with estimated correlation of 0.95 per sample (Fig. S2 A).
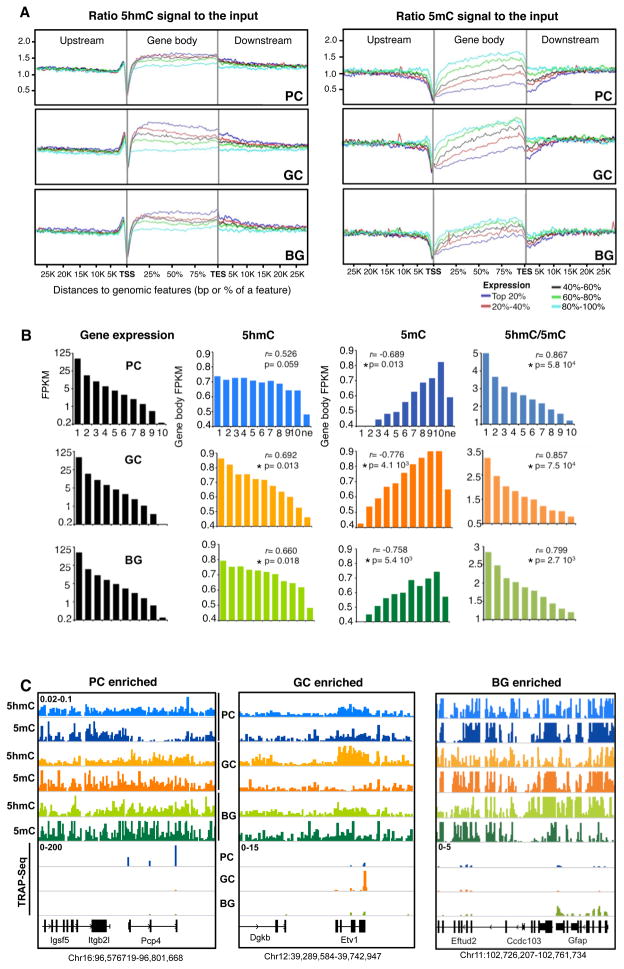
The chromosomal content of these epigenetic marks in each cell type was unremarkable except for the low 5hmC levels in the X chromosome, as previously reported (Szulwach et al., 2011) (Fig S2 C). In general, the distribution of 5hmC across the genome in these cell types was consistent with previous studies of brain tissue (Fig S2 B) (Song et al., 2010; Szulwach et al., 2011). Thus, 5hmC is preferentially enriched over the entire transcription unit of expressed genes, and depleted from both the TSS and intergenic regions (Fig. S2D). Several additional general features are revealed if gene body 5hmC and 5mC are plotted relative to expression level in each of these cell types (Fig. S2D). First, the patterns of 5hmC and 5mC are clearly complementary. Second, for highly expressed genes it appears that 5mC is depleted over the gene bodies. Third, the levels of 5hmC enrichment and 5mC depletion vary between cell types.
To interrogate further the relationships between gene expression and cytosine modification status in each cell type, genes were subdivided into groups based on the cell specific expression rank, and plotted against metagene centric features (Fig 3A). These plots confirmed the depletion of 5hmC at the TSS, and identified a characteristic peak of 5hmC just 900 bp 5′ of the TSS that showed no correlation with the expression state of the genes. Genes in the highest expression percentiles tend to have more 5hmC and less 5mC over their gene bodies than ones which are in the lowest expression percentiles, when 5hmC and 5mC levels in the gene body reach the genome average and 5mC drops below (Fig 3A, Table S1A). However, that these relationships vary significantly between cell types becomes readily apparent when calculating correlations between expression and cytosine modification for each cell type (Fig. 3B). For example, inspection of the 5hmC and 5mC histograms for deciles of genes ranked on expression demonstrates that for GC there are highly significant relationships between gene expression, elevated 5hmC levels (Pearson correlation, r=0.692; p=0.013) and depleted 5mC levels (r=0.776; p=4.1x10−3) within the gene body. Significant relationships of this type are also evident in the BG datasets (5hmC r=.660; p=0.018, 5mC r=0.758; p=5.4x10−3). However, while a clear relationship between gene expression and 5mC depletion is evident in PCs (r=0.689; p=0.013), the relationship between elevated gene body 5hmC and gene expression is much less clear (r=0.526; p=0.059). Next we considered the possibility that it is the ratio or 5hmC to 5mC within the gene body that is most informative with regard to gene expression. Support for this idea comes from the fact that in all cell types, the r coefficients are highly significant and increased if calculated on the basis of the 5hmC/5mC ratio (PC, r= 0.867; GC, r=0.857; BG, r=0.799). An inverse correlation between abundance of 5mC and 5hmC over gene bodies is indicated by the fact that 5hmC/5mC ratio had the best relation to gene expression (Fig 3B). This is expected because hydroxylation of 5mC results in 5hmC (Tahliani et al, 2009) and both of these marks cannot exist on one base. However, it was surprising to see the depletion of 5mC signal correlating better with gene expression than presence of 5hmC, which is especially obvious in PC (Fig 3A, B). We considered the possibility that the low 5mC signal in addition to the evident increase of 5hmC could indicate the presence of unmodified C, 5-formylC (5fC) or 5-carboxylC (5caC). Since 5fC and 5caC are too low to be confidently detected in the whole brain (data not shown and Ito et al., 2011) we reasoned that some sites may have cell type specific gain of unmodified Cs within potential modification sites. Bisulfite sequencing of selected loci demonstrated the increase of unmodified CpGs within the regions displaying cell type specific decrease of 5mC signal (Fig S3A). The unmodified CpGs were ranging from 91% in Diras2 gene to 53% in Foxp4 gene in a cell type showing low 5mC, and 0 % to 3 % respectively in a cell type with high 5mC signal (Fig S3A). These results illustrate the fact that both the loss of cytosine modification at the specific CpG residues and the accumulation of 5hmC within the gene body can contribute substantially to expression.
Figure 3. The relationship between cytosine methylation status and gene expression levels is cell specific.
(A) Metagene profiles of 5hmC and 5mC. Each line represents percentage of genes ranked according the expression levels.
(B) Genes were ranked by expressionand grouped in deciles (from 1, higher to 10, lower). FPKM of gene expression, and their correspondent FPKM of 5hmC, 5mC and the ratio between the two, averaged from single genes, are shown per cell type. Last column of 5hmC and 5mC histograms show averaged FPKM values of non-expressed (ne) genes. Pearson correlation coefficient (r) between expression and feature and p values are shown.
(C) Representative examples of individual genes and their values of 5hmC, 5mC and gene expression from the three cell types. Left panel shows Pcp4, enriched in PCs. Middle panel shows Etv1, enriched in GCs and right panel shows Gfap, enriched in BG. In each panel, values from PCs are colored in blue, GCs in orange and BG in green. First line of each group indicates 5hmC normalized values and second, 5mC. Below, TRAP-Seq normalized values are shown per each cell type. The last line in black represents the gene bodies located in this particular region of the genome.
Cell type specific relationships between cytosine methylation status and gene expression are also apparent when examining the distributions of 5hmC and 5mC in individual highly expressed genes (Fig 3C, S3B, Table S1A). For example, from inspection of the Pcp4 locus it is evident that this gene is expressed at elevated levels in PCs and that the Pcp4 gene body is depleted in 5mC specifically in PCs, but that the level of 5hmC within the Pcp4 gene does not vary visibly between cell types. In contrast, in GCs and BG many genes that are expressed at elevated levels display both significant enrichment of 5hmC within the gene body, and modest depletion of 5mC (e.g. Etv1, Gfap). Strikingly, we have observed individual instances where a differentially modified region predicted the presence of a transcribed gene that is present only in the most recent annotations of the genome. These data clearly illustrate a strong and cell type specific relationship between cytosine modification status and gene expression for individual genes.
MeCP2 is an abundant 5mC and 5hmC binding protein in the mouse brain
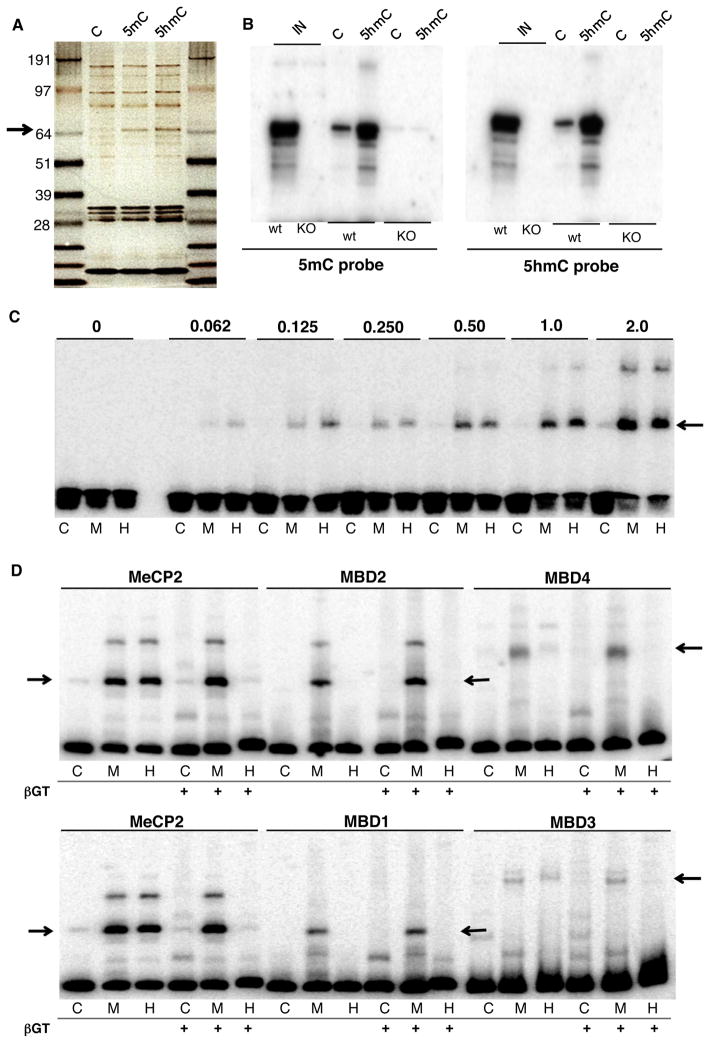
The enrichment of 5hmC and the depletion of 5mC throughout the bodies of expressed genes in terminally differentiated neural cell types, and the fact that 5hmC is at least ten times more abundant in neurons that in ES cells, suggests that the proteins decoding epigenetic information in the brain may be different from those present in ES cells. To identify these factors, nuclear extracts prepared from rodent brain (Klose and Bird, 2004) were incubated with magnetic beads coated with DNA containing unmodified C, 5mC or 5hmC DNA in the presence of excess of non-specific DNA competitor, followed by isolation of the beads and visualization of proteins after elution and SDS PAGE. As shown in Figure 4A, these experiments revealed a band of ~70 kDa that was pulled down with both 5mC and 5hmC, but was not present in the proteins eluted from beads coated with DNA containing unmodified C. This band was excised from a preparative gel of this type, and the protein was identified by mass spectroscopy as MeCP2 (Fig S4A). To confirm this result, similar affinity purifications were repeated from brain nuclear extracts of wild type (WT) and Mecp2 KO (KO) animals using beads coated with DNA containing C or 5hmC, and assayed using Southwestern analysis (Campoy et al., 1995). Thus, membrane bound re-natured proteins were probed with 32P labeled DNA containing either 5mC (Fig 4B left panel) or 5hmC (right panel), revealing a protein of the correct molecular weight that can bind both 5mC and 5hmC containing probe DNAs, and that is not present in samples prepared from KO animals. To our surprise, no other protein with high specificity for 5hmC DNA was revealed in these studies, even in the absence of MeCP2.
Figure 4. MeCP2 is the major protein that binds 5hmC.
(A) Silver stained SDS-PAGE gel of nuclear proteins from frozen rat cerebella that bound beads coated with unmodified C, 5mC or 5hmC DNA. Arrow pointing at the band that was excised and identified by MS from a Coomassie stained replica gel.
(B) Southwestern blot of the nuclear proteins from cerebella of WT or KO mice (IN) that bound to C or 5hmC- coated beads probed with radioactive 5mC or 5hmC DNA.
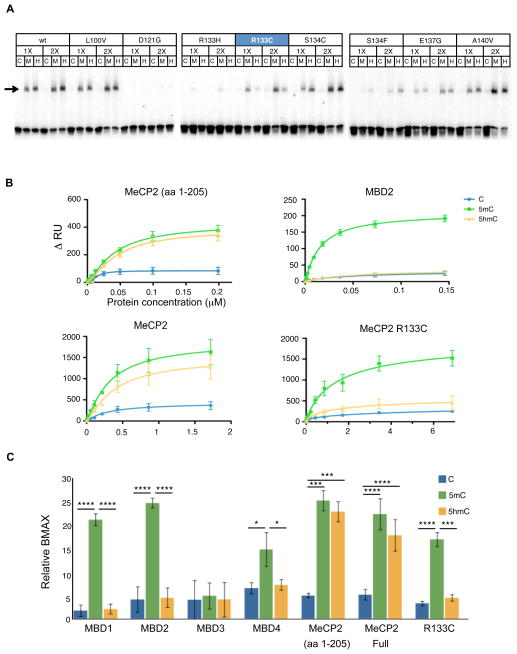
(C) EMSA of C, 5mC (M) or 5hmC (H) probes with increasing concentrations (0 to 2 pmol) of recombinant human MeCP2. The arrow points at the MeCP2-dependent low-mobility complexes.
(D) EMSA of C, 5mC (M) and 5hmC (H) probes in presence of 1 pmol of MeCP2 (aa 1-205), 1.2 pmol of MBD1, 0.5 pmol of MBD2, 50 pmol of MBD3 or 2.5 pmol of MBD4. The probes were either reacted with βGT prior to labeling or not (βGT−). Arrows point at protein-dependent DNA complexes.
Recombinant human MeCP2 binds 5mC and 5hmC in vitro
The identification of MeCP2 as a major 5hmC binding protein in rodent brain is surprising given previous in vitro studies reporting that it binds 5mC containing DNA much more avidly than 5hmC containing DNA (Bostick et al., 2007; Valinluck et al., 2004). To address this issue directly, an N-terminal (NT) fragment of human MeCP2 containing its MBD (residues 1-205) was produced in E. coli, and used in electrophoretic mobility shift assays (EMSA) to measure binding to 5mC, 5hmC or unmodified DNA. At all concentrations tested, the MeCP2 NT failed to bind the unmodified probe, while avidly binding both the 5mC and 5hmC probes (Fig. 4C). As an additional control, EMSA probes were reacted with T-4 phage β-glucosyltransferase (βGT) and uridine diphosphoglucose (UDP-glucose), which results in the specific glucosylation of 5hmC containing DNA without affecting 5mC and C containing probes (Szwagierczak et al., 2010). Binding other MBD family proteins (Fig. 4D) were also analyzed. MeCP2 NT bound specifically to unreacted 5mC and 5hmC probes. Glucosylation of 5hmC probe blocked binding, whereas binding to the 5mC probe was retained as 5mC is refractive to glucosylation. MeCP2 binding to 5hmC was not sequence specific since the binding properties of MeCP2 to a variety of probes selected from the mouse genome did not vary significantly (Fig S4C). In contrast MBD1, 2 and 4 all bound strongly to 5mC containing DNA, and did not bind avidly to 5hmC containing probes. As previously reported (Yildirim et al., 2011) weak and glucosylation-sensitive binding of MBD3 was observed to both 5mC and 5hmC DNAs, and the mobility of the MBD3/5hmC complex was slightly retarded relative to the MBD3/5mC complex.
R133C, an MeCP2 residue mutated in Rett Syndrome, alters preferentially binding to 5hmC DNA
If binding of MeCP2 to 5hmC is critical for its role in the regulation of neuronal nuclear function and gene expression, then it is possible that a subset of the MeCP2 mutations that cause RTT disrupt 5hmC binding without strongly impacting 5mC interaction. To determine if this is the case, binding of MeCP2 MBDs (aa 1-205) carrying a variety of previously characterized RTT mutations were assayed (Kudo et al., 2003). To represent two extreme cases of DNA binding activity three mutations were selected: D121G, that abolishes 5mC binding, and L100V and A140V that don’t disrupt 5mC binding. The rest of the RTT-causing mutations in the MBD were chosen because they showed no or little disruption of nuclear localization or 5mC-binding. Although the general effect of these mutations was to inhibit binding to both 5mC and 5hmC was indistinguishable, we observed a pronounced decrease in the interaction with 5hmC relative to 5mC DNA with the MeCP2 NT carrying the R133C substitution (Fig. 5A). To provide independent analytical data to support the conclusions of the EMSA assays presented above, surface plasmon resonance (SPR) assays were used to measure the binding of full length MeCP2, the MeCP2 NT, other MBD proteins, and the MeCP2 carrying R133C mutation. 5′-biotinylated C, 5mC or 5hmC probes were immobilized on parallel flow cells (Fc) of a streptavidin-coated sensor chip to their saturation level. The steady-state SPR response of each Fc at serial dilutions of above mentioned proteins is shown in Fig. 5B. As predicted, MeCP2 (both NT and full length, Fig. 5B) showed specific binding to both 5mC and 5hmC containing DNA that was strongly dependent on protein concentration, whereas binding to C containing DNA plateaued at very low protein concentrations, consistent with nonspecific binding. In contrast, MBD2 bound strongly to 5mC containing probes and showed nonspecific binding to both C and 5hmC. Interestingly, binding characteristics of the MeCP2 R133C mutant to 5hmC was similar to those of nonspecific binding.
Figure 5. R133C mutation of MeCP2 preferentially disrupts its binding to 5hmC.
(A) The EMSA showing 5hmC binding characteristics of several MeCP2 point mutations (0.5 (1X) or 1 (2X) pmol) observed in RTT patients. Arrow shows the expected position of theMeCP2-dependent complexes.
(B) Steady-state binding curves of MeCP2 (aa 1-205), MBD2, MeCP2 and MeCP2 R133C to C, 5mC and 5hmC reported as SPR response of Fcs immobilized with indicated probes upon application of serial dilutions of proteins. MeCP2 binding to both 5mC and 5hmC (upper left panel) shows specificity. MBD2 is chosen to represent the characteristic curve of 5mC-specificity (upper right panel). Full-length MeCP2 also binds 5hmC and 5mC with similar affinities and R133C mutation of MeCP2 shows nonspecific binding to 5hmC.
(C) The extracted Bmax values, normalized for the protein mass, , of MBD family of proteins, MeCP2 (1-205), full length MeCP2 and R133C mutants. (n=4, SD). **** p-val< 0.0001; *** p-val<0.001; ** p-val<0.01; * p-val< 0.05 in F test to compare variances.
To further assess these results, the maximum binding capacity (Bmax) of each protein was calculated for each probe from steady-state binding curves (Figs 5C, S5A). As expected, MBD1, 2 and 4 showed highly significant specific binding to 5mC DNA. Both the MeCP2 MBD and the full length protein bound 5mC and 5hmC specifically, consistent with the pull down experiments, the Southwestern results and the EMSA data presented above (Figs. 4, 5, S5). No significant difference was observed in the Bmax of MeCP2 binding to 5mC and 5hmC. The most interesting and unexpected data revealed by these SPR assays (Fig. 5, S5) is that R133C MeCP2 mutant retained most of its 5mC binding capability (mean Bmax = 76% of WT, p=0.77) despite loss of specific binding to 5hmC (mean Bmax = 25% of WT, p = 0.0029) (Fig. S5). The fact that this single substitution in the MeCP2 MBD can strongly and preferentially impact the substrate binding properties of MeCP2 is important because identification of MeCP2 mutations that retain WT 5mC binding in the R133C variant yet retain severely diminished 5hmC binding can provide an important avenue for assessing the role of MeCP2 binding to 5hmC in the pathophysiology of RTT. Furthermore, these data demonstrate that small changes the structure of MeCP2 may influence its relative binding properties to 5mC and 5hmC, raising the interesting possibility that the posttranslational modifications to MeCP2 that have been shown to occur in response to a variety of stimuli (Chen et al., 2003; Tao et al., 2009; Rutlin et al., 2011; Adkins et al., 2011; Gonzales et al., 2012) could alter its its substrate specificity and downstream functions.
The distribution of genomic 5hmC does not change in Mecp2 knockout neurons
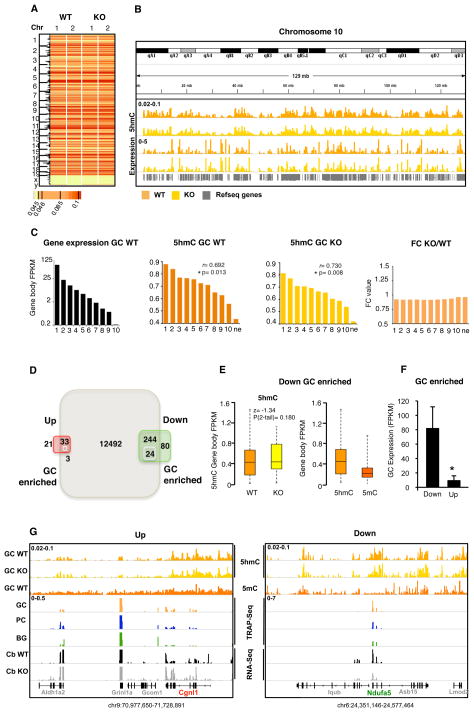
Given the demonstration that MeCP2 binds strongly to 5hmC containing DNA, and the strong positive correlation between gene body 5hmC levels and gene expression observed in GCs, it was of interest to determine whether the MeCP2 helps to establish the levels of 5hmC in expressed genes, or whether 5hmC acts upstream of MeCP2 in its relationship to gene expression. To investigate this issue, the distribution of 5hmC in GC genomes purified from WT and KO mice was mapped and its relationship to gene expression analyzed. Inspection of these data reveals no significant differences in the distribution of 5hmC as a result of loss of MeCP2 (Fig. 6A, B). Consequently, the strong positive correlation between GC gene expression and 5hmC gene body content (r=0.692, p=0.13) is maintained in the absence of MeCP2 (r=0.730, p=0.008) (Fig. 6C, Table S1C). We note, however, that a small but significant decrease in gene body 5hmC levels was evident for expressed genes across all deciles in the MeCP2 KO granule cells (Fig. 6C,S6B). To determine whether this reflected active transcription, we also analyzed the levels of gene body 5hmC in non-expressed genes in the knockout animals. Again, in the KO granule cells a significant, small difference in 5hmC levels was observed. Although we do not know the origin of this finding, the fact that it is occurring in genes irrespective of their expression levels argues strongly that it is not the result of transcriptional activity.
Figure 6. 5hmC levels over gene bodies do not change in MeCP2 KO.
(A) Heatmap of 5hmC FPKM over the chromosomes. Both replicas from wild type (WT) and Mecp2-null (KO) animals are shown.
(B) Chromosome 10 showing the levels of 5hmC and RNA-Seq in WT and KO animals.
(C) Genes were ranked by expression and grouped in deciles. FPKM of gene expression in GC, and their correspondent FPKM of 5hmC in the GC WT, in the GC KO and the average fold change (FC) of each decile are shown. Pearson correlation coefficient (r) between expression and feature and p values are shown. * p-val <0.05
(D) Venn diagram of the dysregulated genes in KO. 36 genes from the upregulated genes are expressed in GC, 3 are enriched (> 2 FC) over PC and BG. 268 genes are downregulated and expressed in GC, 24 of them are enriched (> 2 FC) over PC and BG.
(E) Box and whisker plots of 5hmC and 5mC levels of the 24 downregulated genes enriched in GC (left panel) and 5hmC in WT and KO GC (right panel). Wilcoxon signed-rank test coefficient (z) and p values are shown. * p-val <0.05.
(F) Average of expression of the 24 downregulated genes enriched in GC and 3 upregulated genes enriched in GC. SE is shown. * p-val <0.05 in t-test.
(G) Examples of dysregulated genes in the KO and enriched in GCs. 5hmC, 5mC and gene expression values are represented. Left panel shows Cgnl1 in red upregulated in KO (RNA-Seq values from WT in black and KO in grey). Right panel shows Ndufa5 in green downregulated in KO. In each panel, values from PCs are colored in blue, GCs in orange and BG in green. First line of each panel indicates GC WT 5hmC, second GC KO 5hmC, third, 5mC. Below, TRAP-Seq are shown per each cell type.
To identify genes whose expression is altered in KO GCs, and determine whether the cytosine modification status of this class is altered as a result of loss of MeCP2, RNA-Seq data was collected from cerebella of WT and KO animals (Table S1B). Consistent with previous results (Ben-Shachar et al., 2009), the majority of genes whose expression is altered in the cerebellum in response to loss of MeCP2 were down regulated (Fig. 6D, Table S2C, D). To determine whether genomic 5hmC levels changed within this class of genes in the KO, we restricted our analysis to the 24 genes that are expressed preferentially in GCs because of the cell type specific relationships between expression and cytosine modification documented above. Loss of MeCP2 had no effect on the level or distribution of GC gene body 5hmC for these genes (Fig. 6E, G). As expected, this class of genes was expressed at significantly higher levels than the few upregulated genes identified in the RNA-Seq experiments, and they were enriched in 5hmC and depleted in 5mC (Fig. 6E, F, G). We conclude based on these data that the distribution of 5hmC is determined by mechanisms that are independent of MeCP2, and that 5hmC must act upstream of MeCP2 to facilitate transcription.
5hmC, MeCP2 and the organization of neuronal chromatin
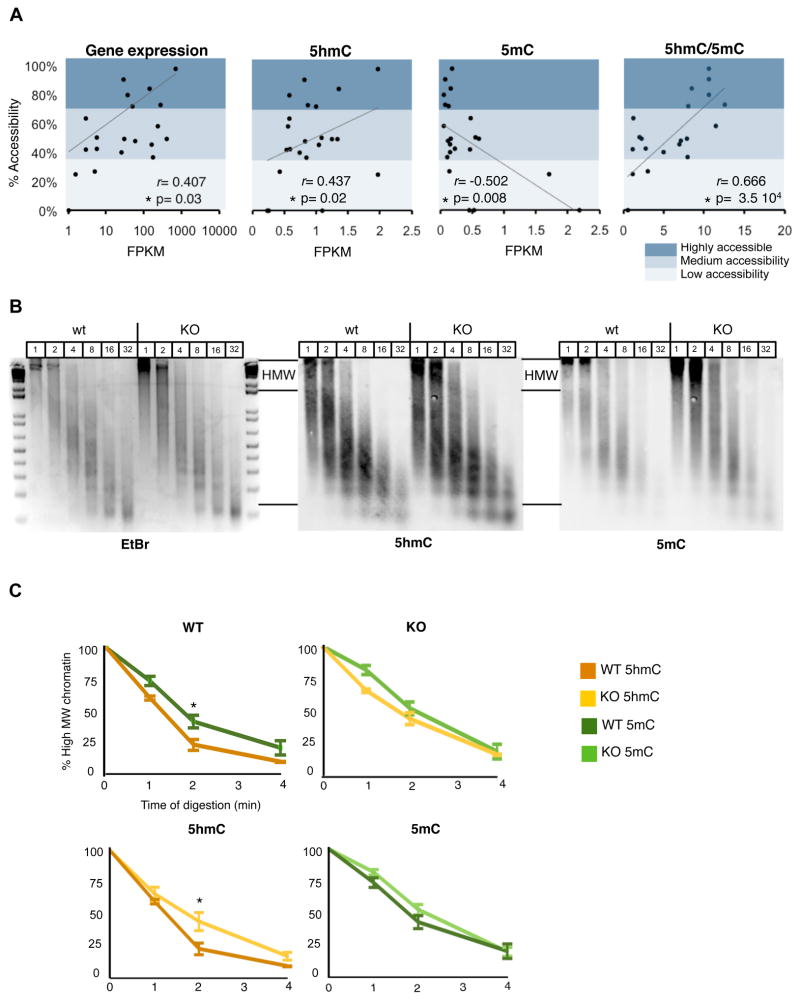
Evidence from a wide variety of studies support a general model in which MeCP2 binding to 5mC at CpG dinucleotides throughout the genome plays an important role in transcriptional repression (Guy et al, 2011). However, the observations that in brain nuclei a large fraction of MeCP2 is localized within highly nuclease accessible regions (Thambirajah et al., 2012), that loss of MeCP2 can lead to downregulation of expressed genes (Ben-Shachar et al, 2009; this study), and that 5hmC is enriched in the gene bodies of highly expressed genes (Song et al. 2010, this study) suggests that MeCP2 binding to 5hmC may also play a role in facilitating gene expression. If this is the case, expressed genes that have a high 5hmC/5mC should be enriched in highly accessible chromatin. To test this prediction, we first measured the relationship between chromatin accessibility and cytosine modification status in the cerebellar nuclei (Fig. 7A, S7). Nuclei were isolated, treated with increasing concentrations of micrococcal nuclease (MNase), and chromatin sensitivity of specific genes assayed. We observed that genes with high 5hmC/5mC values were lost from nuclei at low MNase concentrations, indicating their presence in accessible chromatin. Genes resistant to low concentrations of MNase were preferentially enriched in 5mC and depleted in 5hmC (Fig. S7B). As expected, genes that are not expressed and have high levels of 5mC were resistant to MNase digestion.
Figure 7. 5hmC and MeCP2 and the organization of neuronal chromatin.
(A) Nuclei were digested with MNase for 5 min,and analyzed by qPCR. The correlation between the chromatin accessibility of individual genes and expression, 5hmC, 5mC and 5hmC/5mC (fourth panel) are shown (n=3)
(B) Ethidium Bromide (EtBr) stained 2% agarose gel showing DNA fragments at different time points during MNase digestion (1–32 min) of nuclei from cerebella of WT or KO mice. The blotted gel that was probed with 5hmC or 5mC antibodies.
(C) The precent intensity in high molecular weight DNA (HMW) in (B) was plotted as a function of time. Upper panels show the comparison in 5hmC and 5mC in the WT (left panel) and in MeCP2 KO (right panel). Lower panels show the comparison between WT and MeCP2 KO in 5hmC and 5mC. * p-val <0.05 in an unpaired T-test.
Given the high abundance of MeCP2 in the brain (Fig S4B) (Guy et al., 2001a; Skene et al., 2010; Thambirajah et al., 2012), and our demonstration that MeCP2 binds avidly to 5hmC containing DNA in vitro, we were next interested in assessing its potential role in global regulation of chromatin accessibility. To do so, cerebellar nuclei were isolated from five week old WT and KO mice (Guy et al., 2001b). For each sample, a time course of MNase digestion was performed, and the release of 5hmC and 5mC enriched DNA was assayed with antibodies against 5mC and 5hmC on a Southern blot (Fig 7B, C). The signal from the high molecular weight (HMW), nuclease resistant fraction was measured in four independent cohorts of WT and KO mice, its percentage was calculated to total signal in the lane, and the data at each time of digestion plotted (Figure 7B, C). Two interesting results were obtained. First, we observed that 5hmC enriched DNA is released readily from chromatin by MNase digestion, whereas 5mC containing chromatin is significantly more resistant to digestion (Fig. 7C). This is consistent with the analysis of individual genes shown in Fig 7A, and confirms previous studies demonstrating the 5mC enriched DNA is present MNase resistant compact structures (Karymov et al., 2001). Second, in KO mice a significant, small delay in digestion of 5hmC containing DNA was observed, whereas no reproducible difference in the sensitivity of 5mC containing DNA to MNase was evident (Fig. 7B,C). These data demonstrate that MeCP2 regulates the accessibility of 5hmC containing DNAto MNase, supporting a model in which MeCP2 binding to 5hmC within highly expressed genes may facilitate transcription through its effects on chromatin organization.
Discussion
The data presented here identify a role for MeCP2 in the regulation of chromatin structure, and support a model for the organization of chromatin and gene expression that is of particular importance for CNS. This model depends on three major factors: depletion of 5mC within the bodies of expressed genes, accumulation of high levels of 5hmC within these gene bodies, and occupation of 5hmC binding sites by the abundant and CNS-enriched protein MeCP2. The contributions of each of these factors to gene expression vary between cell types, suggesting that each of them can be regulated independently. Based on our data, and the fact that both 5hmC and MeCP2 are at least an order of magnitude more abundant in CNS than in the periphery (Kriaucionis and Heintz, 2009; Skene et al., 2010), we propose that binding of 5hmC by MeCP2 plays a central role in the epigenetic regulation of neural chromatin and gene expression. Advances in our understanding of the pathophysiology of RTT will require further investigation of this new role for MeCP2 in facilitating gene expression when bound to 5hmC in the context of the traditional repressive functions it elicits upon its binding to 5mC (Guy et al, 2011).
Although a mechanism by which MeCP2 binding to 5hmC could regulate chromatin accessibility remains to be determined, several inferences can be drawn from the existing data. First, the distribution of 5hmC throughout the transcription unit of highly expressed genes distinguishes this mechanism from the established roles of MeCP2 and other MBD family proteins in the organization of repressive chromatin complexes at promoters and enhancers (Guy et al,2011; Yildirim et al., 2011). Our data support the idea that the action of MeCP2 is more akin to a linker histone (Skene et al., 2010), occupying expressed genes through its binding to 5hmC. They are also consistent with the observations that MeCP2 stably associates with nucleosomes (Chandler et al., 1999), that it can compete with histone H1 for nucleosome binding sites (Ghosh et al., 2010), and that the levels of MeCP2 and histone H1 are inversely correlated in neurons (Skene et al., 2010). However, our observations that MeCP2 binds with high affinity to 5hmC and that 5hmC is enriched in expressed genes that are nuclease sensitive forces a reevaluation of the role of MeCP2 binding to chromatin in neural cell types. We propose that binding of MeCP2 to 5hmC in expressed genes facilitates transcription through organization of dynamic chromatin domains. This model provides a mechanistic explanation for the recent demonstration that MeCP2 can also activate gene expression, as some genes are both downregulated upon loss of MeCP2 and upregulated in mice with increased MeCP2 gene dosage (Ben-Shachar et al., 2009; Fig. 6).
Second, our data suggest that both depletion of gene body 5mC and MeCP2 binding to 5hmC are important to establish chromatin domains that facilitate transcription. Thus, there is a strong inverse correlation between gene expression and gene body 5mC. It seems probable that this reflects both the biochemical nature of 5mC binding by MBD proteins, and the consequences of their action. For example, it has recently been shown that in the brain two populations of MeCP2 are present: one in chromatin regions that are enriched in nucleosomes and the other that is loosely bound to highly accessible chromatin domains (Thambirajah et al., 2012). Given our demonstration that genes enriched in 5hmC are also preferentially present in these MNase sensitive domains, it seems likely that this loosely bound MeCP2 is associated with 5hmC rather than 5mC. This suggests that the interaction of MeCP2 with 5hmC establishes a dynamic state of chromatin that would be quite sensitive over time to the presence of much more stable complexes established within that domain by binding of MeCP2 or other less abundant MBD family proteins to 5mC (Lopez-Serra et al., 2006). A cell-specific and dynamically regulated gene expression pattern might be explained by a three-dimensional chromatin structure established by regulating levels of 5mC, 5hmC, MeCP2 and other MBD proteins. Changes in the level or activity of MeCP2 would disrupt this balance, resulting alterations in chromatin structure and, consequently, gene expression. Since in each cell type the levels of 5hmC, 5mC and the proteins that bind them vary, the phenotypic consequences of changes in the function of MeCP2, whether as a result of mutation (Adkins et al., 2011; Tao et al., 2009; Amir et al., 1999) or postranslational modification (Rutlin et al., 2011; Gonzales et al., 2012), will be cell type and circuit specific.
Third, our understanding of the pathophysiology of RTT must now encompass both the role of MeCP2 binding to 5mC in the repression of gene expression (Chahrour and Zoghbi, 2008), and present results supporting a model in which MeCP2 binds to 5hmC within active transcription units. For example, the observations that the distribution of 5hmC, 5mC and their relationship to gene expression vary depending on cell type, and that disease causing mutations of MeCP2 can impact 5hmC binding preferentially (e.g. R133C), could lead to important insights into the specific phenotypes associated with altered MeCP2 function. Our data both support previous genetic studies demonstrating that the consequences of MeCP2 loss in different neural cell types differ both quantitatively and qualitatively (Ben-Shachar et al., 2009), and suggest that the specific biochemical properties of mutant MeCP2 proteins may inform our understanding of their clinical consequences. For example, it is well documented that patients carrying the R133C mutation have a milder form of RTT that is characterized by delayed onset regression, with improved speech and motor skills (Bebbington et al., 2008). However, for many other characteristics, including breathing abnormalities, sleep problems, mood disturbances, and epilepsy prevalence, no significant differences are evident between patients bearing R133C or other mutations (Bebbington et al., 2008). Does this mean that these latter clinical features of RTT are associated with loss of its 5hmC binding capacity, and that they reflect differences in the relative importance of 5hmC versus 5mC binding in different cell types? Is it possible that 5hmC plays a role in the phenotypes that result in categorization of RTT as an Autism Spectrum Disorder? We cannot presently answer these questions, although generation of mouse models with “improved” MeCP2 mutations that continue to strongly impact 5hmC binding yet retain WT 5mC interaction offers an important avenue toward investigation of these issues.
Finally, while we believe binding of MeCP2 to 5hmC is a major step in decoding 5hmC in the CNS, many issues remain to be addressed. We have not, for example, assessed the influence of activity dependent mechanisms (Cohen et al., 2011) on the interactions of MeCP2 with 5mC or 5hmC containing DNA. We have not yet had the opportunity to analyze the relationships between gene expression, 5mC and 5hmC in other glial cell types that have been shown recently to play important roles in mouse models of RTT (Derecki et al., 2012; Lioy et al., 2012). We do not understand the relative importance of the mechanism described here and the recent observation that MBD3 can bind to 5hmC containing DNA (confirmed here), and that it is co-localized with Tet1 at 5hmC containing promoters in ES cells (Yildirim et al., 2011). And we do not know if 5hmC mediated demethylation plays a role in the dynamic control of epigenetic regulation of specific CNS cell types (Cortellini et al. 2011; Ito et al., 2011). Investigation of these and other issues in specific neuronal and glial cell types will be essential if we are to decipher the role of 5hmC in CNS, and understand its contributions to the pathophysiology of RTT.
Experimental procedures
TRAP-Seq
RNA from translating polysomes was extracted as described before (Heiman et al., 2008)(see extended experimental procedures). We obtained more than 30 million 50-bp single-end reads per sample (Figure S1 A) that were separatedly aligned to the mouse genome (mm9) downloaded from UCSC. TopHat software (version 1.3.1) was used for processing reads. Segment size was set to 25bp with two mismatches to the reference allowed, and the minimum anchor size was set to 10bp with no mismatches allowed. The resulting aligned data in bam format were assembled into transcripts using Avadis NGS 1.3.0 (Strand Scientific Intelligence,San Francisco,CA,USA). Annotated transcripts were obtained from Ensembl transcripts (2010.10.07) (http://www.ensembl.org). Transcript abundance was measured in Fragments Per Kilobase of exon per Million fragments mapped (FPKM) similarly to RPKM used in (Mortazavi et al., 2008). Finally, differentially expressed genes were identified by performing a negative binomial test using the DESeq package (Anders and Huber, 2010) of R/Bioconductor (Gentleman et al., 2004). Our conditions were selected and qualitatively validated by comparing the differential expression results with in situ hybridization data from Allen Brain Atlas (see extended experimental procedures). RNA-Seq from MeCP2 KO and WT cerebella was done following the same protocol as inputs of TRAP-Seq
5hmC pull down and MeDIP sequencing
Sorted nuclei from the three cell types were manipulated in parallel during the procedure. 5hmC was pulled down as described (Song et al., 2010) (see extended protocols). After purification DNA was amplified as described in TruSeq DNA Sample kit. MeDIP was done as described in (Weber et al., 2005) with the indicated modifications. 1-0.5 μg of DNA was used for each experiment. Sonicated DNA was end-repaired following by ligation to Illumina paired end sequencing adapters (Illumina, PE- 102- 1003). Enrichment was done using anti-methylC antibody (Eurogentech, BI-MECY-0100), following by amplification with Illumina primers and size selection on an agarose gel. Input samples were produced for each cell types in both procedures
Both 5hmC and 5mC enriched were then sequenced using Illumina platform obtaining more than 50 x10−6, 36-bp single-end reads per sample. Reads were aligned to mm9 mouse genome assembly using Bowtie v0.12.7 (-m1 --best). Further analysis was done using Bioconductor v2.9 using packages chipseq, biomaRt, rtracklayer, MEDIPS and custom scripts. Two biological MeDIP-Seq replicas were done for each of the cell type
5hmC affinity pull-down
1 ug of 5′-biotinylated C, 5mC or 5hmC BDNF probe was immobilized on Dynabeads M-280 Streptavidin (Invitrogen) following manufacturer’s recommendations to pull down 5hmC binding proteins from brain nuclear extract incubated with 2 mg of nuclear extract (see extended protocols). Isolated proteins were analyzed by mass spectrometry (MS)
For more experimental details, please see the supplemental experimental procedures, and Table S3
Supplementary Material
Research HIghlights.
Genome-wide analysis of 5hmC, 5mC and expression in distinct CNS cell types in vivo
5hmC is enriched in active genes in neurons
Identification of MeCP2 as a major binding protein for 5hmC in brain cells
A Rett Syndrome mutation in MeCP2 preferentially inhibits 5hmC binding
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (NH), Simons Foundation Autism Research Initiative (NH), Conte Center PHS MH090963 (NH) Ludwig Institute for Cancer Research (SK), and Spanish MECD (MAM). We wish to thank Chunxiao Song and Chuan He for kindly providing 5hmC pull down reagents, Jim Selfridge and Adrian Bird for providing MECP2-null mouse brains and Brian Lang at GE Healthcare. We would further like to thank Beatriz López and Betsy Gauthier for their assistance, and Jean-Pierre Roussarie, Anne Schaeffer, Emmanuelle Jordi and Ron Gejman for their advice. We also thank Connie Zhao, Christina Caserio and Wenxiang Zhang from the Rockefeller University Genomics Resource Center; Svetlana Mazel, Selamawit Tadesse, Xiao Li and Stanka Semova from the Rockefeller University Flow Cytometry Resource Center and Henrik Molina, Joseph Fernandez, Milica Tesic Mark and Susan Powell from The Rockefeller University Proteomics Resource Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Adkins NL, Georgel PT. MeCP2: structure and functionThis paper is one of a selection of papers published in a Special Issue entitled 31st Annual International Asilomar Chromatin and Chromosomes Conference, and has undergone the Journal, As usual peer review process. Biochemistry and Cell Biology. 2011;89(1):1–11. [Google Scholar]
- Amir RE, I, Van den Veyver B, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebbington A, Anderson A, Ravine D, Fyfe S, Pineda M, de Klerk N, Ben-Zeev B, Yatawara N, Percy A, Kaufmann WE, et al. Investigating genotype-phenotype relationships in Rett syndrome using an international data set. Neurology. 2008;70:868–875. doi: 10.1212/01.wnl.0000304752.50773.ec. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, et al. Quantitative Sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at Single-Base Resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Campoy FJ, Meehan RR, McKay S, Nixon J, Bird A. Binding of histone H1 to DNA is indifferent to methylation at CpG sequences. J Biol Chem. 1995;270:26473–26481. doi: 10.1074/jbc.270.44.26473. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chandler SP, Guschin D, Landsberger N, Wolffe AP. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–7018. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003 Oct 31;302(5646):885–9. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, Kipnis J. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol Cell Biol. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ML, Adams S, Dunaway KW, LaSalle JM. Phosphorylation of Distinct Sites in MeCP2 Modifies Cofactor Associations and the Dynamics of Transcriptional Regulation. Molecular and Cellular Biology. 2012;32(14):2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2001a;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001b;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Research. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karymov MA, Tomschik M, Leuba SH, Caiafa P, Zlatanova J. DNA methylation-dependent chromatin fiber compaction in vivo and in vitro: requirement for linker histone. FASEB J. 2001;15:2631–2641. doi: 10.1096/fj.01-0345com. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J Biol Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo S, Nomura Y, Segawa M, Fujita N, Nakao M, Schanen C, Tamura M. Heterogeneity in residual function of MeCP2 carrying missense mutations in the methyl CpG binding domain. J Med Genet. 2003;40:487–493. doi: 10.1136/jmg.40.7.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange W. Cell number and cell density in the cerebellar cortex of man and some other mammals. Cell Tissue Res. 1975;157:115–124. doi: 10.1007/BF00223234. [DOI] [PubMed] [Google Scholar]
- Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, et al. A role for glia in the progression of Rett’s syndrome. Nature. 2012;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66:8342–8346. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay V Springer-Verlag, editor. Cerebellar cortex: cytology and organization. New York, USA: 1974. [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutlin M, Nelson SB. MeCP2: Phosphorylated Locally, Acting Globally. Neuron. 2011;72(1):3–5. doi: 10.1016/j.neuron.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2010;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Wu H, et al. Chapter 7 - Deciphering Rett Syndrome with Mouse Genetics, Epigenomics, and Human Neurons. International Review of Neurobiology. Academic Press. 2009;89:147–160. doi: 10.1016/S0074-7742(09)89007-7. [DOI] [PubMed] [Google Scholar]
- Thambirajah AA, Ng MK, Frehlick LJ, Li A, Serpa JJ, Petrotchenko EV, Silva-Moreno B, Missiaen KK, Borchers CH, Adam Hall J, et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2012;40:2884–2897. doi: 10.1093/nar/gkr1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Research. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD Complex Regulates Expression of 5-Hydroxymethylcytosine Marked Genes in Embryonic Stem Cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, et al. Base-Resolution Analysis of 5-Hydroxymethylcytosine in the Mammalian Genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.