Abstract
Genetic variation in the IL28B region has been associated with sustained virological response (SVR) rates in chronic hepatitis C (CHC) patients treated with peginterferon-α and ribavirin. We hypothesized that IL28B polymorphism is associated with intrahepatic expression of interferon-stimulated genes (ISGs), known to influence treatment outcome. IL28B genotyping (rs12979860) and whole-genome RNA expression were performed using liver biopsies from 61 North American CHC patients. After correction for multiple testing (false discovery rate < 0.10), 164 transcripts were found to be differentially expressed by IL28B-type. The interferon signaling pathway was the most enriched canonical pathway differentially expressed by IL28B-type (p < 10−5), with most genes showing higher expression in livers of individuals carrying the poor-response IL28B-type. In 25 patients for which treatment response data were available, IL28B-type was associated with SVR (p = 0.0054). ISG expression was also associated with SVR; however, this was not independent of IL28B-type. Analysis of miR-122 expression in liver biopsies showed reduced miR-122 levels associated with poorer treatment outcome, independently of IL28B-type. No association was observed between IL28B-type and levels of liver IL28B or IL28A mRNA expression. IL28B protein sequence variants associated with rs12979860 were therefore investigated in vitro: no differences in ISG induction or inhibition of HCV replication were observed in Huh7.5 cells.
Conclusion
The good response IL28B variant was strongly associated with lower level ISG expression. The results suggest that IL28B genotype may explain the relationship between hepatic ISG expression and HCV treatment outcome, and this is independent of miR-122 expression. IL28B-type was not associated with intrahepatic IL28B mRNA expression in vivo. Further investigation of the precise molecular mechanism(s) by which IL28B genetic variation influences HCV outcomes is warranted.
Keywords: interferon lambda, hepatitis C virus, gene expression
Three initial independent genome wide association studies (GWAS) have recently identified variation in the region of the IL28B gene on chromosome 19 to be a key predictor of peginterferon-α (pegIFN) and ribavirin (RBV) treatment response in patients chronically infected with genotype 1 hepatitis C virus(1–3). This region was subsequently also identified to be important for spontaneous clearance of HCV infection (4, 5), suggesting a role in the regulation of endogenous as well as exogenous type 1 IFN responses. However, the causal variant and the biological mechanism responsible for this association have yet to be unraveled.
In addition, in a GWAS analysis of pretreatment HCV viral load, the paradoxical observation was made that the good response variant was associated with a higher level of circulating HCV RNA(1). Although the differences in median viral load between the respective IL28B genotypes were small and perhaps of limited clinical significance, the finding was surprising, given that most therapeutic studies have noted an association between high viral load and reduced response to pegIFN and RBV treatment(6). A reasonable hypothesis might involve differential levels of intrahepatic interferon-stimulated gene (ISG) expression according to IL28B genotype, where the good response variant was associated with lower levels of intrahepatic ISG expression, and therefore slightly higher levels of circulating serum HCV RNA.
There has been recent interest in the relationship between intrahepatic ISG expression in patients chronically infected with HCV, and response to IFN-based treatment regimens. A growing body of evidence, in both humans and the chimpanzee model, supports a paradigm in which IFN non-response is associated with pre-activated, perhaps maximally or perhaps inappropriately stimulated intrahepatic ISG expression(7–11). In contrast, quiescent liver ISGs pre-treatment have been associated with sensitivity to exogenous IFN therapy and viral eradication. The mechanistic basis for this association is unknown, and it remains unclear whether it is driven by the host, the virus, or both.
We hypothesized that these patterns of intrahepatic ISG expression reflect differences in host innate antiviral immunity, and that a key determinant is the recently identified IL28B polymorphism. The current analysis summarizes results of global liver gene expression profiling according to IL28B genotype, in a well-characterized cohort of North American chronic hepatitis C patients, and in vitro assays of cytokine potency according to IL28B genotype.
Materials and Methods
Patient Population
Sixty-one patients were recruited for this study; all attended the Liver Clinic at Beth Israel Deaconess Medical Center (Harvard University, Boston, MA, USA) for the management of HCV mono-infection, and had consented to storage of liver biopsy specimens and clinical data for research purposes. Liver biopsy in all patients was performed at this institution, and liver specimens were immediately placed in RNA later (Qiagen, Valencia, CA, USA) before storage at −80°C. Forty-one patients were treatment-naïve at the time of liver biopsy, and 20 were non-responders (NR) to prior IFN-based treatment regimens (standard / pegIFN ± RBV). Detailed description of initial viral response was not available for all patients, and since sample size was small, we grouped non-responders and relapsers as treatment failures and compared them to patients who attained SVR (responders). All patients had stopped antiviral therapy at least 6 months prior to biopsy. Written informed consent was obtained from all patients. The protocol was approved by the Institutional Review Board at Beth Israel Deaconess Medical Center.
At the time of liver biopsy, all patients had blood taken for serum liver function tests, a quantitative measure of serum/plasma HCV RNA performed using the Roche PCR assay, with a lower limit of detection of 50 IU/mL. HCV genotyping was performed using the line probe assay in all patients. All biopsy samples were evaluated by a single hepatopathologist and scored for stage of fibrosis and grade of inflammation using the METAVIR scoring system(12).
DNA / RNA extraction
Total RNA and genomic DNA were extracted from liver biopsies using the QIAgen RNeasy Mini Kit (QIAgen, Valencia, CA, USA) with a modification of the manufacturer’s protocol for DNA extraction after elution of RNA (http://www.natureprotocols.com/2009/08/17/a_useful_procedure_to_isolate.php).
IL28B genotyping
The region on chromosome 19 associated with HCV treatment response contains multiple SNPs in linkage disequilibrium around the IL28B gene(1–3). We selected the most strongly associated SNP, rs12979860, which is a bi-allelic polymorphism (CC, CT, TT) that has been associated with treatment response in both Caucasians and African Americans (1). Genotyping was performed using the ABI TaqMan allelic discrimination kit and the ABI7900HT sequence Detection System (Applied Biosystems, Carlsbad, CA, USA) in 5uL reaction volume using standard Taqman Universal PCR conditions. Primer and probe sequences were: Forward Primer 5’-GCCTGTCGTGTACTGAACCA-3’, Reverse Primer 5’-GCGCGGAGTGCAATTCAAC-3’, Probe (C allele) 5’-VIC-TGGTTCGCGCCTTC-3’, Probe (T allele) 5’-FAM-CTGGTTCACGCCTTC-3’. Genotyping was conducted in a blinded fashion relative to HCV treatment status and other patient or treatment response characteristics.
Expression Analysis
Approximately 200 ng total RNA was amplified and transcribed to cRNA, and hybridized to Illumina HT-12 Expression BeadChip per the manufacturer’s protocol (Illumina Inc., San Diego, CA, USA), targeting around 25,000 annotated genes with more than 48,000 probes.
Quantitative PCR for Targeted Gene Expression Measurements
A custom Taqman MGB assay for gene expression was designed to achieve 1000-fold specificity for IL28B vs. the close homolog IL28A (see Supplemental Figure S1). Primer and probe sequences are provided in the Supplemental Materials. Applied Biosystems Taqman Assays-on-Demand were used to detect expression of selected ISGs: IFI6, IFI27, and ISG15. miR-122 and RNA U6 determination was made using a specific Taqman MicroRNA Assay(Applied Biosystems, Foster City, CA, USA). Approximately 1 μg of total RNA from liver biopsies was reverse transcribed using the High-Capacity cDNA Synthesis Kit (Applied Biosystems, Foster City, CA, USA), using either an miRNA-specific primer (for miR-122 and U6) or random hexamer primers. Approximately 100 ng cDNA was used per reaction in a total volume of 10 μL, and duplicate reactions were performed for each sample. Data were transformed using the ΔΔCT method normalizing to GAPDH or beta-actin as reference genes(for miR-122, U6 RNA was used as a reference), and further normalized to the lowest-expressing sample for comparison.
In Vitro Characterization of Coding IL28B Variants
Recombinant proteins were produced by cloning expression-optimized (Blue Heron Biotechnology) IL28B cDNA sequences corresponding to the two alternative alleles at the rs8103142 IL28B coding variant into pGEX-6P-2. GST fusion proteins were expressed in BL-21 cells and purified using glutathione-sepharose according to the manufacturer (GE Healthcare). Protein integrity and concentration were determined by SDS-PAGE.
For purification from human cells, the IL28B cDNA sequences corresponding to the two alternative alleles at the rs8103142 variant were cloned into pcDNA5/FRT/TO and stably integrated into Flp-in-Trex 293 cells(Invitrogen, Carlsbad, CA, USA). After 24 hours of culture of confluent stable cells in serum-free media containing 1 μg/ml doxycycline, the supernatant was collected and concentrated using centricon devices (Millipore, Billerica, MA , USA). Total protein was measured by the Bradford method, and serial dilutions of concentrate were used to treat Huh7.5 cells hosting an HCV replicon as described previously (13). HCV replicon RNA measurement was performed using a custom Taqman real-time PCR method, normalizing to GAPDH mRNA expression. Primer and probe sequences for HCV RNA determination were: Forward Primer 5’-TCAATAGGGTGGCTTCATGCCTCA-3’, Reverse Primer 5’-TGGAGTGAGTTTGAGCTTGGTCCT-3’, Probe 5’-VIC-TGGCAAGTACCTCTTCAACTGGGCA-3’. Measurement of MX1 expression was performed in parallel using a specific Taqman Gene Expression Assay (Applied Biosystems, Foster City, CA, USA). Kinetics of HCV inhibition and MX1 stimulation were estimated by least-squares regression using GraphPad Prism v5(GraphPad Software, Inc., La Jolla, CA, USA).
Statistical Analysis
Microarray expression data were analyzed using the Partek Genomics Suite v6.4 (Partek Inc., St. Louis, MO, USA). Expression data were log2-transformed and normalized by quantile normalization prior to statistical analysis. Outlier samples identified by principal components analysis were removed from the study. A linear mixed model analysis was performed using ANCOVA. IL28B genotype, gender, age and ethnicity were included in the model. A false discovery rate threshold of 0.1 was used to correct for multiple testing, corresponding to a p-value< 4.3 × 10−4. Baseline HCV viral load was added to the linear model to determine viral load-independent relationships between IL28B genotype and gene expression. Enrichment of differentially expressed genes in particular biological pathways was determined using Ingenuity Pathway Analysis v8.0 (Ingenuity Systems Inc., Redwood City, CA, USA).A composite ISG metric was calculated as the average expression level of IFI6, IFI27, and ISG15 after linear (anti-log2) transformation and tested for association with SVR, IL28B genotype, and miR-122 expression by ANOVA. Differences in potency between IFN-λ3 variants were assessed by comparison of the 95% confidence intervals of the IC50 or EC50 parameter estimates.
Results
Patient Characteristics
The characteristics of the 61 patients included in the study are described in Table 1. Patients represented multiple ethnicities including African American, Caucasian and Hispanic, with the majority infected with genotype 1 HCV(Table 1). Most patients had mild-to-moderate hepatic necro-inflammatory activity and fibrosis. The distribution of IL28B genotype is listed in Table 1. The C allele frequency was 0.55 and these genotypes were in Hardy-Weinberg equilibrium(P=0.76).
Table 1. Clinical characteristics and genotype distributions in the study cohort (n = 61).
Data represent median (25 – 75 th centile) unless otherwise indicated.
| Parameter | N=61 | |
|---|---|---|
| Age (years) | 51 | 43 – 56 |
| Male gender | 41 | 67.21% |
| Ethnicity | ||
| Caucasian | 38 | 62.30% |
| Hispanic | 5 | 8.20% |
| Asian | 5 | 8.20% |
| African-American | 12 | 19.67% |
| HCV RNA ( log10 IU/mL) | 6.08 | 5.61 – 6.62 |
| HCV genotype 1 | 55 | 90.16% |
| ALT (IU/mL) | 63 | 46 – 87 |
| METAVIR activity A2–3 (n,%) | 32 | 53.33% |
| METAVIR fibrosis F3–4 (n,%) | 2 | 3.33% |
| IL28B genotype | ||
| CC | 19 | 31.15% |
| CT | 29 | 47.54% |
| TT | 13 | 21.31% |
| C allele frequency | 0.55 | |
Gene Expression Profile vs IL28B genotype
Based on previous reports suggesting modest differences between CT and TT genotypes at rs12979860 in relation to SVR (1–3), genotypes were collapsed into a recessive model and comparisons were performed as CC vs. CT or TT (non-CC) genotypes. After applying a false discovery rate threshold of <0.1 to minimize false positive associations, a total of 164 genes were found to be differentially expressed between genotype groups. A full list of differentially expressed genes and the corresponding fold-change in expression and p-value is provided in Supplemental Table S1.
Further filtering to those genes with FDR <0.1 and absolute expression differences of at least ± 1.5-fold between CC vs. non-CC individuals gave a subset of 32 genes highly differentially expressed by genotype. Hierarchical clustering of this subset showed strong discrimination by IL28B genotype (see Figure 1), with a large number of ISGs having significantly lower expression in CC vs non-CC genotypes, including biologically relevant ISGs such as MX1 (2.6-fold), OAS1 (1.9-fold), OAS2 (2.2-fold), OAS3 (2.1-fold), IFIT1 (2.3-fold), IFIT2, (1.7-fold), IFIT3 (1.8-fold) and ISG15 (3.9-fold). Notably, three genes in this subset showed the reverse effect, with higher expression in CC vs non-CC individuals: FCN1 (1.8-fold), CXCL9 (3.3-fold) and CCL8 (1.6-fold).
Figure 1.
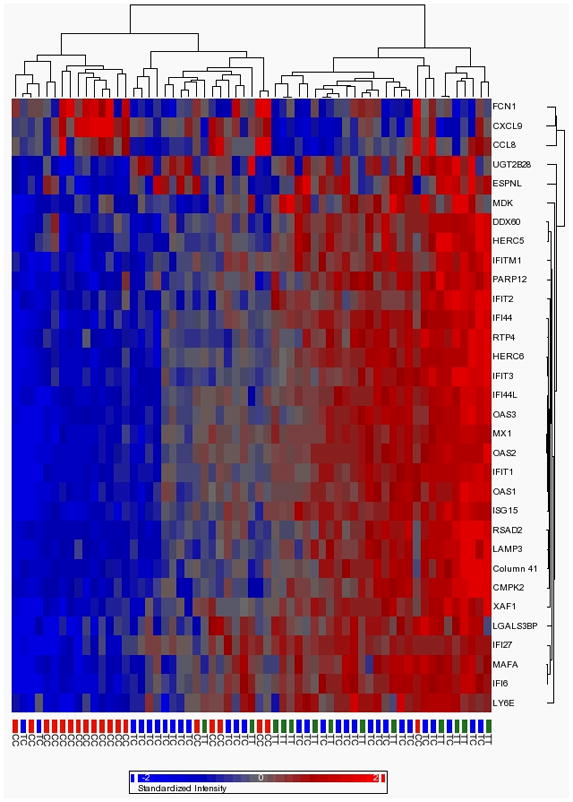
Hierarchical clustering of samples according to genes showing the greatest differences in expression between IL28B genotypes. For clarity, only probes with FDR-corrected p-value <0.1 and a fold-change of >1.5 or of <−1.5 were included in the dendogram. IL28B-CC samples have higher expression than CT and TT samples when probe is red and lower expression when probe is blue.
Genes showing significant association with IL28B genotype were then analyzed for overrepresentation in known biological pathways using Ingenuity software. A number of immunological pathways were found to be significantly enriched for IL28B-associated genes, including interferon signaling as well as viral recognition pathways and a variety of adaptive and innate immune response pathways (see Figure 2). Further analysis of enrichment for biological functions showed a similar enrichment for inflammatory and antimicrobial response genes, among other functions such as cell-to-cell signaling (Supplemental Figure S2).
Figure 2.
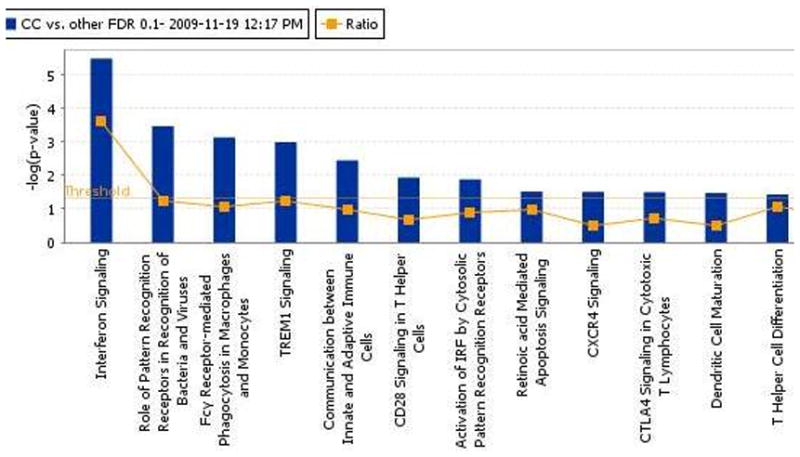
Pathway analysis showed that genes showing differential expression by IL28B-type clustered in the canonical interferon signaling pathway (FDR P value threshold=0.1). Several additional immune-related pathways reach significance in this analysis.
Tests for independence of IL28B-associated and viral load-associated ISG expression
It was hypothesized that high baseline viral load may induce ISG expression in a manner unrelated to IL28B genotype. Thus, further analysis was performed to determine whether particular genes may be associated with IL28B genotype independently of viral load. Inclusion of baseline viral load in the ANOVA model revealed essentially the same set of genes associated with IL28B genotype, i.e., that the differences in gene expression by IL28B genotype were largely independent of HCV viral load at baseline. Hierarchical clustering of gene expression showed a similar pattern of low ISG expression in CC individuals, with only FCN1 and CXCL9 showing higher expression in the protective CC genotype group (see Supplemental Figure S3).
Tests for independence of IL28B genotype and ISG expression in prediction of treatment response
Data on pegIFN and RBV treatment response (sustained viral response, or SVR) was available for a small subset of patients (n=25, including five responders and twenty non-responders). 17 were prior non-responders to pegIFN and RBV therapy (all treatment had been stopped at least 6 months prior to liver biopsy); 8 were treated after the liver biopsy, and of these patients, 5 were responders and 3 were non-responders. This allowed for an exploratory analysis of the relationship between ISG expression and SVR conditioned on IL28B genotype. Despite the small sample size, a significant association between CC genotype and SVR was observed in this subset (p = 0.0054, CC vs. non-CC). In the same subset of samples, tests for association between SVR and 3 ISGs previously shown to be relevant to treatment outcome (ISG15, IFI27 and IFI6) were shown to be significant. These genes were chosen on the basis of previous data linking them to SVR(7), and all 3 showed at least a 1.5-fold difference in expression between responders and non-responders (p-value < 0.05) in the current dataset, and were confirmed by real-time quantitative PCR. A composite ISG expression variable based on the average value of the real-time PCR-based expression measures for these three ISGs similarly showed a significant association with SVR. Due to limitations of sample size, we lacked sufficient power to discriminate whether ISG expression or IL28B genotype had stronger association with SVR; however, an exploratory analysis including both IL28B genotype and mean ISG expression in the regression model suggested that IL28B genotype may be the predominant factor (see Supplemental Table S2).
miR-122 Expression and IL28B Genotype
Recent evidence indicates a necessary role of miR-122 expression in HCV replication (14). Therefore, we evaluated the relationship between IL28B type and intrahepatic expression levels of miR-122 using a targeted real-time PCR assay (miRNA expression levels were not tested on the microarray). miR-122 expression showed no correlation with rs12979860 genotype (see Supplemental Figure S4). In relation to SVR, miR-122 expression showed significantly lower expression levels in liver biopsies from non-responders compared with responders (Figure 3), as described previously (15). Further, although there was a high degree of correlation among the targeted ISGs IFI6, IFI27 and ISG15, these ISGs were not correlated with miR-122 expression (Supplemental Figure S5), suggesting that the association between IL28B-type, ISG expression levels and SVR is independent of miR-122.
Figure 3.
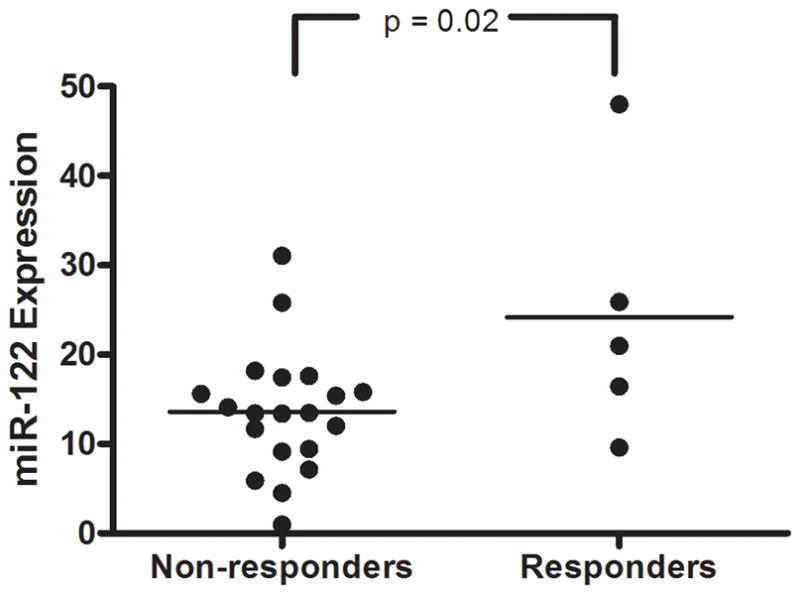
Relationship between hepatic miR-122 expression and SVR. Real-time PCR measurement of miR-122 expression in liver biopsies from patients with known treatment outcome showed improved response rate in individuals with higher miR-122 expression levels (p = 0.024 by ANOVA).
Intrahepatic Expression of IL28B
Expression of IL28A and IL28B mRNA was detectable in HCV infected liver tissue; however, expression of these transcripts showed no significant relationship with IL28B genotype. Due to the lack of specificity in the probe sequences for IL28A vs IL28B on the microarray, and the superior quantitative power of targeted real-time PCR assays, a specific quantitative PCR assay was designed for determination of IL28B mRNA expression (Supplemental Figure S1). Consistent with the microarray results, IL28B mRNA expression as determined using the targeted assay showed no relationship with IL28B genotype (Figure 4). Additionally, no relationship was observed between IL28B genotype and liver IL28A mRNA expression (Supplemental Figure S6).
Figure 4.
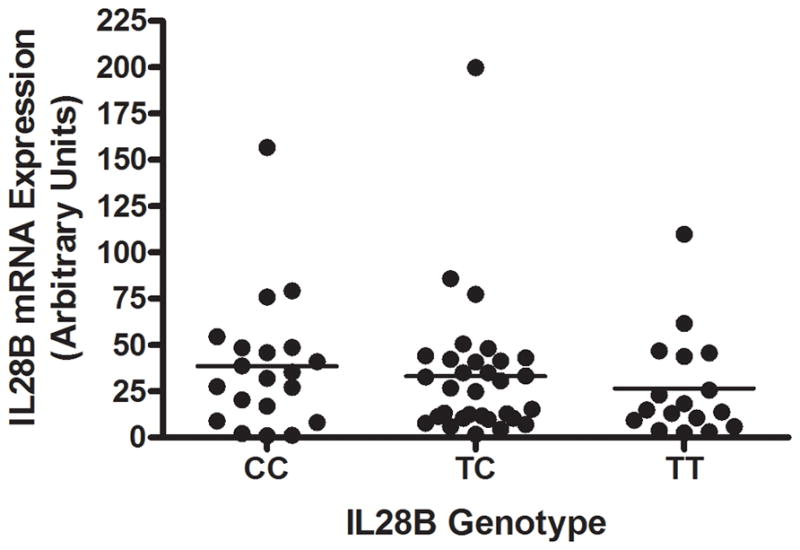
Relationship between IL28B genotype and IL28B mRNA expression in liver tissue. A real-time PCR assay specific for IL28B demonstrated the absence of association between IL28B genotype and IL28B mRNA expression (p = 0.58 by one-way ANOVA), consistent with the microarray expression data.
Comparison of Variant IL28B Potencies In Vitro
We have previously identified through fine mapping strategies that the rs12979860 polymorphism is on a common haplotype block with a non-synonymous coding variant in exon 2 of IL28B, rs8103142, the only coding polymorphism in IL28B (1). As described previously, the set of polymorphisms identified in the IL28B gene region are highly correlated (r2 > 0.85 for all pairwise comparisons), and therefore it is not possible through genetic association alone to determine which if any of these variants is causal for the association, given current sample sizes. Since no significant differences were detected in IL28B mRNA expression levels in CC vs. non-CC individuals (Figure 4), we reasoned that the single conservative amino acid change (Lys70Arg, rs8103142) correlating with IL28B genotype may be responsible for differential anti-viral potency and induction of ISGs. To test this hypothesis, the recombinant IFN-λ3 variants were expressed in E. Coli as GST-fusion proteins and purified for functional assays. Treatment of Huh7.5 hepatoma cells harboring the HCV-Con1 subgenomic replicon (13)with either IFN-λ3 variant caused similar dose-dependent declines in viral RNA abundances, with maximum inhibitory concentrations of ~10 ng/mL (Figure 5). Corresponding analysis of MX1 mRNA levels demonstrated potent MX1 induction after treatment with either variant, suggesting that the Lys to Arg amino acid difference does not affect IFN-λ3 potency.
Figure 5.
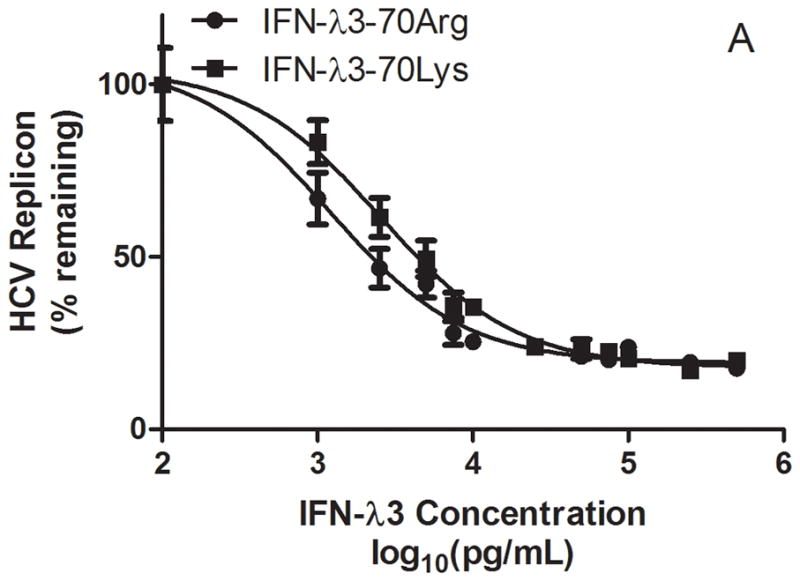
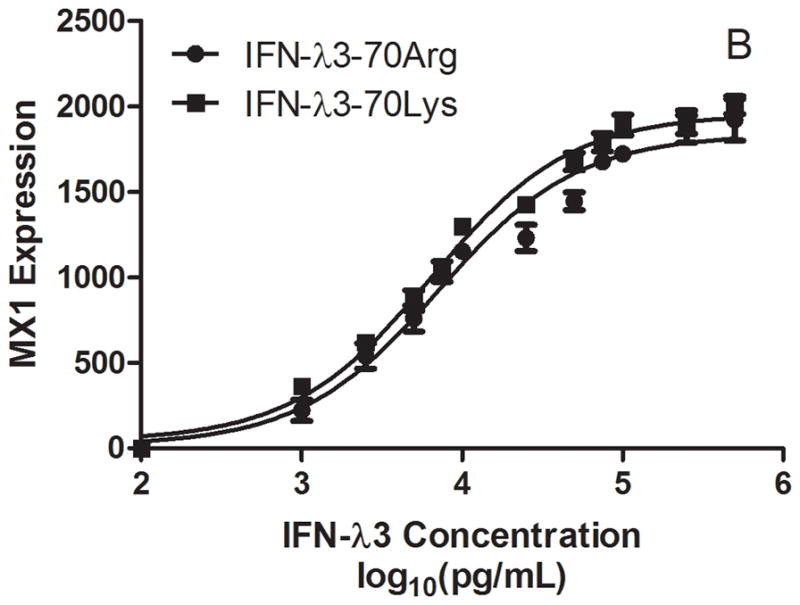
Kinetics of inhibition of HCV replication (A) and stimulation of MX1 expression (B) by recombinant variant IFN-λ3 proteins in HCV replication-competent Huh7.5 cells.
IFN-λ3 is not known to undergo post-translational modifications that would be absent in recombinant proteins produced in E. Coli. Nevertheless, we wished to exclude the possibility that a hypothetical difference in potency due to Lys70Arg would be obscured due to expression in bacteria. We therefore established stable HEK-293 cell lines that inducibly express each IL28B variant and secrete the IFN-λ3 proteins into cell culture media. In order to test functionality of these factors, media from cells expressing IL28B were harvested and concentrated and, as a negative control, media from parental cells lacking a stable IL28B trans-gene was similarly concentrated. Western blot analysis revealed that IFN-λ3 was only detectable in media from over-expressing cells (data not shown). HCV replicon-harboring cells were treated with varying amounts of IFN-λ3 or control media and measurements of HCV and MX1 RNA levels were again performed. As with results obtained with E. Coli-produced proteins, each IFN-λ3 variant generated in human cell culture significantly repressed HCV RNA levels and induced MX1 expression with indistinguishable potencies(data not shown). Taken together, these data suggest that that other non-coding variant(s) dictate phenotypic response to anti-HCV therapy.
Conclusions
We have investigated the relationship between IL28B genotype and patterns of intrahepatic gene expression in an ethnically diverse U.S.-based cohort of patients chronically infected with HCV. Previous reports have shown that high baseline ISG expression in liver tissue is associated with a poor response to treatment with IFN-based therapy (7–11). Here, we show that IL28B genotype is strongly associated with intrahepatic ISG expression, with the poor-response (non-CC) IL28B genotypes exhibiting higher ISG levels compared to patients with the favorable CC IL28B genotype. In a subgroup, viral clearance was associated with both IL28B genotype and lower level ISG expression. We also show that liver expression levels of IL28B mRNA are not associated with IL28B genotype, suggesting that the polymorphism might have an effect in biological potency of IL28B signaling. However, experimental analysis of the non-synonymous coding variant rs8103142 did not demonstrate any differences in anti-HCV or ISG induction potency between protein sequence variants.
The analysis of the intrahepatic transcriptome in patients with the poor response CT / TT IL28B-types compared to CC patients identified differential expression of > 150 genes. The list was dominated by canonical ISGs, including MX1, OAS1/2/3, IFIT1/2/3, IFI6/27/44 and ISG15; all were up-regulated > 1.5-fold in CT/TT patients compared to CC patients. These genes have previously been shown to be among the main effector pathways of the IFN-mediated antiviral response (16). Pathway analysis confirmed that genes showing differential expression by IL28B-type clustered in the canonical interferon signaling pathway. That baseline induction of high level ISG expression is associated with poor therapeutic response to exogenous IFN therapy, perhaps by exhausting the IFN response pathway, has been suggested previously (7–9, 11). The data therefore confirms, in a North American population, the recent results from a study in a Japanese patient population reporting an association between intahepatic levels of ISG expression and pegIFN/RBV response as well as IL28B genotype (rs8099917)(17).
Among the transcripts whose expression was highly differentiated by IL28B genotype, only three genes (CCL8, CXCL9, and FCN1) were shown to be significantly up-regulated in the protective CC genotype group. The chemotactic cytokine (CC) pathway, and CXCL9 in particular, has previously been implicated in the control of HCV infection (18–21). The ficolin-1 (FCN1) gene, though not previously associated with anti-HCV activity, is suspected to play a role in innate immunity and also represents a plausible candidate for control of HCV (22, 23). The explanation for the discordant expression of these 3 genes relative to the general pattern of low level ISG expression remains unclear. A key question is whether the pattern of intrahepatic ISG expression is driven directly by IFN-λ3, or is a secondary effect of IFN-λ3 modulating a type 1 IFN effect, and this will require further investigation.
miR-122 has recently been identified to promote HCV replication, and an antisense inhibition of miR-122 has been shown to inhibit HCV (24). miR-122 is also involved in cholesterol metabolism, and inhibition profoundly reduced serum cholesterol levels in chimpanzees(24). IL28B type has recently been associated with serum LDL levels in CHC patients (25). It was therefore plausible and important to investigate whether the IL28B polymorphism was associated with miR-122 expression levels. However, miR-122 was not observed to be associated with either IL28B type or ISG expression. Furthermore, in the treatment subgroup, we observed a significant relationship between miR-122 levels and treatment outcome, that was independent of IL28B genotype. This suggests that these two pathways may act independently to influence response to IFN-based treatment of HCV infection. The finding suggests that future treatment modalities for HCV infection may potentially take advantage of both IFN-dependent and miR-122-dependent modes of viral eradication, which may produce a synergistic effect on viral clearance.
The current study tests two of the most obvious hypotheses regarding the mechanism for the effect of IL28B genotype on HCV clearance: 1) differential intrahepatic expression of IL28B mRNA based on IL28B genotype, and 2) differential antiviral potency between protein sequence variants of the IL28B gene product, IFN-λ3. The relationship between IL28B genotype and IL28B mRNA in the literature has been controversial(1–3).Previous studies have suggested that the effect of IL28B genotype on HCV treatment response may occur through differential expression of IL28B by genotype, with the low-response genotype showing lower expression of IL28 in whole blood from healthy volunteers (2), or whole blood from HCV infected patients (3). More recently, Honda and colleagues showed that expression levels were similar in HCV-infected livers according to IL28B – type(17). Here, we also show, using a highly specific assay for IL28B mRNA, that IL28B genotype is not associated with IL28B expression in liver biopsies from HCV infected patients. This suggested that the biological effect of the IL28B polymorphism might be to effect potency, which would require a non-synonymous causal variant. Previous work has suggested a possible causal role for a polymorphism in exon 2 (rs8103142, Arg70Lys). However, we rigorously investigated the potential for Arg70Lys to impact anti-viral potency of IL28B and found no discernible difference between variants. As fine mapping studies have not demonstrated any other coding variants to be highly linked with the discovery SNP, this suggests that the mechanism likely involves an explanation other than signaling potency. Chen and colleagues have recently shown that ISG expression levels may differ between different cell populations in the liver, where ISG upregulation was more pronounced in hepatocytes in non-responders to IFN therapy, and in Kupffer cells in responders (26). As microarray analysis does not differentiate the cell of origin of the observed ISG expression, it is possible that the IL28B polymorphism has cell-specific effects on expression that may not have been detected in the current study. Alternatively, genotype-specific polymorphisms could conceivably impact post-transcriptional regulation of IL28B expression in liver(i.e. mRNA splicing, translation, etc.) that would be missed by microarray and RT-PCR analyses.
The results presented here provide a paradox. The non-CC genotypes, associated with higher level ISG expression in the setting of chronic hepatitis C infection, have previously been associated with lower rates of spontaneous clearance. In contrast, the CC genotypes, in which low level ISG expression is observed, have been associated with high rates of spontaneous clearance (4, 5). It may be that the differential pattern of gene expression is specific to chronic HCV, and cannot be extrapolated to acute HCV. Alternatively, a more integrative hypothesis is that the effect of the poor-response IL28B variant is to ‘sabotage’ the liver’s response to type 1 IFN – perhaps by exhausting the common signaling pathway. In contrast, in the setting of the good-response CC variant, the liver remains relatively dormant, and ‘primed’ for type 1 IFN response. However, this hypothesis is speculative given current knowledge. Little is known about the interaction between type 1 and type 3 IFNs in vivo. Cell culture models of HCV replication suggest an additive antiviral effect of IFN-α and IFN-λ1/2, but no data is available yet for IFN-λ3; furthermore, these experimental models do not account for the host immune milieu. The precise pattern of interaction between type 1 IFN and type 3IFN signaling therefore remains unclear.
In conclusion, these data provide novel insights into the host genetic regulation of IFN signaling in the livers of patients chronically infected with HCV. The IL28B polymorphism is associated with differential intrahepatic gene expression profiles, where the good response IL28B variants are associated with lower levels of hepatic ISG expression. IL28B genotype was not associated with differences in intrahepatic IL28B gene expression, and protein sequence variants of IFN-λ3 do not appear to explain the differences in ISG expression or anti-HCV response by IL28B genotype. Thus, the biological mechanism underlying the influence of the IL28B polymorphism on IFN treatment response remains unclear. Further examination of the noncoding polymorphisms in the IL28B gene region, and of the dynamics of type 1 IFN sensitivity in the setting of sustained IFN-λ activity, both in vitro and in vivo, may provide a fuller understanding of the role of IFN-λ in natural clearance of HCV and its pharmacological treatment.
Supplementary Material
Acknowledgments
Financial Support:
Part of this work was supported by the National Institutes of Health grant NIH U19 AI066313 “Hepatitis C Cooperative Research Centers” to D.S. and N.H.A.
Abbreviations
- IL28B
Interferon 28B
- SVR
sustained virological response
- CHC
chronic hepatitis C
- ISG
interferon-stimulated gene
- IL28B
interferon 28A
- GWAS
genome-wide association study
- HCV
hepatitis C virus
- IFN
interferon
Footnotes
Disclosure: Authors T.J.U., A.J.T., J.F., K.V.S., J.G.M., and D.B.G. are listed as inventors on a patent application related to this work.
Bibliography
- 1.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 2.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 4.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. 1345, e1331–1337. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 5.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asselah T, Bieche I, Narguet S, Sabbagh A, Laurendeau I, Ripault MP, Boyer N, et al. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516–524. doi: 10.1136/gut.2007.128611. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 9.Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanford RE, Guerra B, Bigger CB, Lee H, Chavez D, Brasky KM. Lack of response to exogenous interferon-alpha in the liver of chimpanzees chronically infected with hepatitis C virus. Hepatology. 2007;46:999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 13.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 15.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- 16.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Nakamura M, et al. Hepatic Interferon-Stimulated Genes Expression Is Associated With Genetic Variation in Interleukin 28B and the Outcome of Interferon Therapy for Chronic Hepatitis C. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 18.Butera D, Marukian S, Iwamaye AE, Hembrador E, Chambers TJ, Di Bisceglie AM, Charles ED, et al. Plasma chemokine levels correlate with the outcome of antiviral therapy in patients with hepatitis C. Blood. 2005;106:1175–1182. doi: 10.1182/blood-2005-01-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florholmen J, Kristiansen MG, Steigen SE, Sorbye SW, Paulssen EJ, Kvamme JM, Konopski Z, et al. A rapid chemokine response of macrophage inflammatory protein (MIP)-1alpha, MIP-1beta and the regulated on activation, normal T expressed and secreted chemokine is associated with a sustained virological response in the treatment of chronic hepatitis C. Clin Microbiol Infect. 2010 doi: 10.1111/j.1469-0691.2010.03206.x. [DOI] [PubMed] [Google Scholar]
- 20.Zeremski M, Petrovic LM, Chiriboga L, Brown QB, Yee HT, Kinkhabwala M, Jacobson IM, et al. Intrahepatic levels of CXCR3-associated chemokines correlate with liver inflammation and fibrosis in chronic hepatitis C. Hepatology. 2008;48:1440–1450. doi: 10.1002/hep.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu J, Le Y, Kon OL, Chan J, Lee SH. Biosynthesis of human ficolin, an Escherichia coli-binding protein, by monocytes: comparison with the synthesis of two macrophage-specific proteins, C1q and the mannose receptor. Immunology. 1996;89:289–294. doi: 10.1046/j.1365-2567.1996.d01-732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Tay PN, Kon OL, Reid KB. Human ficolin: cDNA cloning, demonstration of peripheral blood leucocytes as the major site of synthesis and assignment of the gene to chromosome 9. Biochem J. 1996;313 ( Pt 2):473–478. doi: 10.1042/bj3130473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JH, Lao XQ, Tillmann HL, Rowell J, Patel K, Thompson A, Suchindran S, et al. Interferon-lambda genotype and low serum low-density lipoprotein cholesterol levels in patients with chronic hepatitis C infection. Hepatology. 2010;51:1904–1911. doi: 10.1002/hep.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, Anand N, et al. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123–1133. e1121–1123. doi: 10.1053/j.gastro.2009.10.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


