Abstract
Host cell invasion by a major periodontal pathogen, Porphyromonas gingivalis, has been proposed as an important mechanism involved in host–pathogen interactions in periodontal and cardiovascular diseases. The present study sought to gain insight into the underlying mechanism(s) involved in previously demonstrated fusobacterial modulation of host cell invasion by P. gingivalis. An immortalized human gingival cell line Ca9-22 was dually infected with P. gingivalis ATCC 33277 and Fusobacterium nucleatum TDC 100, and intracellular invasion was assessed by scanning electron microscopy (SEM) and confocal scanning laser microscopy (CSLM). SEM observation showed that P. gingivalis and F. nucleatum formed consortia and were in the process of penetrating into Ca9-22 by 30–60 min after infection. In CSLM, Ca9-22 cells that contained both P. gingivalis and F. nucleatum were frequently observed after 2 h, although cells that contained exclusively P. gingivalis were also found. Infection by P. gingivalis and/or F. nucleatum revealed evident colocalization with a lipid raft marker, GM1-containing membrane microdomains. In an antibiotic protection assay, depletion of epithelial plasma membrane cholesterol resulted in a significant reduction of recovered P. gingivalis or F. nucleatum (~33% of untreated control; p < 0.001). This inhibition was also confirmed by CSLM. Sequential infection experiments showed that timing of infection by each species could critically influence the invasion profile. Co-infection with F. nucleatum significantly enhanced host cell invasion by P. gingivalis 33277, its serine phophatase SerB mutant and complemented strains, suggesting that the SerB does not play a major role in this fusobacterial enhancement of P. gingivalis invasion. Thus, the interaction between F. nucleatum and host cells may be important in the fusobacterial enhancement of P. gingivalis invasion. Collectively, these results suggest that lipid raft-mediated process is at least one of the potential mechanisms involved in fusobacterium-modulated host cell invasion by P. gingivalis.
Keywords: Polymicrobial infection, Host invasion, Porphyromonas gingivalis, Gingival epithelial cells, Periodontitis, Fusobacterium nucleatum
1. Introduction
Periodontitis is a polymicrobial infection-driven inflammatory disease of the oral cavity, characterized by chronicity and destruction of the tooth-supporting tissues [1]. Porphyromonas gingivalis, one of the predominant etiologic agents in periodontitis, expresses a diverse spectrum of virulence factors. The ability to invade host cells, replicate intracellularly and spread from cell to cell is a hallmark of virulence for many pathogenic microorganisms [2]. Host cell invasion by P. gingivalis has been proposed as a possible mechanism of pathogenesis in periodontal and cardiovascular diseases [3–5].
Fusobacterium nucleatum is a Gram-negative anaerobe associated with various human infections [6]. It is ubiquitous in the oral cavity and is implicated as a causative agent in periodontal diseases. F. nucleatum initially adheres to early colonizers such as Gram-positive cocci, and, along with other organisms such as Strepotococcus gordonii, enhances the adherence of other periodontopathic bacteria including P. gingivalis [7,8]. F. nucleatum also coaggregates with a number of other microbial species in the oral cavity, thus playing a critical role in periodontal biofilm formation. F. nucleatum also binds to and invades different types of host cells [9,10]. Residence within host cells provides these periodontal pathogens with a nutrient-rich, generally reducing environment that is partially protected from the host immune system [11].
There is increasing evidence in the literature for the importance of polymicrobial infections in which selected microorganisms interact in a synergistic or antagonistic fashion, impacting pathogenesis of periodontal disease [12,13]. Studies have suggested that amplified virulence may emerge during interactions between F. nucleatum and P. gingivalis [12,14]. In previous studies, we showed that P. gingivalis invasion of host cells was enhanced by co-infection with F. nucleatum [15,16]. However, information is still lacking on how F. nucleatum interacts with P. gingivalis or host cells in a way that influences P. gingivalis invasion.
Formation of membrane microdomains, called lipid rafts, segregates specific molecular effectors into functional units for efficient signaling and sorting processes [17]. Lipid rafts act as platforms in protein sorting and signal transduction [18]. The critical role of the host cell plasma membrane in response to pathogens was demonstrated by studies indicating that lipid rafts are the preferred entry sites for several invasive pathogens including Salmonella, Shigella, Listeria, Chlamydia and Campylobacter [19–23]. The lipid-raft route may afford protection from the intracellular degradative lysosomal pathway [24]. Actin and microtubule remodeling, the recruitment of lipid raft components, and host cell phosphorylation activities are considered to be required for cellular invasion by P. gingivalis [3,25–27]. Traditional studies on cellular invasion by periodontal pathogens have generally involved monocultures of bacteria without regard for the polymicrobial nature of oral bacterial communities. Given the importance of lipid rafts in microbe–host cell interaction and the polymicrobial nature of periodontitis, it was of particular interest to investigate if lipid rafts are involved in a periodontal polymicrobial interactions with epithelial cells.
SerB, a haloacid dehydrogenase family phosphoserine phosphatase enzyme is present in the outer membrane of P. gingivalis, and contact with gingival epithelial cells induces secretion into the extracellular milieu [28]. Cell associated and extracellular SerB impacts host cell signal transduction pathways that control the cytoskeletal architecture, and treatment of epithelial cells with SerB induces actin microfilament and tubulin microtubule rearrangements [29,30]. While SerB is important for P. gingivalis invasion and intracellular survival, its contribution to host cell invasion in polymicrobial infection has not been addressed.
The purpose of the present study was to gain insight into the mechanism of fusobacterium-modulated invasion of human gingival epithelial cells by P. gingivalis. In particular, the role of lipid rafts was investigated.
2. Materials and methods
2.1. Bacterial strains and growth conditions
P. gingivalis ATCC 33277 and P. gingivalis W83 (ATCC BAA-308), F. nucleatum TDC100 (a clinical isolate and working strain in our laboratory [15]) were routinely maintained on tryptic soy agar (Difco Laboratories, Detroit, MI) supplemented with 10% defibrinated horse blood, hemin (5 μg/ml) and menadione (0.5 μg/ml) at 37 °C under anaerobic conditions.
An isogenic serB-defective mutant of P. gingivalis ATCC 33277, SerB [30], and complemented cSerB strains were maintained anaerobically at 37 °C on blood agar plates as described previously [30].
2.2. Cells and culture conditions
An established human gingival epithelial cell line, Ca9-22, was purchased from Health Science Research Resources Bank (Osaka, Japan). The Ca9-22 cells were maintained in Eagle’s minimal essential medium (MEM) supplemented with glutamine (0.6 mg/ ml), heat-inactivated 10% fetal calf serum, and gentamicin (10 μg/ ml)/amphotericin B (0.25 μg/ml) (Cascade Biologics, Portland, OR, USA) at 37 °C in 5% CO2 in humidified air.
2.3. Scanning electron microscopy (SEM)
Semi-confluent Ca9-22 cells on 12-mm-diameter glass coverslips were cocultured with P. gingivalis and/or F. nucleatum in MEM without antibiotics for different time period ranging 30–60 min. The multiplicity of infection (MOI) was calculated based on the number of cells per well at confluence. We used MOI 100 for all SEM experiments. After the pre-set time, cells were washed with phosphate buffered saline (PBS) and then fixed in 1.5% glutaraldehyde in 0.1 M Na-cacodylate buffer (pH7.4) for 1 h. They were stained with aqueous osmium tetroxide for 30 min, washed with distilled water, dehydrated with graded alcohols, critical-point dried, and sputter-coated with gold by ESC-101 (ELIONIX, Tokyo, Japan). Ca9-22 cells exposed to each bacterial strain were visualized with SEM (JSM-6340F, JEOL, Tokyo) to directly observe bacterial interaction with host cells.
2.4. Confocal scanning laser microscopy
2.4.1. Mono- or polymicrobial infection
In order to visualize internalization of P. gingivalis or F. nucleatum into Ca9-22, confocal scanning laser microscopy (CSLM) was performed. A dual labeling technique, based on the method described by Inagaki et al. [31], was used to discriminate intracellular from extracellular bacteria. Ca9-22 cells were grown on coverslips in six-well tissue culture plates and infected with P. gingivalis or F. nucleatum for 2 h. Cells were fixed in 4% paraformaldehyde in PBS (Wako Pure Chemical Industries, Osaka, Japan) for 10 min. After washing three times with PBS, any excess of reactive groups paraformaldehyde were quenched with 50 mM NH4Cl in PBS for 10 min at RT. After washing, cells were incubated with a rabbit polyclonal anti-P. gingivalis or anti-F. nucleatum serum diluted 1:500 in PBS–0.5% BSA for 60 min. Following incubation, coverslips were washed three times with PBS and incubated with Alexa Fluor 488 (green fluorescent dye)-conjugated goat anti-rabbit immunoglobulin G (Molecular Probes, Eugene, OR, USA) diluted 1:500 for 30 min to visualize attached bacteria. Internalized bacteria were then stained by first permeabilizing Ca9-22 cells by dipping coverslips in 0.4% Triton X-100 solution for 5 min, then staining with the rabbit anti-P. gingivalis antiserum or anti-F. nucleatum antiserum followed by Alexa Fluor 568 (red fluorescent dye)-coupled goat anti-rabbit IgG (Molecular Probes) diluted 1:500 as described above. Actin filaments were stained with Alexa Fluor 647 (blue fluorescent dye) conjugated to phalloidin (Molecular Probes) for 30 min according to the manufacturer’s recommendations to visualize the cellular cytoskeleton and confirm internalization. P. gingivalis could be distinguished from F. nucleatum on the basis of cellular morphology. Coverslips mounted in an antifading mounting medium (VECTA-SHEILD, Vector Laboratories, Burlingame, CA, USA) were examined by confocal scanning laser microscopy (CSLM) using an LSM5 DUO microscope (Carl Zeiss MicroImaging, Göttingen, Germany) with a 63× oil immersion objective. A series of 20–25 Z-stack images was scanned in increments using excitation wavelengths of 488, 561 and 633 nm. Images were analyzed using ZEN 2008 software (Carl Zeiss).
Where appropriate, Z stacks of the X–Y sections of CLSM were processed to render a three-dimensional (3D) image using ‘Iso Surface’ function of Imaris 7.0.0 (Bitplane AG; Zurich, Switzerland) software. Imaris ‘Spot Detection’ algorithm was applied to allow P. gingivalis to be differentially labeled.
2.4.2. Colocalization with GM1
Confocal microscopy was also used to investigate a possible role of lipid rafts in P. gingivalis internalization of gingival epithelial cells. Ca9-22 cells were infected with P. gingivalis or P. gingivalis with F. nucleatum (MOI = 100:1), and infection was allowed to proceed for 15 or 30 min at 37 °C. Unattached bacteria were removed by washing. After fixation, P. gingivalis was stained with Alexa Fluor 568 goat anti-rabbit IgG following incubation with anti-P. gingivalis or anti-F. nucleatum polyclonal antisera. A lipid raft marker, GM1 ganglioside, was stained with Alexa Fluor 647-labeled cholera toxin subunit B (20 μg/ml, Molecular Probes) for 30 min. The cells were imaged as above, and colocalization of bacteria with GM1 was assessed using Imaris ‘Coloc’ function.
2.4.3. Sequential infection of F. nucleatum and P. gingivalis
For sequential infection experiments, Ca9-22 cells were initially infected by F. nucleatum for 1 h, and then P. gingivalis was added and further incubated for 1 h. To analyze invasion profile, a Z-series with 0.3-μm intervals was scanned and images of the X–Z and Y–Z planes were reconstructed with the orthogonal section tool of the Zen 2008 software. To quantify internalized P. gingivalis or F. nucleatum by CSLM, intracellular bacteria were counted manually. At least five fields and ~200 cells were analyzed for each experiment, and three independent experiments were done. The results were normalized by the control experiments.
2.5. Manipulation of plasma membrane cholesterol
To test the effect of pre-treatment by specific lipid raft disrupter on bacterial invasion, Ca9-22 cells were incubated with 10 mM methyl-β-cyclodextrin (MβCD, Sigma–Aldrich, St. Lois, MO) for 30 min at 37 °C, prior to infection and throughout the period of infection.
2.6. Antibiotic protection assay
Invasion of bacteria was quantitated by a standard antibiotic protection assay as described previously [3]. Briefly, epithelial cells were seeded in 12-well flat-bottom culture plates at a cell density of 2.0 × 105 cells per well. Prior to infection, the cells were washed twice with PBS and incubated further 2 h in MEM without antibiotics. The bacteria were harvested by centrifugation, washed with PBS, and resuspended in MEM at desired concentrations. The bacterial suspensions were added to confluent Ca9-22 monolayers and incubated at 37 °C in 5% CO2 for 2 h. After incubation, unattached bacteria were removed following washing of the monolayers three times with PBS. External adherent cells were then killed by incubating the infected monolayers with MEM containing 200 μg/ml of metronizazole and 300 μg/ml of gentamicin for 1 h. After exposure to antibiotic, monolayers were washed twice with PBS, and lysed in 1 ml of sterile distilled water per well. Cells were incubated for 30 min, during which they were disrupted by repeated pipetting. Lysates were serially diluted and plated on blood agar plates supplemented with hemin and menadione, and incubated anaerobically at 37 °C for 10 days. Colony forming units of invasive organisms were then enumerated. Invasion efficiency was expressed as the percentage of the initial inoculum recovered after antibiotic treatment and Ca9-22 lysis. P. gingivalis and F. nucleatum are distinguishable by color and morphology of their colonies.
In our previous experiments, we found that there was a modest increase in the invasion efficiencies of P. gingivalis with increasing MOI. Invasion reached a plateau at about 100 MOI [15]. Thus, we used MOI of 100 in all subsequent experiments.
Polymicrobial infection of Ca9-22 cells was performed as described previously [15,16]. Briefly, polymicrobial inocula were prepared by mixing equal volumes (1 × 107 cells per well) of bacterial suspensions and were incubated for 5 min at RT prior to initiating the infection. For the monomicrobial control infection, bacterial suspensions were mixed with an equal volume of MEM.
2.7. Data and statistical analyses
All experiments were performed in duplicate or triplicate for each condition and repeated at least three times. Statistical comparisons were performed using a software package (InStat 3.0, GraphPad Software, La Jolla, CA, USA). P-values less than 0.05 were considered statistically significant.
3. Results
3.1. Interaction of P. gingivalis and F. nucleatum with gingival epithelial cells
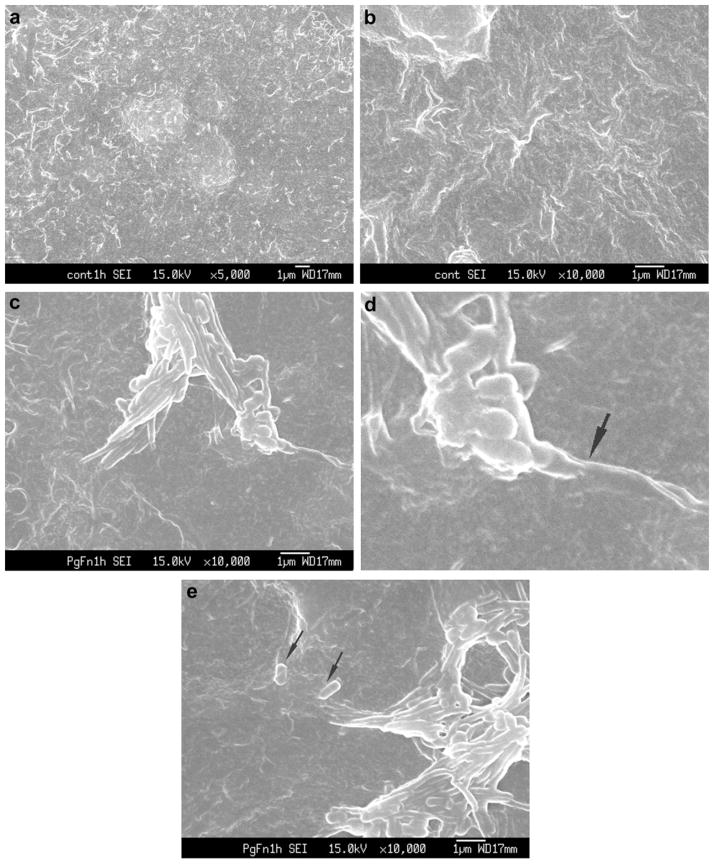
SEM revealed P. gingivalis and F. nucleatum adhering to the surface of Ca9-22 cells after 30–60 min co-infection. P. gingivalis and F. nucleatum consortia partially penetrating the host cell surface were also observed (Fig. 1). F. nucleatum appeared to invade Ca9-22 cells in a polar manner (Fig. 1c, d), rather than along the length of the bacterium. Although P. gingivalis was often closely associated with F. nucleatum, some P. gingivalis cells were independently located in close proximity to the entry site of F. nucleatum (Fig. 1e).
Fig. 1.
(a) and (b) SEM of control uninfected Ca9-22 cells demonstrated that cellular projections (surface roughness) were not due to the presence of bacteria. ×5000 and ×10,000. (c) Ca9-22 co-infected with P. gingivalis ATCC 33277 and F. nucleatum TDC 100 for 1 h. P. gingivalis and F. nucleatum were commonly forming consortia and appear to be penetrating into the host cell. ×10,000 (d) Enlarged view; membrane blebbing was observed (arrow). (e) Near the entry site of F. nucleatum–P. gingivalis consortia, independent P. gingivalis (arrow) was observed.
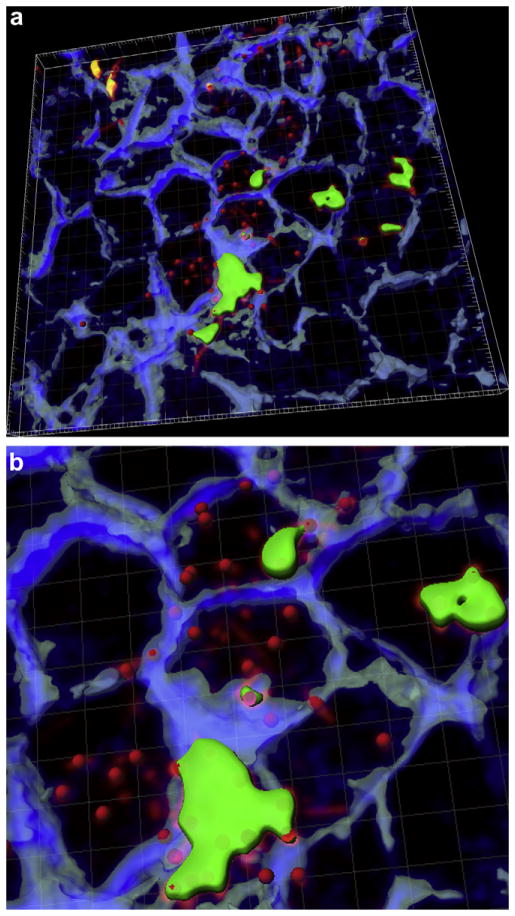
Internalization of P. gingivalis and F. nucleatum was also confirmed by CSLM using dual labeling technique. It was commonly observed that Ca9-22 cells contained both P. gingivalis and F. nucleatum (Fig. 2). This co-internalization was even more pronounced with P. gingivalis W83 (data not shown). However, in case of co-infection with P. gingivalis 33277, some of the host cells solely contained P. gingivalis.
Fig. 2.
(a) Invasion of P. gingivalis or F. nucleatum into gingival epithelial cells. Z stacks of the X–Y sections of CSLM were processed to render a 3D image using ‘Iso Surface’ functions of Imaris 7.0.0 software. Internalized P. gingivalis ATCC 33277 and F. nucleatum TDC 100 (shown as longer, fusiform shape) were stained red, while extracellular bacteria were shown green-yellow. Anti-P. gingivalis and anti-F. nucleatum antisera were used. ‘Spot Detection’ algorithm of Imaris software was used to further differentiate P. gingivalis from F. nucleatum. The host cell cytoskeleton stained with phalloidin appeared blue. (b) Enlarged view. (For interpretation of color referred in this figure legend, the reader is referred to web version of the article.)
3.2. Colocalization of P. gingivalis and F. nucleatum with lipid rafts
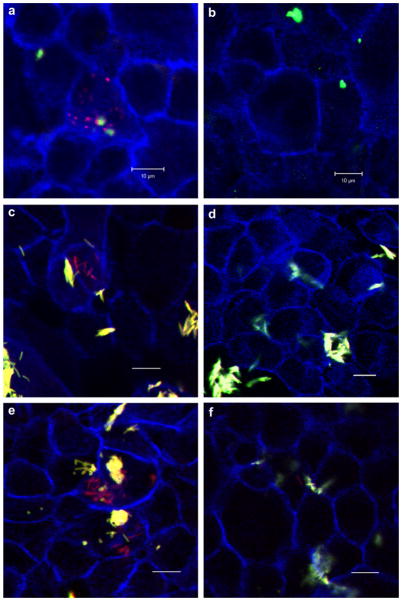
P. gingivalis or F. nucleatum (appearing green) attached to Ca9-22 cells, and revealed evident colocalization between GM1-containing membrane microdomains (red) and bacteria (yellow) (Fig. 3). We found that approximately 60% of P. gingivalis and/or F. nucleatum colocalized with GM1. This was the case in almost all the ~20 fields analyzed in three independent experiments.
Fig. 3.
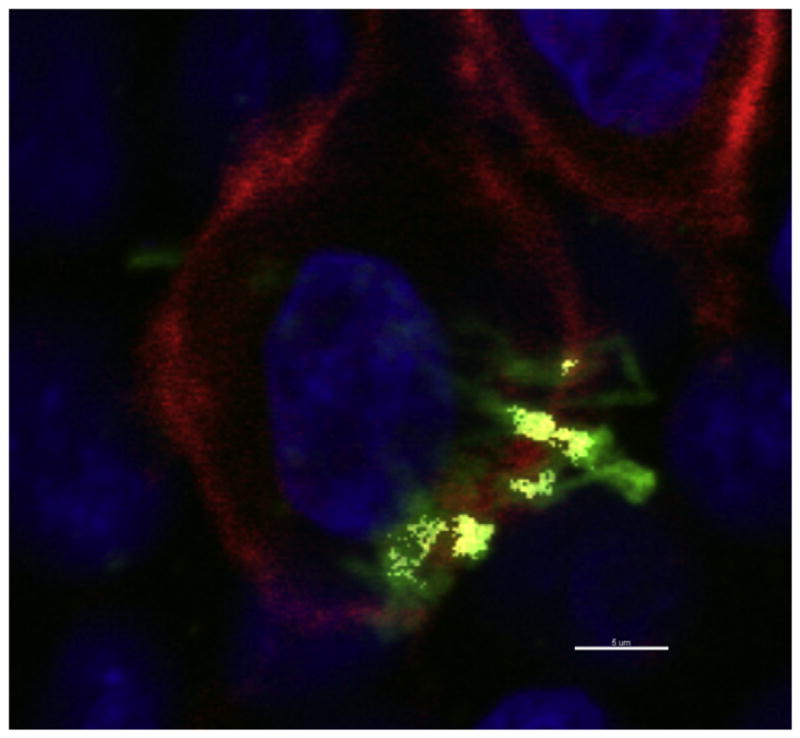
P. gingivalis and F. nucleatum colocalizes with lipid rafts. Ca9-22 cells, cultured on glass coverslips, were exposed to P. gingivalis ATCC 33277 and F. nucleatum TDC 100 (MOI = 100:1). Dual species infection was allowed to proceed for 15 min and unattached bacteria were removed by washing, followed by cell fixation and staining for bacteria (anti-P. gingivalis and anti-F. nucleatum antibodies + Alexa Fluor 488- conjugated goat anti-rabbit IgG; green), and GM1 (lipid raft marker) with Alexa Fluor 594-labeled CTB (red). CSLM revealed apparent colocalization between P. gingivalis and F. nucleatum (appearing yellow) and GM1-containing membrane microdomains (red). DAPI (Blue). Bar = 5 μm.
Some bacteria showed no apparent colocalization with GM1, suggesting that gingival epithelial cell – P. gingivalis and F. nucleatum interactions may also involve non-raft membrane regions.
3.3. Effect of cholesterol depletion on the bacterial invasion
In order to further assess the involvement of lipid rafts in P. gingivalis invasion in co-infection with F. nucleatum, methyl-beta-cyclodextrin (MβCD)was used to deplete Ca9-22 cells of cholesterol and disrupt lipid rafts. MβCD-treated, MβCD-untreated cells were then infected with P. gingivalis and/or F. nucleatum (MOI = 100:1) and viable internalized bacteria (colony forming units, CFU) were enumerated using an antibiotic protection-based recovery assay. With MβCD treatment, P. gingivalis or F. nucleatum CFU recovered was reduced to 13–33% of untreated control (Table 1). The MβCD treatment did not significantly affect the cell viability.
Table 1.
Effect of MβCD on bacterial invasion of Ca9-22.
| Infection | Invasion of Ca9-22 (% of untreated control) |
|---|---|
| Mono-infection | |
| P. gingivalis | 33.4 ± 18.4* |
| F. nucleatum | 13.3 ± 14.5* |
| Dual-infection | |
| P. gingivalis | 14.4 ± 11.4* |
| F. nucleatum | 27.2 ± 15.9* |
Ca9-22 cells were infected by P. gingivalis 33277 and/or F. nucleatum TDC 100 for 2 h in vitro. An antibiotic protection assay was used to quantify intracellular bacteria. In order to disrupt lipid rafts, cells were pretreated for 30 min with 10 mM MβCD. Values given as means ± standard deviations of triplicate independent determinations from a typical experiment.
Statistically significantly different from untreated control (p < 0.001) by ANOVA with Bonferroni multiple comparisons test.
These results correlated with CSLM observations; pre-treatment of Ca9-22 cells significantly abrogated bacterial internalization in monomicrobial infections (Fig. 4a–d) and in dual-infections (Fig. 4e, f).
Fig. 4.
Effect of cholesterol depletion on the bacterial invasion assessed by CSLM. Ca9-22 cells were incubated with DMSO (control) or 10 mM methyl-beta-cyclodextrin (MβCD) or for 30 min at 37 °C, prior to infection. (a) P. gingivalis 33277 control, (b) P. gingivalis + MβCD, (c) F. nucleatum TDC 100 control, (d) F. nucleatum TDC 100 + MβCD, (e) P. gingivalis + F. nucleatum control, (f) P. gingivalis + F. nucleatum + MβCD. Bar = 10 μm. Internalized bacteria were stained red, while extracellular bacteria were shown green-yellow. For dual-infection, anti-P. gingivalis and anti-F. nucleatum antisera were used. The host cell cytoskeleton stained with phalloidin appeared blue. (For interpretation of color referred in this figure legend, the reader is referred to web version of the article.)
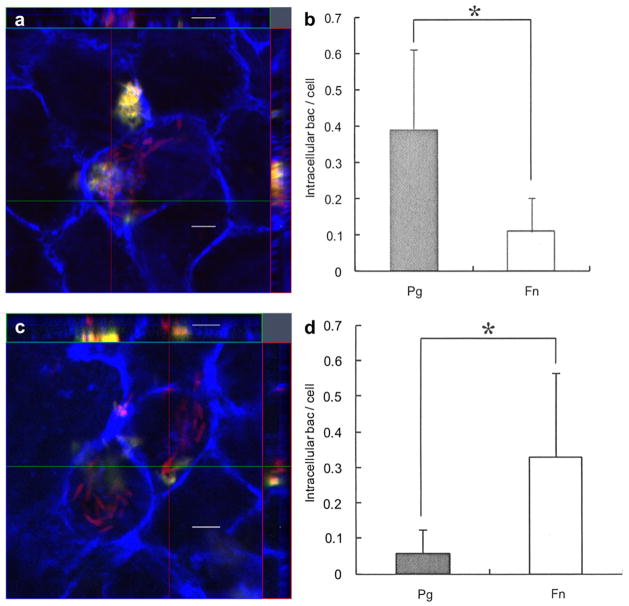
3.4. Effect of F. nucleatum pre-incubation on P. gingivalis invasion
To assess the effect of F. nucleatum pre-incubation on P. gingivalis invasion, Ca9-22 cells were sequentially infected by each species and examined by CSLM. In contrast to simultaneous infection (Fig. 5a, b), pre-infection of Ca9-22 with F. nucleatum TDC 100 resulted in approximately 80% reduction in P. gingivalis invasion (Fig. 5c, d). In this case, the intracellular bacteria were predominantly F. nucleatum.
Fig. 5.
Effect of F. nucleatum internalization on P. gingivalis invasion. (a) Ca9-22 cells were co-infected with F. nucleatum TDC 100 and P. gingivalis 33277 for 2 h. It was common to find both P. gingivalis and F. nucleatum intracellulary, but some cells exclusively contained P. gingivalis (data not shown). Internalized bacteria were stained red, while extracellular bacteria were shown green-yellow. The host cell cytoskeleton stained with phalloidin appeared blue. (b) Quantification of intracellular bacteria. Pg: P. gingivalis 33277, Fn: F. nucleatum TDC 100. Values are shown as means ± standard deviations. At least five fields and ~200 cells were analyzed for each experiment, and three independent experiments were done. Statistical significance between invasion levels was measured by using Mann–Whitney U test. *p < 0.05. (c) Ca9-22 cells were first infected with F. nucleatum TDC 100 for 1 h, then P. gingivalis 33277 was added and further incubated for another hour. In this sequential infection experiment, intracellular bacteria were predominantly F. nucleatum. Internalized bacteria were stained red, while extracellular bacteria were shown green-yellow. Anti-P. gingivalis and anti-F. nucleatum antisera were used. The host cell cytoskeleton stained with phalloidin appeared blue. (d) Quantification of intracellular bacteria. Values are shown as means ± standard deviations. *p < 0.05. (For interpretation of color referred in this figure legend, the reader is referred to web version of the article.)
3.5. Effect of P. gingivalis SerB on dual infection
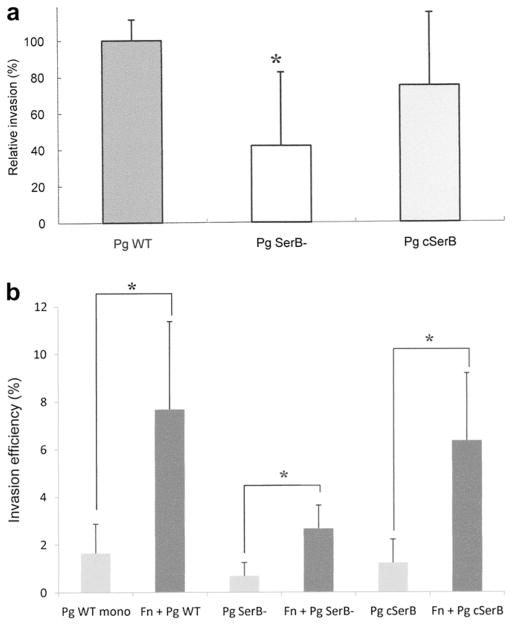
In order to test the effect of a key effector of P. gingivalis on dual infection, an antibiotic protection assay was performed using the SerB mutant and complemented strains of P. gingivalis 33277 [30]. The invasion efficiency for P. gingivalis SerB mutant strain was approximately 40% of that for P. gingivalis wild type strain (Fig. 6a).
Fig. 6.
(a) Invasion of gingival epithelial cells (Ca9-22) by P. gingivalis 33277 wild type strain, its isogenic SerB mutant and complemented strains. Statistical significance between invasion levels was measured by using Mann–Whitney U test. *p < 0.05. Pg WT: P. gingivalis 33277, Pg SerB-: P. gingivalis SerB mutant, Pg cSerB: Complemented SerB mutant. (b) Effect of co-infection with F. nucleatum on invasion of gingival epithelial cells by P. gingivalis 33277, its isogenic mutant and complemented strains. Statistical significance between invasion levels was measured by using Kruskal–Wallis test with Dunn’s multiple comparisons test. *p < 0.001. Fn: F. nucleatum TDC 100.
In a dual infection experiment, co-infection with F. nucleatum TDC100 significantly enhanced host cell invasion by P. gingivalis 33277, its SerB mutant and complemented strains (Fig. 6b).
4. Discussion
In order to gain insight into interactions between periodontal pathogens and host cells, we set out to investigate the host cell invasion by P. gingivalis in prototype polymicrobial consortia. Previous studies from our laboratory showed that F. nucleatum enhances P. gingivalis invasion of human gingival epithelial and aortic endothelial cells [15,16]. In this study, we further examined the underlying mechanism(s) involved in modulation by looking specifically at the role of membrane microdomains. Here we demonstrate that a lipid raft-dependent mechanism is involved in fusobacterium-modulated P. gingivalis internalization into gingival epithelial cells.
There are at least two advantages to bacterial entry through lipid rafts: avoidance of the intracellular degradative pathway, which would lead to disintegration of bacteria, and triggering of the cell signaling that leads to membrane ruffling and rearrangement of the cytoskeleton, which are both required for entry of bacteria [32]. P. gingivalis utilizes actin and microtubule remodeling, the recruitment of lipid raft components, and host cell phosphorylation activity for cellular invasion [3,26,27,32,33]. Once inside the cell, P. gingivalis evades lysosomes and survives in the cytoplasm, where it remains viable for extended periods of time and even replicates [3,34,35]. P. gingivalis may co-operate with F. nucleatum for more efficient entry into host cells, and lipid rafts play an important role in this interaction, although intracellular fate of P. gingivalis involved in polymicrobial infection remains to be investigated. In CSLM experiments, some bacteria did not show any colocalization with GM1 containing microdomains. Moreover, the treatment with MβCD, a drug that sequesters membrane cholesterol and disrupts lipid rafts, did not completely abrogate the bacterial internalization in the antibiotic protection assay. It is important to note that MβCD has pleiotropic effects; the treatment may also affect non-raft-associated proteins as a result of the presence of cholesterol in non-raft domains [36,37]. It is difficult to draw a definite conclusion since we did not assess the efficiency of cholesterol depletion in the present experiment. However, taken together, these data suggest that the gingival epithelial cell – P. gingivalis and F. nucleatum interactions may also involve non-raft membrane regions.
Our SEM observation showed multiple F. nucleatum with adherent P. gingivalis cells in the process of host cell penetration. This may suggest that enhanced P. gingivalis internalization occur via adhesion to invading F. nucleatum. The ability of F. nucleatum to directly transport other bacteria into oral epithelial cells has been shown in a previous study [38]. In case of enteropathogens, Campylobacter jejuni was shown to promote the internalization of non-invasive bacteria into enterocytes via a lipid raft-mediated transcellular process [39]. However, in this case, it was suggested that Campylobacter did not directly ‘shuttle’ non-invasive bacteria into the host cells. In our CSLM experiments, it was often observed that Ca9-22 cells contained both P. gingivalis and F. nucleatum (Fig. 2). This co-internalization was even more pronounced with P. gingivalis W83 (data not shown). We have previously reported that co-infection with F. nucleatum strains resulted in 2–10-fold increase in invasion of host cells by P. gingivalis W83, which is minimally invasive in monomicrobial infection [15]. However, in case of co-infection with P. gingivalis 33277, some of the host cells contained P. gingivalis exclusively. Also, in the SEM observation, some P. gingivalis cells were independently located in close proximity to the entry site of F. nucleatum. At this time, we cannot precisely determine whether F. nucleatum internalization is a perquisite for enhanced P. gingivalis internalization. It is possible that mechanism(s) involved in fusobacterial enhancement of host cell invasion is different for these two species.
F. nucleatum was shown to invade human oral epithelial cells via a “zipper” mechanism and require the involvement of actins, microtubules, signal transduction, protein synthesis, and energy metabolism of the epithelial cell, as well as protein synthesis by F. nucleatum [9]. We reasoned that, if invasion mechanism(s) exploited by F. nucleatum is different from that of P. gingivalis, preincubation of F. nucleatum would possibly interfere with that process. To test that possibility, Ca9-22 cells were preincubated with F. nucleatum 1 h, followed by co-incubation with P. gingivalis for another hour. The Ca9-22 cells were then processed for visualization by confocal microscopy. Compared to the controls simultaneously incubated with P. gingivalis and F. nucleatum, preincubation with F. nucleatum inhibited P. gingivalis invasion by ~80%. This suggested that a strategy for cellular invasion utilized by F. nucleatum is different from that by P. gingivalis. Predominant F. nucleatum entry may turn off the signals needed for subsequent P. gingivalis entry.
The P. gingivalis serine phosphatase, SerB, is involved in internalization and survival in gingival epithelial cells [28,30]. SerB is functional within epithelial cells, interacting with cytoplasmic GAPDH and HSP90, and modulating microtubule dynamics. SerB also regulates the dynamics of the actin cytoskeleton and inhibits the production of IL-8 [29]. Given the importance of SerB in P. gingivalis-host cell interaction, it was of interest to delineate the role of SerB in the fusobacterial enhancement of P. gingivalis invasion. In an antibiotic protection assay, co-infection with F. nucleatum TDC 100 significantly enhanced host cell invasion by P. gingivalis 33277, its SerB mutant and complemented strains. These results suggested that the P. gingivalis serine phosphatase, SerB, does not play a major role in this fusobacterial enhancement of P. gingivalis invasion. Possible role of P. gingivalis FimA remains to be investigated.
It is possible that the interaction between F. nucleatum and host cells plays an important role in the fusobacterial enhancement of P. gingivalis invasion. The question still remains as to which factor(s) of F. nucleatum is responsible for this modulation. Our previous experiments using metabolic inhibitors and antisera revealed that the invasion profile is different between mono-infection and polyinfection [15]. We also suggested that physical interactions between the co-infectants and/or the host cells are necessary, since co-incubation with methanol-fixed or heat-killed F. nucleatum appeared to partially enhance the P. gingivalis invasion [16]. It was postulated that F. nucleatum may possess lectin-like and nonlectin-like adhesions for binding to various partners [40–43]. Several putative adhesion molecules have been suggested for F. nucleatum for involvement in binding to other microbial species [6,42,44–48]. Further studies are necessary to clarify the mechanism(s) by which F. nucleatum modulates P. gingivalis invasion of host cells.
In summary, we conclude that a lipid raft-dependent entry platform is at least one of the potential mechanisms involved in fusobacterium-modulated P. gingivalis internalization into gingival epithelial cells. This study increases our understanding of intracellular invasion strategies utilized by selected periodontal pathogens and gives new insight into intricate and significant interactions that take place between microbial species as they coexist in the host and during the infectious processes.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research (C) 22592317 from Japan Society for Promotion of Science (to AS), Oral Health Science Center Grant hrc 8 from Tokyo Dental College, and a Project for Private Universities: matching fund subsidy from Ministry of Education, Culture, Sports, Science and Technology of Japan, 2010–2012, and NIH R01DE11111 (to RL).
References
- 1.Hajishengallis G. Porphyromonas gingivalis–host interactions: open war or intelligent guerilla tactics? Microbe Infect. 2009;11:637–45. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Michel R, Cohen J, DeCarlo A, Kozarov E. Intracellular survival and vascular cell-to-cell transmission of Porphyromonas gingivalis. BMC Microbiol. 2008;8:26. doi: 10.1186/1471-2180-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–85. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshpande RG, Khan MB, Attardo Genco C. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun. 1998;66:5337–43. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Invasion of human coronary artery cells by periodontal pathogens. Infect Immun. 1999;67:5792–8. doi: 10.1128/iai.67.11.5792-5798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000–9. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 7.Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect Immun. 2001;69:5736–41. doi: 10.1128/IAI.69.9.5736-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191:6804–11. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han YW, Shi W, Huang GTJ, Kinder Haake S, Park N-H, Kuramitsu H, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–6. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabija-Wolter G, Cimpan M, Costea DE, Johannessen AC, Sørnes S, Neppelberg E, et al. Fusobacterium nucleatum enters normal human oral fibroblasts in vitro. J Periodontol. 2009;80:1174–83. doi: 10.1902/jop.2009.090051. [DOI] [PubMed] [Google Scholar]
- 11.Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontology 2000. 2010;52:68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feuille F, Ebersole JL, Kesavalu L, Stepfen MJ, Holt SC. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect Immun. 1996;64:2094–100. doi: 10.1128/iai.64.6.2094-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kesavalu L, Sathishkumar S, Bakhavatchalu V, Mattews C, Dawson D, Steffen M, et al. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007;75:1704–12. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebersole JL, Feuille F, Kesavalu L, Holt SC. Host modulation of tissue destruction caused by periodontopathogens: effects on a mixed microbial infection composed of Porphyromonas gingivalis and Fusobacterium nucleatum. Microb Pathog. 1997;23:23–32. doi: 10.1006/mpat.1996.0129. [DOI] [PubMed] [Google Scholar]
- 15.Saito A, Inagaki S, Kimizuka R, Okuda K, Hosaka Y, Nakagawa T, et al. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2008;54:349–55. doi: 10.1111/j.1574-695X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 16.Saito A, Inagaki S, Ishihara K. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb Pathog. 2009;47:329–33. doi: 10.1016/j.micpath.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Seveau S, Pizarro-Cerda J, Cossart P. Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbe Infect. 2007;9:1167–75. doi: 10.1016/j.micinf.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–72. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 19.Knodler L, Vallance B, Hensel M, Jäckel D, Finlay BB, Steele-Mortimer O. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol Microbiol. 2003;49:685–704. doi: 10.1046/j.1365-2958.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 20.Wooldridge KG, Williams PH, Ketley JM. Host signal transduction and endocytosis of Campylobacter jejuni. Microb Pathog. 1996;21:299–305. doi: 10.1006/mpat.1996.0063. [DOI] [PubMed] [Google Scholar]
- 21.Seveau S, Bierne H, Giroux S, Prevost M, Cossart P. Role of lipid rafts in E-cadherin- and HGFRHGFR/Met-mediated entry of Listeria monocytogenes into host cells. J Cell Biol. 2004;166:743–53. doi: 10.1083/jcb.200406078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jutras I, Abrami L, Dautry-Varsat A. Entry of the lymphogranuloma venereum strain of Chlamydia trachomatis into host cells involves cholesterol-rich membrane domains. Infect Immun. 2003;71:260–6. doi: 10.1128/IAI.71.1.260-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafont F, Abrami L, van der Goot FG. Bacterial subversion of lipid rafts. Curr Opin Microbiol. 2004;7:4–10. doi: 10.1016/j.mib.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Wang M, Hajishengallis G. Lipid raft-dependent uptake, signaling, and intracellular fate of Porphyromonas gingivalis in mouse macrophages. Cell Microbiol. 2008;10:2029–42. doi: 10.1111/j.1462-5822.2008.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamont RJ, Yilmaz O. In or out: the invasiveness of oral bacteria. Periodontol 2000. 2002;30:61–9. doi: 10.1034/j.1600-0757.2002.03006.x. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe K, Yilmaz O, Nakhjiri SF, Belton CM, Lamont RJ. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect Immun. 2001;69:6731–7. doi: 10.1128/IAI.69.11.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuda K, Furuta N, Inaba H, Kawai S, Hanada K, Yoshimori T, et al. Functional analysis of alpha5beta1 integrin and lipid rafts in invasion of epithelial cells by Porphyromonas gingivalis using fluorescent beads coated with bacterial membrane vesicles. Cell Struct Funct. 2008;33:123–32. doi: 10.1247/csf.08012. [DOI] [PubMed] [Google Scholar]
- 28.Bainbridge B, Verma RK, Eastman C, Yehia B, Rivera M, Moffatt C, et al. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response and induction of alveolar bone resorption in rats. Infect Immun. 2010;78:4560–9. doi: 10.1128/IAI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasegawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect Immun. 2008;76:2420–7. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci U S A. 2006;103:11027–32. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect Immn. 2006;74:5023–8. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manes S, del Real G, Martinez- AC. Pathogens: raft hijackers. Nat Rev Immunol. 2003;3:557–68. doi: 10.1038/nri1129. [DOI] [PubMed] [Google Scholar]; Sandros J, Madianos PN, Papapanou PN. Cellular events concurrent with Porphyromonas gingivalis invasion of oral epithelium in vitro. Eur J Oral Sci. 1996;104:363–71. doi: 10.1111/j.1600-0722.1996.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 33.Yilmaz O, Young PA, Lamont RJ, Kenny GE. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology. 2003;149:2417–26. doi: 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 34.Papapanou PN, Sandros J, Lindberg K, Duncan MJ, Niederman R, Nannmark U. Porphyromonas gingivalis may multiply and advance within stratified human junctional epithelium in vitro. J Periodontal Res. 1994;29:374–5. doi: 10.1111/j.1600-0765.1994.tb01237.x. [DOI] [PubMed] [Google Scholar]
- 35.Madianos PN, Papapanou PN, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalis FDC 381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–4. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrami L, van der Goot FG. Plasma membrane microdomains act as concentration platforms to facilitate intoxication by aerolysin. J Cell Biol. 1999;147:175–84. doi: 10.1083/jcb.147.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafont F, Goot FGVD. Bacterial invasion via lipid rafts. Cellular Microbiol. 2005;7:613–20. doi: 10.1111/j.1462-5822.2005.00515.x. [DOI] [PubMed] [Google Scholar]
- 38.Edwards AM, Grossman TJ, Rudney JD. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun. 2006;74:654–62. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalischuk LD, Douglas Inglis G, Buret AG. Campylocacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog. 2009;1:2. doi: 10.1186/1757-4749-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangan DF, Novak MJ, Vora SA, Mourad J, Kriger PS. Lectinlike interactions of Fusobacterium nucleatum with human neutrophils. Infect Immun. 1989;57:3601–11. doi: 10.1128/iai.57.11.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuttle RS, Strubel NA, Mourad J, Mangan DF. A non-lectin-like mechanism by which Fusobacterium nucleatum 10953 adheres to and activates human lymphocytes. Oral Microbiol Immunol. 1992;7:78–83. doi: 10.1111/j.1399-302x.1992.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 42.Shaniztki B, Hurwitz D, Smorodinsky N, Ganeshkumar N, Weiss E. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect Immun. 1997;65:5231–7. doi: 10.1128/iai.65.12.5231-5237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss EI, Shaniztki B, Dotan M, Ganeshkumar N, Kolenbrander PE, Metzger Z. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol Immunol. 2000;15:371–7. doi: 10.1034/j.1399-302x.2000.150606.x. [DOI] [PubMed] [Google Scholar]
- 44.Murray PA, Kern DG, Winkler JR. Identification of a galactose-binding lectin on Fusobacterium nucleatum FN-2. Infect Immun. 1988;56:1314–9. doi: 10.1128/iai.56.5.1314-1319.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufman J, DiRienzo JM. Isolation of a corncob (coaggregation) receptor polypeptide from Fusobacterium nucleatum. Infect Immun. 1989;57:331–7. doi: 10.1128/iai.57.2.331-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinder SA, Holt SC. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J Bacteriol. 1993;175:840–50. doi: 10.1128/jb.175.3.840-850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo M, Han YW, Sharma A, Nardin ED. Identification and characterization of human immunoglobulin G Fc receptors of Fusobacterium nucleatum. Oral Microbiol Immunol. 2000;15:119–23. doi: 10.1034/j.1399-302x.2000.150208.x. [DOI] [PubMed] [Google Scholar]
- 48.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. Identification and characterization of a novel adhesin unique to oral Fusobacteria. J Bacteriol. 2005;187:5330–40. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]