Abstract
RNA virus exploration within the field of medical virology has greatly benefited from technological developments in genomics, deepening our understanding of viral dynamics and emergence. Large-scale first-generation technology sequencing projects have expedited molecular epidemiology studies at an unprecedented scale for two pathogenic RNA viruses chosen as models: influenza A virus and dengue. Next-generation sequencing approaches are now leading to a more in-depth analysis of virus genetic diversity, which is greater for RNA than DNA viruses because of high replication rates and the absence of proofreading activity of the RNA-dependent RNA polymerase. In the field of virus discovery, technological advancements and metagenomic approaches are expanding the catalogs of novel viruses by facilitating our probing into the RNA virus world.
Keywords: dengue, influenza, intrahost diversity, metagenomics, molecular epidemiology, next-generation sequencing, viral genomics
Since the sequencing of the first bacteriophage genomes, MS2 in 1976 [1] and φX174 in 1977 [2], the decoding of viruses has informed research in the fields of genetics, ecology, epidemiology, and, more recently, computational biology. After 37 years since the introduction of chain-termination dideoxy sequencing by Sanger [3], we continue to regard the principles of this technique as the gold standard for virus sequencing, and rely on them heavily. Over the years, automation of this method culminated in capillary sequencing on the Applied Biosystems (ABI; CA, USA) 3730xl platform. Since then, virus research has been riding the genomics wave, benefiting greatly from comparative genomic analyses of viral strains. The RNA virus genomics field, however, has lagged slightly behind. The extra step required to convert the RNA to DNA delayed RNA virus exploration just when the rest of the genomics field was adopting the whole-genome shotgun approach at the core of its high-throughput sequencing workflows. More recently, second-generation sequencing tools and methodological improvements have, in some ways, leveled the playing field. Nevertheless, virus genomics is still in its early stages; as such, it holds the promise of new and exciting discoveries.
This review focuses on a few RNA viruses, with an emphasis on the influenza A virus, and some of the breakthroughs made with large-scale sequencing. Its purpose is to highlight how sequencing technology is leading to important discoveries and thus expanding our understanding of viral dynamics and emergence.
Large-scale molecular epidemiology of emerging & circulating RNA viruses
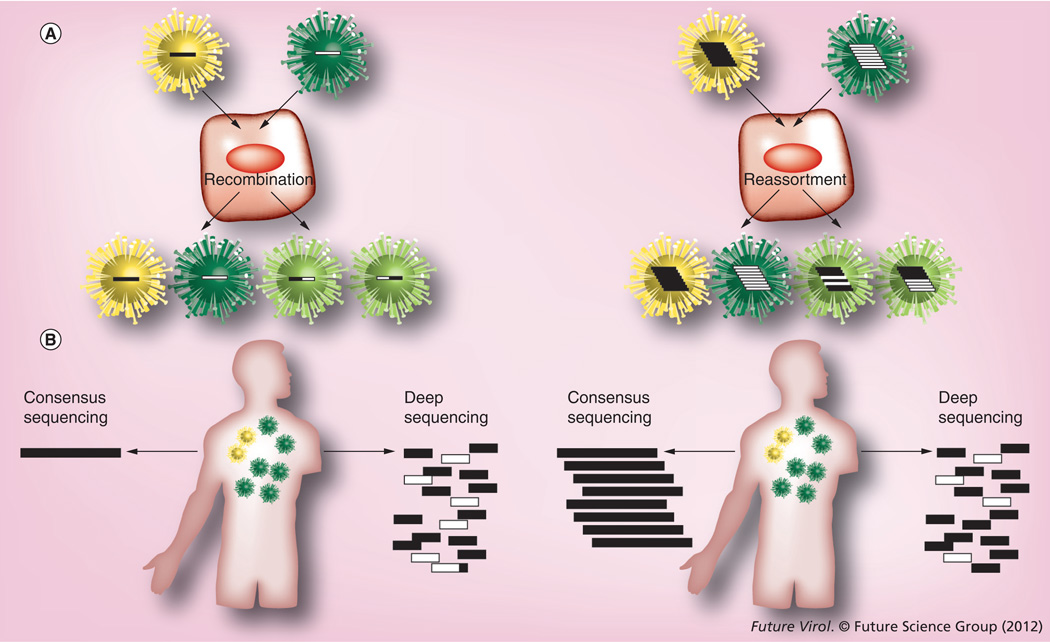
A characteristic of RNA viruses is their great genetic diversity, attributable to the absence of proofreading activity of the RNA-dependent RNA polymerase [4] and their high replication rates, with one genomic mutation expected to be introduced at every replication cycle. As well as mutations introduced by the error prone RNA-dependent RNA polymerase, viruses like influenza A and dengue also undergo a genomic mixing process leading to genetic diversity, which has a major bearing on adaptation: influenza’s 13.5-kb negative-sense, segmented RNA genome allows for genomic reassortment during mixed infections and coinfection of cells, while dengue’s 1.1-kb positive-sense unique segment can recombine across serotypes [5], a process not believed to occur for influenza (Figure 1A) [6]. The rapidity of mutation, replication and reassortment/recombination means that each infected host is likely to carry viral populations with high genetic diversity. To better understand RNA viral evolution, epidemiology and pathogenesis from the genomic information, widespread efforts are underway to sequence circulating and newly emerging viruses collected around the world. In 2004, in the USA, the National Institute of Allergy and Infectious Diseases of the NIH launched whole-genome sequencing projects to study historic and currently circulating viral strains of the influenza and dengue viruses. To date, these projects have generated whole-genome sequence data for more than 2500 dengue viruses and 8200 influenza viruses from human populations and various mammalian species around the globe. This large-scale sequencing effort represents one of the most developed models for applying genomics at the epidemiologic level for infectious diseases. It also gives a new outlook on the dynamics of negative-sense (influenza) and positive-sense (dengue) RNA viruses, which are responsible for some of the most alarming diseases emerging around the world.
Figure 1. Genetic mechanisms of viral diversity and genomic sequencing strategies.
(A) Example of recombination between ssRNA virus genomes (i.e., dengue) and reassortment between segmented RNA virus genomes (i.e., influenza A) coinfecting the same cell. (B) Schema of how consensus sequencing provides a homogeneous representation of a virus population while deep sequencing generates sequence reads that capture genetic diversity, but that may not be unambiguously assembled into individual genomic segments.
Human influenza A virus
Whole-genome Sanger sequencing and subsequent phylogenetic analyses of complete human seasonal A/H1N1 and A/H3N2 influenza viruses circulating around the world gave us the first comprehensive picture of influenza viral evolution, including its pattern of transmission through human populations. As a result, several novel observations were made, including the prevalence of gene segment exchanges (reassortment) and adaptive evolution of multiple, cocirculating viral lineages, demonstrating the dynamic nature of seasonal influenza transmission and evolution [7]. Large-scale genomic studies of seasonal influenza viruses have also characterized the occurrence of reassortment and periodic selective sweeps between the northern and southern hemispheres, suggesting a ‘sink-source’ model of viral ecology by which new lineages are seeded from a reservoir located in the tropics to populations in temperate regions [8]; however, more recent genomic analyses have challenged this model [9]. By comparing the phylogeography of complete A/H3N2 viral genomes from multiple continents throughout several influenza seasons, A/H3N2 influenza viruses appear to exist as a migrating metapopulation, such that viruses from multiple locales – but not the tropics – are sources of epidemics [9].
Genomic studies aimed at discerning trends in influenza viral evolution during pandemics and historic accounts of viral evolution has also made valuable contributions to our current understanding of influenza evolutionary dynamics. Within an epidemic, multiple clades of seasonal A/H1N1 and A/H3N2 viruses cocirculated and expressed novel patterns of complex spatial spread [10]. A retrospective study discovered that since the Spanish influenza pandemic of 1918, intrasubtype reassortment of seasonal and pandemic A/H1N1 viruses appeared to be a more important evolutionary process than previously realized, and may have played a role in the antigenic shifting of the virus [11]. The emergence of the 2009 A/H1N1 influenza pandemic presented the unique opportunity to conduct similar, extensive genomic epidemiological studies on a pathogen that had immediate public health implications on a global scale. The emergence of the A/H1N1 pandemic of 2009 was shown to be the result of notable reassortment events [12]. Large-scale whole-genome Sanger sequencing and phylogeographic analyses indicated that within months of introduction into the USA, the virus diversified into distinct lineages, with defined spatial patterns in wave 1 of the pandemic, followed by extensive viral migration and mixing in its second wave [13,14]. At the lower, community level, this pandemic A/H1N1 virus exhibited remarkable spatial fluidity, with multiple independent introductions, indicating that community-based methods for infection control are not likely to prevail in future pandemics [15]. Similarly, in Canada, the spread of the A/H1N1 pandemic virus was marked by numerous introductions of several lineages, but with limited genetic diversity [16]. Whole-genome sequencing and analysis of A/H3N2 influenza viruses collected in 2009 between regular seasonal seasons illustrated the introduction, widespread transmission and cocirculation of seasonal influenza viruses during a time-frame when influenza transmission was uncommon. This provides additional evidence of the complexity of viral ecology, and emphasizes the importance of molecular surveillance year-round [17].
Avian influenza A virus
Wild aquatic birds are the reservoir hosts for nearly all known subtypes of influenza A viruses (hemagglutinin, H1–H16; neuraminidase, N1–N9); they have been implicated in stable host-switching events, leading to novel, emerging influenza lineages within various species of domestic animals, as well as past human pandemics. Yet it was not until the outbreak in humans of the highly pathogenic A/H5N1 avian influenza virus (AIV) in southeast Asia in 2003, which subsequently spread to Europe in 2004, coupled with the lack of publicly available AIV reference genomes, that large-scale, complete AIV genome sequencing efforts began. Similar to large genomic studies of human seasonal influenza viruses, a high-throughput, Sanger sequencing approach was also employed to generate complete AIV genomes. This effort led to the sequencing of the first complete non-Asian-origin A/H5N1 avian influenza genomes [18] and gave epidemiologic and evolutionary evidence of a novel Euro–African A/H5N1 viral lineage separate from the common ancestor Eurasian-origin lineage, which was responsible for fatal A/H5N1 human infections. Furthermore, constant reassortment and independent evolution of this lineage were observed, helping to elucidate the epidemiology and evolution of this rapidly spreading zoonotic virus [18]. Large-scale AIV genomics studies more than doubled the number of existing wild-bird-origin influenza genomes within public databases including the National Center for Biotechnology Information, Influenza Research Database and The Global Initiative on Sharing All Influenza Data EpiFlu database, and, furthermore, identified cocirculating viral lineages and high levels of reassortment. Evolutionary analyses of the largest and most complete data set of wild-bird-origin AIV genomes, available in 2008, proposed that AIV in wild birds forms ‘transient genome constellations’, whereas the eight gene segments of AIV are continually reshuffled by reassortment [19]. Consequently, a multitude of large-scale, AIV genomic studies were initiated to survey wild birds in the Pacific northwest region of the USA (namely, Alaska), in order to observe intercontinental movement of AIV gene segments via wild migratory birds. Alaska is a unique region where long-distance migratory birds from both the Asian and North American migratory flyways congregate on resting grounds. It has been hypothesized that this overlap of intercontinental flyways provides an opportunity for the introduction of Eurasian-origin AIV, including the highly pathogenic A/H5N1, into the North American wild-bird population. These studies have taken a species-specific approach to the genomic epidemiology of low-pathogenic AIV, focusing on long-distance migrators such as northern pintails, shorebirds, gulls and more resident species, including mallard ducks, which copopulate this region. Low-pathogenic AIV from Alaskan northern pintails and mallards each possessed a higher level of Eurasian-origin gene segments than previously reported. These findings suggest that a higher degree of intercontinental AIV gene transfer occurs in Alaskan birds, and provide evidence that wild migratory birds transfer AIV to more resident species across phylogenetic boundaries [20,21]. In comparison, Alaskan shorebirds possessed a much lower level of intercontinental gene transfer than previously reported, suggesting that AIV infection in Alaskan shorebirds is either secondary or may involve another migratory species [22]. Interestingly, AIV in Alaskan gulls exhibited high levels of reassortment and intercontinental gene transfer in viral subtypes previously thought to be gull specific; however, AIV in Eurasian gulls showed no evidence of intercontinental gene transfer [23]. Overall, highly similar AIV genome constellations have been observed to persist over time in several different avian species across Alaska; however, to date, an eight-segment Eurasian-origin AIV has not been detected in wild birds in Alaska [24].
Dengue virus
Like influenza virus, dengue virus is a serious threat to public health, with 40% of the world’s population living in endemic regions. Transmitted by Aedes mosquitoes, and characterized by four antigenically distinct serotypes, it is responsible for a spectrum of diseases, from mild to severe. Large-scale sequencing helped characterize strain variation and its effect on epidemic activity, leading to a better understanding of the pathogenesis and re-emergence of this virus. There are currently no vaccines available for dengue because of the complexity of the viral population dynamics. A clear understanding of the spatial and temporal dynamics is necessary for the development of better intervention strategies. Full-length phylogenetic analysis of 187 dengue virus type 3 (denv-3) viruses isolated from Asia and the Americas showed geographical speciation and mutational differences in outbreak years [25]. Whole-genome sequencing of denv-2 samples collected in the south Pacific over a span of 30 years showed that viruses responsible for the outbreaks during that time were the result of a single introduction [26]. The attenuation observed was thus correlated with the accumulation of genetic changes over time in the virus genomes, rather than with the introduction of milder strains to the region. One of the largest studies of dengue evolution comprised the whole-genome sequencing of 751 denv-1 viruses collected in rural and urban Vietnam between 2003 and 2008 [27]. The phylogeographic analysis enabled the determination of patterns of transmission, and demonstrated limited spatial movement, which stands in contrast to what is observed with influenza. In Nicaragua, in a study across epidemic seasons, disease severity was correlated with the sequential replacement of one clade by another. Serotype-specific immunity and viral genetics were both shown to contribute to disease severity [28]. A common observation in dengue evolutionary dynamics is genotype replacement, with a new lineage displacing the circulating lineage. For example, genotype replacement occurred in Cambodia and Thailand with the displacement of denv-2 [29]. In a similar study in Puerto Rico, 22 years of consecutive sampling demonstrated clade replacement of denv-2 viruses before the emergence of denv-3 [30].
Next-generation sequencing & viral genomics
While automation of current sequencing workflows accelerates the pace of viral genome sequencing, its high cost, coupled with reliance on the relatively low base-pair generation of Sanger-based methods, severely limits output and jeopardizes the accurate representation of the true genetic diversity of the viral populations studied. Classical viral genome sequencing approaches rely on PCR-mediated production of overlapping amplified products and cloning, followed by Sanger sequencing. Errors can be introduced at the PCR step, as all DNA polymerases have an intrinsic error rate. The Taq DNA polymerase, for example, introduces approximately one error every 125,000 bases, but some more recently discovered enzymes with proofreading activity claim error rates that are 100-fold lower [31]. Sequence data is then assembled, followed by rigorous editing and finishing steps to ensure the accuracy of viral sequence polymorphisms. In some viral population studies, this approach has led to efficient analyses of viral population diversity – for example, in the large-scale characterization of intrahost dengue diversity of close to 8500 clones from 49 human plasma specimens [32]. This, however, requires considerable effort and money. The cloning step is often skipped if the goal is to obtain a consensus of the virus genome or when the viral population is homogeneous enough that no interference with the sequencing signal occurs. Because this PCR/Sanger-based method provides what is essentially a population overview (or consensus) (Figure 1B) of nucleotide diversity, the genetic complexity of many viral samples prohibits rapid, unambiguous assembly of sequence data. While these approaches have served an important purpose, it is the new sequencing platforms – called next-generation, or NextGen sequencing – that offer vastly higher sequence output, that are ideally suited for viral population genetics and molecular epidemiology.
The most commonly used NextGen platforms at the present time are the Roche GS-FLX/454 (454 Life Sciences, CT, USA), Illumina’s GAIIx and HiSeq/MiSeq (CA, USA) and ABI’s SOLiD. Gaining rapidly in popularity, are ABI’s Ion Torrent and Pacific Biosciences’ PacBio. (For a recent review of the commercially available platforms; see [33]). These technologies produce massive parallelization of clonal amplification of nucleotide sequences, greatly facilitating the identification of individual virus strains in genetically heterogeneous populations (Figure 1B). Because of the very high sequencing redundancy (i.e., depth of coverage) of these new platforms, and the relatively small size of virus genomes, the multiplexing of multiple virus isolates, barcoded so they can be sequenced simultaneously but with their respective sequences characterized individually, is now possible. This approach is also particularly appropriate for the characterization of drug-resistance mutations in a viral population in order to determine the prevalence of the mutations before they become dominant and lead to the emergence of resistant strains. Direct PCR product sequencing (the classical approach) has its limitations, and is usually used to detect prevalence above 20% within an infected host [34]. Sequencing multiple clones is labor intensive and costly, although it has high sensitivity. The NextGen platforms enable the analysis of sequence reads generated from each of the amplified molecules, as well as the identification of very low frequencies of pre-existing mutations that could lead to the emergence of a new phenotype. Studies using GS-FLX or GAIIx to identify rare drug-resistant variants in HIV, HCV and HBV [34–37] demonstrate the strength of this approach to clarify the genetic complexity of these viral populations. In chronic infections such as HCV and HIV, the characterization of genetic diversity within each infected patient is limited by the very high heterogeneity of the population. In a recent study on HIV-I dynamics, deep molecular characterization was used to identify viral populations that persist in the patient under suppressive therapy [38]. Low-level viremia limits the characterization of virions that remain in HIV-1-infected individuals undergoing antiretroviral therapy. But while plasma viral load may be undetectable at less than 50 RNA copies per ml, deep-sequencing data of the selectively amplified reverse-transcriptase genes isolated from episomal and integrated HIV-1 DNA paint a much clearer picture of the existence of reservoirs within the host [39].
NextGen sequencing for viral surveillance
NextGen sequencing is being applied to reveal the evolution and origin of novel, emerging viral pathogens. SARS-coronavirus, for example, emerged in humans in 2002–2003 as the etiologic agent responsible for a disease resulting in severe acute respiratory distress [40–42]. The outbreak spread rapidly around the globe, eventually infecting approximately 8000 people. Thereafter, coronavirus genomics was initiated as an important field of research to track coronavirus evolution and analyze coronaviruses in bats, known reservoirs for SARS-coronavirus-like viruses, as well as many other coronaviruses [43]. Samples of wild-type and mutant viruses that had been grown through multiple passages were sequenced by Sanger and NextGen technologies to show that specific mutations caused substantial reduction in replication fidelity [44]. These results provided compelling evidence for a role for nsp14-ExoN in RNA proofreading during coronavirus replication. Complete genomes of new coronaviruses isolated from mink exhibiting epizootic catarrhal gastroenteritis were shown to be clearly distinct from any other known coronaviruses, with 92% identity to contemporary strains. Phylogenetically distant from human coronavirus strains, it appears that these isolates, together with partially sequenced ferret enteric coronavirus and ferret systemic coronavirus, comprise a new species within the genus Alphacoronavirus [45].
In the case of influenza virus studies, NextGen sequencing is proving to be central to understanding the emergence of human pandemic, antigenic and drug-resistant strains. In addition, it is providing more comprehensive surveillance information about influenza viruses in animal reservoirs. Whole-genome sequencing of the first cohort of seasonal A/H3N2 viruses, collected from Uganda in 2008 and 2009, revealed markers of enhanced transmission and drug resistance, in addition to two different lineages representing antigenic drift [46]. In the UK, surveillance using NextGen sequencing of whole influenza genomes traced specific transmission chains of the A/H1N1 pandemic virus [47]. Comparable to other reports, multiple, independent introductions of genetically complex A/H1N1 pandemic viral lineages were observed during the first wave of the pandemic; yet at least two viral lineages were detected before the first laboratory-confirmed case, and several transmission chains persisted between waves [47]. Although the complete evolutionary history of the A/H1N1 pandemic of 2009 is not fully understood, this virus was genetically determined to be a triple-reassortment strain originating from a well-established lineage circulating in pigs [12,48]. Thus, ongoing surveillance efforts using NextGen sequencing have focused on sequencing retrospective and contemporary influenza viruses from these animals, to gain deeper insight into the origins and evolutionary future of this virus. Complete genome characterization of North American H1 swine isolates collected in 2008 yielded a high level of overall viral diversity, both antigenically and phylogenetically, forming separate clades when compared with the A/H1N1 pandemic virus. This suggests that the pandemic A/H1N1 virus and any closely related progeny viruses were not present in this swine population prior to 2009 [49]. Phylogenetic analysis of A/H1N1 pandemic viruses transmitted from humans to commercial pigs after 2009 showed reassortment between the pandemic A/H1N1 virus and endemic swine influenza viruses that generated seven different A/H1N1 viral genotypes [50]. While no single genotype became dominant within the population, continued surveillance of these and other dynamic and emerging viruses further promotes the utility of NextGen sequencing for this purpose.
NextGen sequencing of wild-bird-origin AIVs has proved to be a more sensitive method of surveillance in comparison to traditional Sanger sequencing. Within a population of sentinel mallards, NextGen sequence data confirmed the presence of mixed AIV infections within individual specimens, represented by sequences generated for more than one hemagglutinin type [51]. The most recent trend being followed to unravel the epidemiology of AIV of low pathogenicity in wild birds is to combine whole-genome NextGen sequencing with phylogeographic modeling of ecologic variables. By modeling gene flow within and between the four major ecological North American flyways, it was discovered that despite increased gene flow of Eurasian and Australasian-origin AIV to the Pacific flyway, the spread of AIV within North American wild birds is constrained by geographical distance and avian flyways. Additional ecologic attributes most likely play a role in shaping low-pathogenic AIV epidemiology in North American birds, and the resultant model could be utilized to estimate the spread of a novel lineage in the wild-bird population [52].
Methodological and statistical modeling studies dependent on NextGen deep sequencing have been developed to characterize complete genomes, glycosylation patterns and antigenic evolution, to detect rare mutations and to explore viral diversity (often referred to as ‘quasispecies’) [53–58]. NextGen sequencing and analyses of the intrahost diversity of multiple influenza genomes isolated from individual patients revealed that humans can harbor antigenically distinct influenza viruses, and viruses possessing different sensitivities to antiviral agents [59]. A similar study used de novo sequencing to detect a heterogeneous population of A/H1N1 pandemic influenza viruses in lung tissue collected from an autopsy sample [60]. Deep sequencing of A/H1N1 pandemic viruses isolated from immunosuppressed patients showed that these individuals may be an important source of viral genetic and phenotypic diversity due to mixed infection and the development of drug resistance [61]. Collectively, these data illuminate previously unknown sources of viral diversity and drug resistance, both of which are vital for pandemic preparedness and patient management in the clinical setting.
NextGen sequencing in virus discovery
NextGen sequencing has applications beyond the molecular epidemiology of known viruses, notably for the discovery and characterization of novel viruses. In particular, the field of metagenomics, also referred to as ‘community genomics’, has profited by advancements in sequencing technology, making it possible to characterize microbes and viruses within samples of collected environmental DNA. Active discovery and cataloging of novel and variant viruses by deep sequencing began with the analysis of environmental samples; it is now being employed for a wide range of biological and clinical samples. The traditional methods for detecting novel viruses, such as cell culture, serology and electron microscopy, are now being used as supportive tools for metagenomics findings [62], whereas NextGen sequencing has become the tool of choice for culture-independent genomic analyses of a wide variety of samples. New emerging viruses or variants of known viruses are recognized as potential agents responsible for new infectious diseases, which, though prevalent, often go unrecognized [63]. NextGen sequencing, in combination with hybridization- and PCR-based techniques, is proving to be a powerful approach for global screening of pathogens and disease surveillance.
Along with technological advances has come a surge of interest in the detection of unknown viruses in various samples, and diagnostic virology has been greatly impacted by viral metagenomics [64]. Owing to its high sensitivity and ability to characterize sequences without previous knowledge, NextGen sequencing is a much more powerful tool than conventional methods in diagnostic virology (reviewed in [65]). Diagnostic validation of this approach has been demonstrated by direct detection of bacterial and viral pathogens from nasal and fecal samples of patients with seasonal influenza infections [66]. Metagenomic sequencing of a collection of clinical isolates, which failed routine diagnostic assays, identified BK polyomavirus, HSV and variants of other viruses [67]. Many other clinical conditions, such as gastroenteritis, diarrhea, respiratory tract infections and cancer, have also been studied by high-throughput sequencing, resulting in the identification of novel viruses [68–72]. Similarly, novel enterovirus genotypes have been identified in patients with acute flaccid paralysis [73]. Metagenomic analyses of live-attenuated vaccines led to the detection of minority variants and adventitious viruses that go undetected in commercial vaccine preparations [74]. With rapidly evolving and cheaper sequencing technologies, high-throughput sequencing approaches could be established in a surveillance system for early detection of viruses from diverse clinically relevant samples.
Another widely studied area is the sequence analysis of viromes of animal populations, which are major reservoirs of viruses with the potential for zoonotic transmission, leading to emergence [75]. Although traditional sequencing of fecal samples from domestic animals such as horses, rabbits and birds has revealed novel alphaherpesviruses and picornaviruses [68,76], NextGen methods allow a more rapid and systematic approach for identifying novel viruses implicated in zoonoses [73,77,78]. For example, a vast amount of data was generated for pig and dog fecal viromes, which have been used for comparative analyses with human fecal flora [77,79]. Methodologies for more specific isolation of virions from specimens have been improving in the past few years [80]. Recently, single-virus genomics has been developed to improve the whole-genome amplification of a single virus particle [81]. Much progress has been made in the field of DNA virus discovery, but large-scale genomic research into RNA viruses lags behind, owing to the problems associated with their genomic nucleic acid content – notably, the stability of the RNA molecules and the necessity to convert RNA to cDNA for sequencing. A few studies have reported the discovery of novel RNA viruses [82–84], most notably Schmallenberg virus, a new species in the genus Orthobunyavirus [85], and more are anticipated as sample preparation methodologies improve [86]. An important aspect of all these virome studies is the identification of unclassified/unknown sequences that do not have any matches in viral databases, suggesting that we are just tapping the surface of the vast ocean of uncharacterized microorganisms.
Conclusion
Whole-genome sequencing and phylogenetic analyses of globally circulating emerging viruses, like dengue and influenza, continue to clarify our understanding of the genetic diversity and viral evolution of pathogens that are responsible every year for high rates of human morbidity and mortality on a global scale. These broad studies illustrate the power of genomics to vastly increase our overall knowledge of viral epidemiology and evolutionary dynamics, which are central to understanding viral ecology. We have observed how large-scale sequencing has answered questions on the global circulation and reassortment of influenza viruses; on dengue virus serotype emergence; and on intrahost diversity in acute infections. We have also seen the value of metagenomic applications in virus discovery. But while NextGen platforms have simultaneously increased our throughput and decreased the costs per base pair, as compared with first-generation sequencing workflows, if we are to take full advantage of the developments in sequencing technologies, a number of variables will have to be optimized to achieve better results. One of the crucial parameters affecting the quality and detection sensitivity of NextGen sequencing is the purification of virions and viral nucleic acids from samples, particularly when dealing with clinical specimens where quantity is an issue and amplification of nucleic acids is necessary. Another is the limitation of short-read length, which on the SOLiD and Illumina HiSeq averages between 50–100 bases, as compared with Sanger-based sequencing, which can routinely give read lengths above 800 bases. This has severe limitations for the identification of coevolving or linked mutations on the same molecule when the nucleotide positions of interest are separated by a distance that exceeds the sequence read length. These are relevant in viral transmission studies, where a clear picture of the true genetic diversity of the virus population is necessary. Furthermore, the rapid evolution of these sequencing technologies is responsible for generating ever-growing volumes of data. As an example, for the determination of intrahost genetic diversity of 50 multiplexed influenza A virus samples, an eighth of a HiSeq2000 sequence run would typically generate 13–15 GB of sequence data; for efficient analysis, 64 GB of RAM and 3 TB of disk space would be required. Significant bioinformatics resources and infrastructure solely dedicated to data management, analyses and tool development are required for nearly all NextGen sequencing platforms, and this presents another challenge within this dynamic, fast-paced field. A well-planned investment in bioinformatics capabilities will undoubtedly become the norm in any team pursuing virus population questions that can only be addressed with NextGen technologies. As advancements in sequencing continue to be made, and biologists learn to exploit the deluge of data generated, NextGen sequencing stands poised to make even more contributions to the ever-growing exploration of the microbial world.
Future perspective
Many viral evolution studies are beyond the capacity of currently available sequencing technologies. As well as the short read length, another limitation is the inability to accurately characterize all individual genomes present in a virus population sampled within a host. PCR amplification is necessary for most second-generation sequencing technologies; the exception is the Helicos Genetic Analysis Platform, the first commercially available machine with single-molecule sequencing capability, which was successfully used for the sequencing of a complete virus genome without first amplifying the DNA [87]. Imaging of individual molecules attached on a surface is performed during extension with fluorescently labeled nucleotides. While protocol development now allows the direct sequencing of RNA molecules on this platform, read length is still very short (average length of 35 bases). Pacific Biosciences developed a third-generation system based on single-molecule real-time sequencing by synthesis that leads to average read lengths of 1000 bases, with maximums reaching more than 10,000 bases [88]. The sequencing accuracy remains low, however, as compared with the second-generation platforms; not a negligible issue when analyzing the genetic diversity of populations of viruses that are themselves error prone. Many emerging viruses have an RNA genome with diverse genomic structures (circular, linear, segmented, double-stranded and single-stranded). There are thus fundamental questions that cannot be fully addressed until direct sequencing of complete RNA molecules, isolated from individual virions, becomes a possibility; questions such as how a virus evolves in a new host, the level of viral genetic diversity before and after transmission, the effect of transmission bottlenecks on virus population structure and the true representation of defective particles in an infective dose. Third-generation sequencing technologies using nanopores or direct imaging of full-length molecules are, however, right around the corner [88]. There is no doubt that even newer platforms, which advance the technology for single-molecule decoding, will emerge in the next few years. Virus discovery, molecular epidemiology and evolutionary dynamic studies on emerging viruses will then enter a new phase of virus exploration and be greatly facilitated by the tremendous amount of sequence data that could be generated from extremely small samples.
Executive summary.
Whole-genome analyses & viral evolutionary dynamics
-
▪
There is now clear evidence of extensive influenza virus migration and mixing.
-
▪
During seasonal epidemics, there is extensive cocirculation and reassortment of influenza viral lineages.
-
▪
Influenza viruses in wild birds form transient genome constellations, with constant reshuffling of gene segments.
Large-scale sequencing to characterize viral emergence
-
▪
There is clear evidence of intercontinental movement of avian influenza virus gene segments.
-
▪
The ‘source–sink’ model for the emergence of influenza viruses from the tropics is being challenged by the observation that multiple locales are sources of seasonal influenza viruses.
-
▪
Whole-genome virus sequencing from clinical specimens showed that strain variations of dengue viruses were associated with epidemic activity.
-
▪
A common occurrence observed in dengue evolutionary dynamics is the displacement of circulating lineages by newly introduced dengue serotypes.
Next-generation sequencing to assess intrahost diversity
-
▪
Complex viral populations can be characterized by using the deep-sequencing methodology.
-
▪
Hosts can be coinfected with antigenically distinct viruses and viruses with different sensitivities to antivirals, and represent important sources of phenotypic diversity, with a potential for emergence in the population.
Virus discovery & next-generation sequencing platforms
-
▪
Diagnostic virology is greatly impacted by next-generation sequencing because of increased sensitivity.
-
▪
The analysis of animal viromes allows the characterization of potential reservoirs of emerging viruses.
-
▪
Metagenomic studies identified novel viruses and variants from the environment, further expanding our ability to characterize emerging pathogens.
Future perspective
-
▪
Progressive understanding of the evolutionary biology of RNA viruses is limited by the current next-generation sequencing technology platforms.
-
▪
RNA virus discovery and full characterization will benefit greatly from the single-molecule sequencing promised by third-generation sequencing platforms.
Acknowledgments
VG Dugan and E Ghedin are funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services under contract number HHSN2722 00900007C.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affifiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Fiers W, Contreras R, Duerinck F, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, Air GM, Barrell BG, et al. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 3.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 4.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122(2):281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 5.Worobey M, Rambaut A, Holmes EC. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl Acad. Sci. USA. 1999;96(13):7352–7357. doi: 10.1073/pnas.96.13.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boni MF, Zhou Y, Taubenberger JK, Holmes EC. Homologous recombination is very rare or absent in human influenza A virus. J. Virol. 2008;82(10):4807–4811. doi: 10.1128/JVI.02683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bragstad K, Nielsen LP, Fomsgaard A. The evolution of human influenza A viruses from 1999 to 2006: a complete genome study. Virol. J. 2008;5:40. doi: 10.1186/1743-422X-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bahl J, Nelson MI, Chan KH, et al. Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc. Natl Acad. Sci. USA. 2011;108(48):19359–19364. doi: 10.1073/pnas.1109314108. ▪▪ A/H3N2 influenza viruses collected from seven globally distinct geographic regions from 2003 to 2006 were investigated, demonstrating that each region was a potential source population.
- 10.Nelson MI, Edelman L, Spiro DJ, et al. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 2008;4(8):e1000133. doi: 10.1371/journal.ppat.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson MI, Viboud C, Simonsen L, et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008;4(2):e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson M, Spiro D, Wentworth D, et al. The early diversification of influenza A/H1N1pdm. PLoS Curr. Influenza. 2009:RRN1126. doi: 10.1371/currents.RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson MI, Tan Y, Ghedin E, et al. Phylogeography of the spring and fall waves of the H1N1/09 pandemic influenza virus in the United States. J. Virol. 2011;85(2):828–834. doi: 10.1128/JVI.01762-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes EC, Ghedin E, Halpin RA, et al. Extensive geographical mixing of 2009 human H1N1 influenza A virus in a single university community. J. Virol. 2011;85(14):6923–6929. doi: 10.1128/JVI.00438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham M, Liang B, Van Domselaar G, et al. Nationwide molecular surveillance of pandemic H1N1 influenza A virus genomes Canada, 2009. PLoS One. 2011;6(1):e16087. doi: 10.1371/journal.pone.0016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghedin E, Wentworth DE, Halpin RA, et al. Unseasonal transmission of H3N2 influenza A virus during the swine-origin H1N1 pandemic. J. Virol. 2010;84(11):5715–5718. doi: 10.1128/JVI.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzberg SL, Kingsford C, Cattoli G, et al. Genome analysis linking recent European and African influenza (H5N1) viruses. Emerg. Infect. Dis. 2007;13(5):713–718. doi: 10.3201/eid1305.070013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dugan VG, Chen R, Spiro DJ, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4(5):e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koehler AV, Pearce JM, Flint PL, Franson JC, Ip HS. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta) Mol. Ecol. 2008;17(21):4754–4762. doi: 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- 21.Pearce JM, Reeves AB, Ramey AM, et al. Interspecific exchange of avian influenza virus genes in Alaska: the influence of transhemispheric migratory tendency and breeding ground sympatry. Mol. Ecol. 2011;20(5):1015–1025. doi: 10.1111/j.1365-294X.2010.04908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearce JM, Ramey AM, Ip HS, Gill RE., Jr Limited evidence of trans-hemispheric movement of avian influenza viruses among contemporary North American shorebird isolates. Virus Res. 2010;148(1–2):44–50. doi: 10.1016/j.virusres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Wille M, Robertson GJ, Whitney H, Bishop MA, Runstadler JA, Lang AS. Extensive geographic mosaicism in avian influenza viruses from gulls in the northern hemisphere. PLoS One. 2011;6(6):e20664. doi: 10.1371/journal.pone.0020664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves AB, Pearce JM, Ramey AM, Meixell BW, Runstadler JA. Interspecies transmission and limited persistence of low pathogenic avian influenza genomes among Alaska dabbling ducks. Infect. Genet. Evol. 2011;11(8):2004–2010. doi: 10.1016/j.meegid.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt DJ, Pickett BE, Camacho D, et al. A phylogenetic analysis using full-length viral genomes of South American dengue serotype 3 in consecutive Venezuelan outbreaks reveals a novel NS5 mutation. Infect. Genet. Evol. 2011;11(8):2011–2019. doi: 10.1016/j.meegid.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steel A, Gubler DJ, Bennett SN. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a retrospective phylogenetic study of events in the South Pacific three decades ago. Virology. 2010;405(2):505–512. doi: 10.1016/j.virol.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghwani J, Rambaut A, Holmes EC, et al. Endemic dengue associated with the co-circulation of multiple viral lineages and localized density-dependent transmission. PLoS Pathog. 2011;7(6):e1002064. doi: 10.1371/journal.ppat.1002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohainle M, Balmaseda A, Macalalad AR, et al. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci. Transl. Med. 2011;3(114):114ra128. doi: 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vu TT, Holmes EC, Duong V, et al. Emergence of the Asian 1 genotype of dengue virus serotype 2 in vietnam: in vivo fitness advantage and lineage replacement in south-east Asia. PLoS Negl. Trop. Dis. 2010;4(7):e757. doi: 10.1371/journal.pntd.0000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mcelroy KL, Santiago GA, Lennon NJ, Birren BW, Henn MR, Munoz-Jordan JL. Endurance, refuge, and reemergence of dengue virus type 2, Puerto Rico, 1986-2007. Emerg. Infect. Dis. 2011;17(1):64–71. doi: 10.3201/eid1701.100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogrefe HH, Hansen CJ, Scott BR, Nielson KB. Archaeal dUTPase enhances PCR amplifications with archaeal DNA polymerases by preventing dUTP incorporation. Proc. Natl Acad. Sci. USA. 2002;99(2):596–601. doi: 10.1073/pnas.012372799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thai KT, Henn MR, Zody MC, et al. High-resolution analysis of intrahost genetic diversity in dengue virus serotype 1 infection identifies mixed infections. J. Virol. 2012;86(2):835–843. doi: 10.1128/JVI.05985-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glenn TC. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11(5):759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 34.Margeridon-Thermet S, Shulman NS, Ahmed A, et al. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J. Infect. Dis. 2009;199(9):1275–1285. doi: 10.1086/597808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedskog C, Mild M, Jernberg J, et al. Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing. PLoS One. 2010;5(7):e11345. doi: 10.1371/journal.pone.0011345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 2009;199(5):693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 37.Nasu A, Marusawa H, Ueda Y, et al. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PLoS One. 2011;6(9):e24907. doi: 10.1371/journal.pone.0024907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buzon MJ, Seiss K, Weiss R, et al. Inhibition of HIV-1 integration in ex vivo-infected CD4 T cells from elite controllers. J. Virol. 2011;85(18):9646–9650. doi: 10.1128/JVI.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Henn MR, Boutwell CL, Charlebois P, et al. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8(3):e1002529. doi: 10.1371/journal.ppat.1002529. ▪▪ Report on an elegant deep-sequencing approach for the detection of low-frequency HIV-1 variants. The majority of the low-frequency mutations were associated with viral adaptations to host cellular immune responses.
- 40.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 41.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 42.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balboni A, Battilani M, Prosperi S. The SARS-like coronaviruses: the role of bats and evolutionary relationships with SARS coronavirus. New Microbiol. 2012;35(1):1–16. [PubMed] [Google Scholar]
- 44.Eckerle LD, Becker MM, Halpin RA, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6(5):e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlasova AN, Halpin R, Wang S, Ghedin E, Spiro DJ, Saif LJ. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J. Gen. Virol. 2011;92(Pt 6):1369–1379. doi: 10.1099/vir.0.025353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byarugaba DK, Ducatez MF, Erima B, et al. Molecular epidemiology of influenza A/H3N2 viruses circulating in Uganda. PLoS One. 2011;6(11):e27803. doi: 10.1371/journal.pone.0027803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baillie GJ, Galiano M, Agapow PM, et al. Evolutionary dynamics of local pandemic H1N1/09 influenza lineages revealed by whole genome analysis. J. Virol. 2011;86(1):11–18. doi: 10.1128/JVI.05347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 49.Lorusso A, Vincent AL, Harland ML, et al. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J. Gen. Virol. 2011;92(Pt 4):919–930. doi: 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ducatez MF, Hause B, Stigger-Rosser E, et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 2011;17(9):1624–1629. doi: 10.3201/1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dugan VG, Dunham EJ, Jin G, et al. Phylogenetic analysis of low pathogenicity H5N1 and H7N3 influenza A virus isolates recovered from sentinel, free flying, wild mallards at one study site during 2006. Virology. 2011;417(1):98–105. doi: 10.1016/j.virol.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lam TT, Ip HS, Ghedin E, et al. Migratory flyway and geographical distance are barriers to the gene flow of influenza virus among North American birds. Ecol. Lett. 2012;15(1):24–33. doi: 10.1111/j.1461-0248.2011.01703.x. ▪ Comprehensive phylogeographic study incorporating wild-bird-origin influenza genomes, spatial distance and avian flyways/geographical barriers to model the avian influenza virus gene flow within North American wild birds.
- 53.Flaherty P, Natsoulis G, Muralidharan O, et al. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res. 2012;40(1):e2. doi: 10.1093/nar/gkr861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoper D, Hoffmann B, Beer M. A comprehensive deep sequencing strategy for full-length genomes of influenza A. PLoS One. 2011;6(4):e19075. doi: 10.1371/journal.pone.0019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoper D, Hoffmann B, Beer M. Simple, sensitive, and swift sequencing of complete H5N1 avian influenza virus genomes. J. Clin. Microbiol. 2009;47(3):674–679. doi: 10.1128/JCM.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoper D, Kalthoff D, Hoffmann B, Beer M. Highly pathogenic avian influenza subtype H5N1 escaping neutralization: more than HA variation. J. Virol. 2011 doi: 10.1128/JVI.00797-11. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kampmann ML, Fordyce SL, Avila-Arcos MC, et al. A simple method for the parallel deep sequencing of full influenza A genomes. J. Virol. Methods. 2011;178(1–2):243–248. doi: 10.1016/j.jviromet.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Roedig JV, Rapp E, Hoper D, Genzel Y, Reichl U. Impact of host cell line adaptation on quasispecies composition and glycosylation of influenza a virus hemagglutinin. PLoS One. 2011;6(12):e27989. doi: 10.1371/journal.pone.0027989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghedin E, Fitch A, Boyne A, et al. Mixed infection and the genesis of influenza virus diversity. J. Virol. 2009;83(17):8832–8841. doi: 10.1128/JVI.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuroda M, Katano H, Nakajima N, et al. Characterization of quasispecies of pandemic 2009 influenza A virus (A/H1N1/2009) by de novo sequencing using a next-generation DNA sequencer. PLoS One. 2010;5(4):e10256. doi: 10.1371/journal.pone.0010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghedin E, Laplante J, Depasse J, et al. Deep sequencing reveals mixed infection with 2009 pandemic influenza A (H1N1) virus strains and the emergence of oseltamivir resistance. J. Infect. Dis. 2011;203(2):168–174. doi: 10.1093/infdis/jiq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu B, Liu L, Huang X, et al. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog. 2011;7(11):e1002369. doi: 10.1371/journal.ppat.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360(25):2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 64.Yang J, Yang F, Ren L, et al. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J. Clin. Microbiol. 2011;49(10):3463–3469. doi: 10.1128/JCM.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barzon L, Lavezzo E, Militello V, Toppo S, Palu G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12(11):7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamura S, Yang CS, Sakon N, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4(1):e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Svraka S, Rosario K, Duizer E, Van Der Avoort H, Breitbart M, Koopmans M. Metagenomic sequencing for virus identification in a public-health setting. J. Gen. Virol. 2010;91(Pt 11):2846–2856. doi: 10.1099/vir.0.024612-0. [DOI] [PubMed] [Google Scholar]
- 68.Blinkova O, Kapoor A, Victoria J, et al. Cardioviruses are genetically diverse and cause common enteric infections in south Asian children. J. Virol. 2009;83(9):4631–4641. doi: 10.1128/JVI.02085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4(2):e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gaynor AM, Nissen MD, Whiley DM, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3(5):e64. doi: 10.1371/journal.ppat.0030064. ▪ Clear example of how a high-throughput strategy can be used to help in the diagnosis of infections of unknown etiology.
- 72.Victoria JG, Kapoor A, Li L, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 2009;83(9):4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li L, Victoria J, Kapoor A, et al. A novel picornavirus associated with gastroenteritis. J. Virol. 2009;83(22):12002–12006. doi: 10.1128/JVI.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Victoria JG, Wang C, Jones MS, et al. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J. Virol. 2010;84(12):6033–6040. doi: 10.1128/JVI.02690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blomstrom AL. Viral metagenomics as an emerging and powerful tool in veterinary medicine. Vet. Q. 2011;31(3):107–114. doi: 10.1080/01652176.2011.604971. [DOI] [PubMed] [Google Scholar]
- 76.Jin L, Lohr CV, Vanarsdall AL, et al. Characterization of a novel alphaherpesvirus associated with fatal infections of domestic rabbits. Virology. 2008;378(1):13–20. doi: 10.1016/j.virol.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 77.Shan T, Li L, Simmonds P, Wang C, Moeser A, Delwart E. The fecal virome of pigs on a high-density farm. J. Virol. 2011;85(22):11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc. Natl Acad. Sci. USA. 2012;109(11):4269–4274. doi: 10.1073/pnas.1116200109. ▪▪ Surveillance using Sanger and nextgeneration sequencing of more than 20 bat species in Guatemala discovered a novel lineage of influenza A virus where the hemagglutinin gene segment was novel (named H17).
- 79.Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 2011;92(Pt 11):2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Volkening JD, Spatz SJ. Purification of DNA from the cell-associated herpesvirus Marek’s disease virus for 454 pyrosequencing using micrococcal nuclease digestion and polyethylene glycol precipitation. J. Virol. Methods. 2009;157(1):55–61. doi: 10.1016/j.jviromet.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 81.Allen LZ, Ishoey T, Novotny MA, Mclean JS, Lasken RS, Williamson SJ. Single virus genomics: a new tool for virus discovery. PLoS One. 2011;6(3):e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Djikeng A, Spiro D. Advancing full length genome sequencing for human RNA viral pathogens. Future Virol. 2009;4(1):47–53. doi: 10.2217/17460794.4.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010;48(12):4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Victoria JG, Kapoor A, Dupuis K, Schnurr DP, Delwart EL. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 2008;4(9):e1000163. doi: 10.1371/journal.ppat.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoffmann B, Scheuch M, Hoper D, et al. Novel orthobunyavirus in cattle, Europe, 2011. Emerg. Infect. Dis. 2012;18(3):469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, Van Dijk AA. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J. Gen. Virol. 2009;90(Pt 6):1423–1432. doi: 10.1099/vir.0.009381-0. [DOI] [PubMed] [Google Scholar]
- 87.Harris TD, Buzby PR, Babcock H, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320(5872):106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 88. Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum. Mol. Genet. 2010;19(R2):R227–R240. doi: 10.1093/hmg/ddq416. ▪ Comprehensive and clear overview of third-generation sequencing platforms and how they compare with first- and second-generation sequencing.