Nuclear transport of large protein cargoes such as Tpr is more sensitive to the alteration of the ratio of nuclear to cytoplasmic Ran that occurs in Progeria.
Abstract
The RanGTPase acts as a master regulator of nucleocytoplasmic transport by controlling assembly and disassembly of nuclear transport complexes. RanGTP is required in the nucleus to release nuclear localization signal (NLS)–containing cargo from import receptors, and, under steady-state conditions, Ran is highly concentrated in the nucleus. We previously showed the nuclear/cytoplasmic Ran distribution is disrupted in Hutchinson-Gilford Progeria syndrome (HGPS) fibroblasts that express the Progerin form of lamin A, causing a major defect in nuclear import of the protein, translocated promoter region (Tpr). In this paper, we show that Tpr import was mediated by the most abundant import receptor, KPNA2, which binds the bipartite NLS in Tpr with nanomolar affinity. Analyses including NLS swapping revealed Progerin did not cause global inhibition of nuclear import. Rather, Progerin inhibited Tpr import because transport of large protein cargoes was sensitive to changes in the Ran nuclear/cytoplasmic distribution that occurred in HGPS. We propose that defective import of large protein complexes with important roles in nuclear function may contribute to disease-associated phenotypes in Progeria.
Introduction
The RanGTPase system plays a central role in regulating nuclear import and export in eukaryotes. Ran regulates import and export through protein interactions that are highly specific for its GDP- and GTP-bound forms (Görlich et al., 1996). RanGDP is recognized in the cytoplasm by NTF2 (nuclear transport factor 2), which mediates its rapid translocation into the nucleus where the Ran guanine nucleotide exchange factor RCC1 (regulator of chromosome condensation 1) mediates a nucleotide exchange reaction that generates RanGTP (Bischoff and Ponstingl, 1991; Ribbeck et al., 1998; Smith et al., 1998). Nuclear RanGTP functions to promote disassembly of import complexes containing Importin-β that have translocated from the cytoplasm to the nucleoplasm and assembly of export complexes containing Crm1 that subsequently translocate from the nucleoplasm to the cytoplasm (Rexach and Blobel, 1995; Fornerod et al., 1997; Stade et al., 1997). Because the engagement of Ran with both import and export complexes occurs in the nucleus and is RanGTP specific, maintaining a sufficient concentration of nuclear Ran via NTF2-dependent import and nucleotide exchange by RCC1 is critical for nuclear transport and represents a mechanism that is conserved across phyla (Ohtsubo et al., 1989; Corbett and Silver, 1996; Paschal et al., 1997; Ribbeck et al., 1998; Smith et al., 1998).
Our laboratory has shown that fibroblasts from patients with Hutchinson-Gilford Progeria syndrome (HGPS) have a defect in the RanGTPase system (Kelley et al., 2011). Cells from these patients show a significant reduction in the nuclear level of Ran, which can be quantified as a reduced nuclear/cytoplasmic concentration of Ran. HGPS is caused by a de novo mutation in LMNA that generates a mutant lamin A protein termed Progerin (Eriksson et al., 2003). Progerin exerts dominant-negative effects on the cell, and although the molecular basis of these effects has not been defined, it is clear that Progerin effects are linked to a defect in its posttranslational processing (Worman et al., 2010). Progerin lacks the proteolytic cleavage site that is used to release lamin A from its lipid anchor at the membrane (Eriksson et al., 2003). Thus, Progerin remains stably attached to the inner nuclear membrane, where it induces changes in nuclear morphology as well as changes in chromatin state and gene expression (Csoka et al., 2004; Shumaker et al., 2006). How Progerin disrupts the Ran system is unclear. The fact that the Ran guanine nucleotide exchange factor RCC1 undergoes reversible chromatin binding as part of the nucleotide exchange cycle (Nemergut et al., 2001), together with Progerin-induced reduction in RCC1 nuclear mobility (Kelley et al., 2011), led us to propose the Ran defects in HGPS might reflect reduced exchange activity by RCC1.
Our current view of the Ran disruption in HGPS cells is that it reduces the nuclear concentration of Ran, but only to a concentration that can be tolerated in terms of nuclear transport levels that are necessary to maintain cell viability. One could envision, therefore, that Ran disruption in HGPS affects all Ran-dependent transport pathways to a limited degree, depending on the abundance of transport receptors and cargoes. An alternative possibility is that certain nuclear transport pathways are more sensitive to changes in the Ran system, based on the affinity of Ran for different nuclear transport receptors and their distinct cargoes.
In our initial description of Ran system changes in HGPS cells, we showed that nuclear import of translocated promoter region (Tpr), a major nucleoporin of the nuclear pore complex (NPC), is inhibited by expression of Progerin (Kelley et al., 2011). Tpr forms the “basketlike” structure on the nuclear side of the NPC, using Nup153 as its NPC anchoring site (Hase and Cordes, 2003; Krull et al., 2004; D’Angelo and Hetzer, 2008; Strambio-De-Castillia et al., 2010; Wente and Rout, 2010). Tpr contains a single nuclear localization sequence near the C terminus, which is necessary and sufficient for its nuclear import (Cordes et al., 1998). Tpr is also known to be the last nucleoporin assembled into the NPC at the end of mitosis (Bodoor et al., 1999b; Burke and Ellenberg, 2002; Qi et al., 2004), showing that the basic NPC function is established before assembly of the basket. With a subunit molecular mass of 267 kD, Tpr is one of the largest nucleoporins, which also is known to assemble into a homodimer (Hase et al., 2001). In this study, we provide formal evidence that the Tpr import defect in HGPS reflects the sensitivity of this pathway to the nuclear concentration of Ran. Detailed analysis of the Tpr import pathway showed that it depends on a bipartite NLS and recognition by KPNA2, a highly abundant import receptor that is capable of binding a variety of import signals (Kelley et al., 2010). Our data indicate that the import defect of Tpr is not linked specifically to its type of NLS, its import receptor, or the ability of the complex to undergo Ran-induced dissociation but rather to its molecular size. Our data suggest that disruption of the Ran gradient in HGPS can reduce the nuclear accumulation of specific proteins, including Tpr, based on the different Ran requirement for nuclear import of large cargoes.
Results
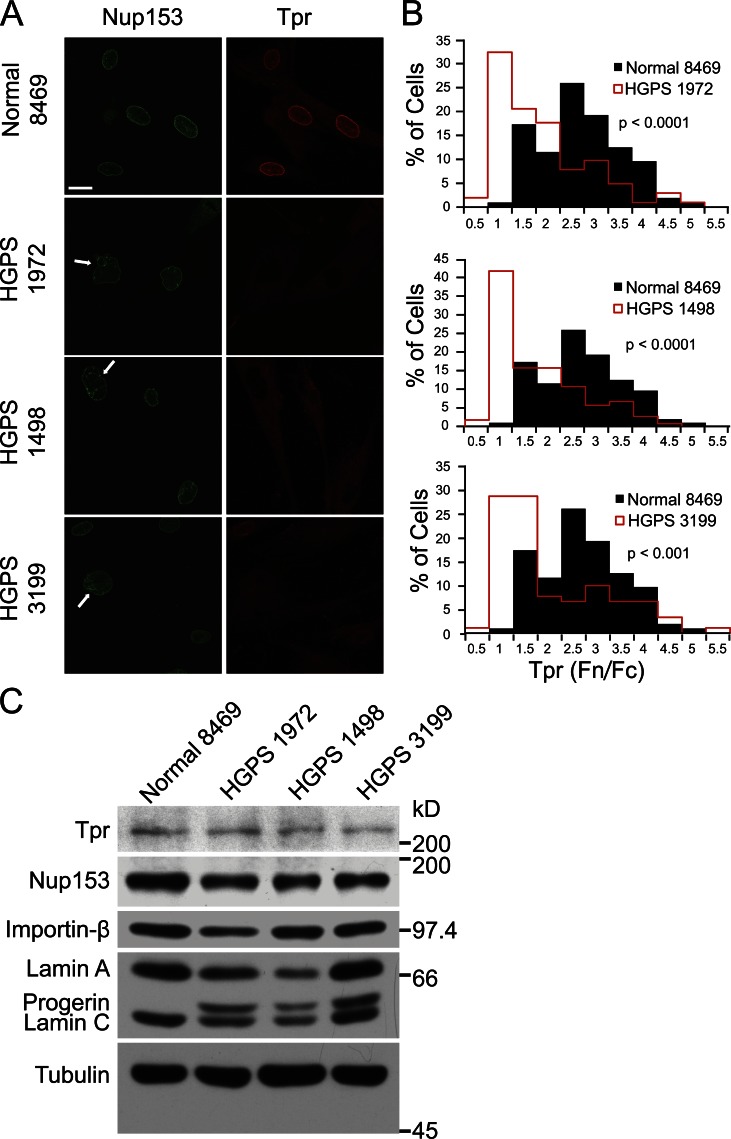
Our laboratory has shown that Tpr, which forms the basket on the nuclear side of the NPC (Krull et al., 2004), is mislocalized to the cytoplasm in HGPS patient cells (Kelley et al., 2011). We set out to define the basis of this defect, first by examining whether the Tpr-anchoring protein Nup153 (Hase and Cordes, 2003) is properly localized at the nuclear envelope in cells from HGPS patients. In primary fibroblasts from three different HGPS patients (HGPS 1972, HGPS 1498, and HGPS 3199), Tpr displayed quantitative defects in nuclear localization (P < 0.0001), as determined by comparing the nuclear/cytoplasmic ratios (fluorescence nucleus/fluorescence cytoplasm [Fn/Fc]) of Tpr in patient (Fig. 1 B, red histograms) and normal (Fig. 1 B, black histograms) cells (Fig. 1, A and B). By double-label immunofluorescence (IF), Nup153 was properly localized in HGPS fibroblasts with strong Tpr import defects (Fig. 1 A), suggesting the nuclear localization defect of Tpr is not a consequence of loss of its anchoring site inside the nucleus. We noted that patient fibroblasts with Tpr defects often displayed irregular nuclear morphology (Fig. 1 A, white arrows), commonly reported for HGPS cells (Goldman et al., 2004). Tpr protein levels in the patient and control fibroblasts were similar (Fig. 1 C), indicating the reduced nuclear levels of Tpr in HGPS cells are not caused by reduced protein expression.
Figure 1.
The nucleoporin Tpr is mislocalized in fibroblasts from HGPS patients. (A) IF microscopy showing the localization of Tpr (red) and its NPC docking partner Nup153 (green) in normal (8469) and HGPS (1972, 1498, and 3199) primary fibroblasts. The white arrows denote examples of nuclei with membrane changes. Bar, 20 µm. (B) Histograms of Tpr Fn/Fc (fluorescence of nucleus/fluorescence of cytoplasm) from control (Normal 8469; n = 208) and patient (HGPS 1972, HGPS 1498, and HGPS 3199; n = 91, 104, and 203, respectively) cells. The IF microscopy is from a single experiment and is representative of nine experiments. (C) Immunoblotting of Tpr, Nup153, Karyopherin-β, lamin A/C, Progerin, and Tubulin in normal and patient fibroblasts.
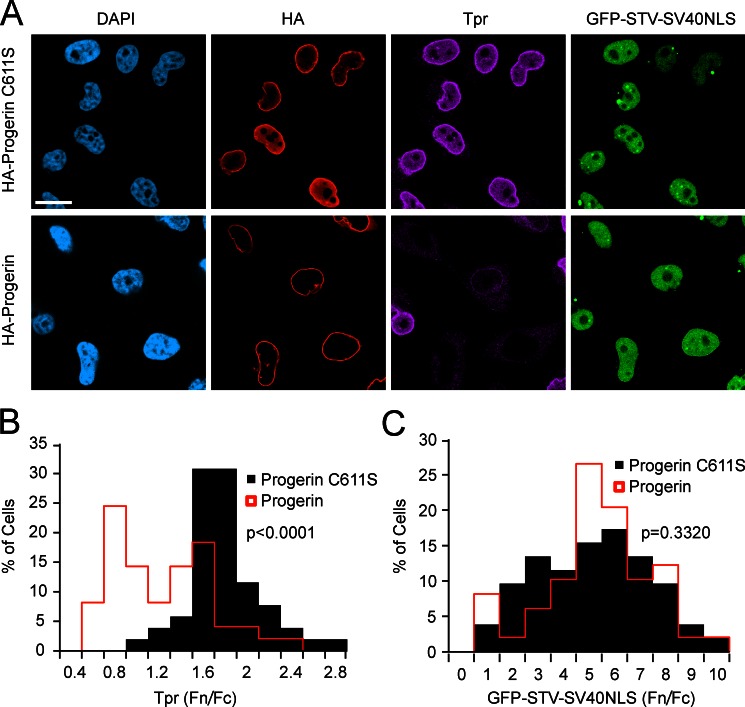
Tpr import can be inhibited by ectopic expression of Progerin in HeLa cells (Kelley et al., 2011). To determine whether Progerin effects are selective for Tpr import or reflect a more global effect on NLS-dependent import, we introduced HA-Progerin into a HeLa cell line that stably expresses a GFP reporter protein containing the SV40NLS (Black et al., 1999). By triple-label IF, HA-Progerin–expressing cells with defects in Tpr import (Fig. 2, A [purple] and B) did not show significant defects in SV40NLS-dependent import (Fig. 2, A and C, compare black bars to Progerin transfection denoted by red lines). To determine whether the inhibitory effect of Progerin on Tpr import requires nuclear membrane attachment, we engineered a form of Progerin that contains a cysteine to serine substitution (C611S) in the C-terminal CAAX motif, which contains the farnesylation site (Weber et al., 1989; Lutz et al., 1992; Sinensky et al., 1994). HA-Progerin C611S was expressed at a level similar to HA-Progerin; however, the C611S substitution prevented the Tpr import defects induced by Progerin. Because Progerin requires a functional CAAX motif to inhibit Tpr import, and Tpr import defects in HGPS fibroblasts can be blocked with a farnesyltransferase inhibitor that blocks Progerin farnesylation (Kelley et al., 2011), we conclude the dominant-negative effect of Progerin on Tpr import is initiated by a stable attachment to the inner nuclear membrane.
Figure 2.
A functional CAAX motif is required for Progerin to inhibit Tpr import. (A) Triple-label IF microscopy of HeLa cells transfected with HA-Progerin (red) and HA-Progerin with a CAAX motif substitution (C611S; red). Endogenous Tpr (purple) and GFP-STV-SV40NLS (green) were also detected. Bar, 20 µm. (B) Histogram of Tpr Fn/Fc from HA-Progerin C611S–expressing cells (n = 52) and HA-Progerin–expressing cells (n = 49; P < 0.0001). The IF microscopy is from a single experiment and is representative of five experiments. (C) Histogram of GFP-STV-SV40NLS Fn/Fc from HA-Progerin C611S (n = 52)– and HA-Progerin–expressing cells (n = 49; P = 0.3320). The IF microscopy is from a single experiment and is representative of 10 experiments.
Tpr import correlates with the nuclear levels of Ran
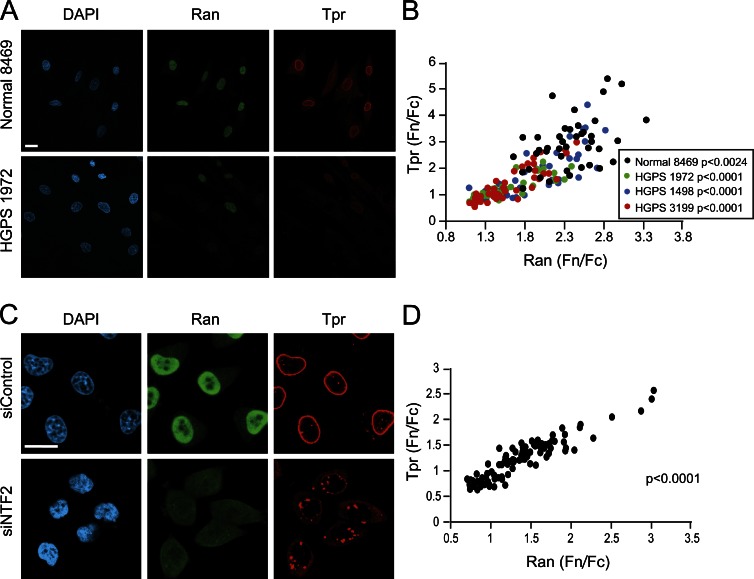
As the Ran gradient is disrupted in HGPS cells in which Tpr import is defective (Kelley et al., 2011), we hypothesized that Tpr localization to the nucleus might be highly sensitive to the nuclear concentration of Ran. We found that nuclear levels of Tpr (Fn/Fc) and nuclear Ran (Fn/Fc) are highly correlated in both normal and patient fibroblasts (Fig. 3, A and B, Spearman P < 0.0024 for Normal 8469, and P < 0.0001 for all HGPS patient lines). The correlation between nuclear Tpr and Ran was also observed in HeLa cells depleted of the Ran import factor NTF2 (Fig. 3, C and D, Spearman P < 0.0001), in which defective import of Tpr induces the accumulation of Tpr aggregates in the cytoplasm. Nuclear levels of Ran are rate limiting for Tpr import in both normal and HGPS patient cells, likely because nuclear RanGTP is required for Tpr dissociation from its import receptor (Ben-Efraim et al., 2009). Disrupting the Ran gradient by NTF2 knockdown resulted in Tpr aggregates in the cytoplasm and perhaps in the nucleus (Fig. 3 C). As Ran system mutants were shown by Ryan et al. (2003) to affect NPC assembly in yeast, it is certainly possible that NTF2 knockdown induces defects beyond the Progerin reduction of Tpr import.
Figure 3.
Tpr localization is correlated with the Ran protein gradient. (A) IF microscopy of Tpr (red) and Ran (green) in Normal 8469 and HGPS 1972 fibroblasts. (B) Tpr Fn/Fc values plotted as a function of Ran Fn/Fc values for control 8469 (n = 41) and HGPS 1972, HGPS 1498, and HGPS 3199 fibroblasts (n = 31, 41, and 43, respectively). Spearman P < 0.0024 for control 8469, and P < 0.0001 for each HGPS line. (C) Tpr (red) and Ran (green) in control and NTF2-depleted HeLa cells. (D) Tpr Fn/Fc values plotted as a function of Ran Fn/Fc values (Spearman P < 0.0001) in control and NTF2-depleted HeLa cells. The correlations shown in C and D are from a single experiment and representative of at least three experiments. siControl, control siRNA. Bars, 20 µm.
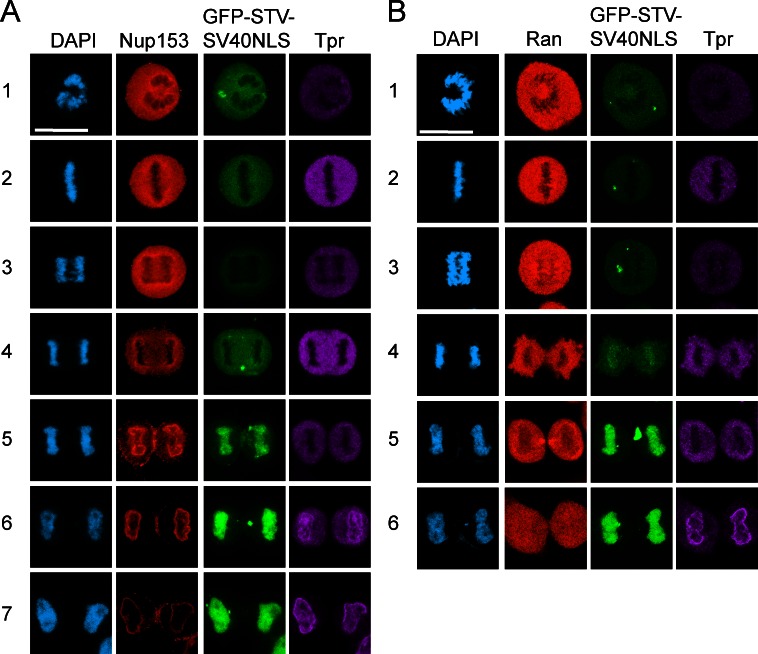
Nuclear envelope disassembly during mitosis results in loss of the nuclear/cytoplasmic Ran gradient protein. Reestablishing the Ran gradient in telophase therefore requires nuclear envelope formation and NPC assembly, which occur in a stepwise process. Tpr requires an NLS for its localization (Cordes et al., 1998) and is the last nucleoporin incorporated to the NPC (Bodoor et al., 1999a,b; Antonin et al., 2008; Dultz et al., 2008; Lince-Faria et al., 2009), indicating that nuclear pores are functional for transport before addition of Tpr to the NPC. We posited the temporal delay in Tpr import described by other groups might reflect the time required to establish a steep nucleocytoplasmic Ran gradient. Using a Nup153 antibody to detect NPC assembly at the nuclear envelope, we observed the accumulation of GFP-streptavidin (STV)-SV40NLS in daughter cell nuclei (Fig. 4 A, rows 5 and 6) before the nuclear localization of Tpr (Fig. 4 A, row 7). Nuclear accumulation of GFP-STV-SV40NLS can be observed before a nuclear/cytoplasmic Ran gradient protein has been established, and at this stage, Tpr is still cytoplasmic (Fig. 4 B, row 5). These data, together with the data from interphase cells (Fig. 3), are consistent with a model wherein the efficiency of nuclear import of two different cargoes, Tpr and the SV40NLS reporter, reflects the Ran sensitivity of the respective transport pathway.
Figure 4.
Tpr import occurs after the Ran gradient is formed during mitosis. (A) IF microscopy for Nup153 (red), GFP-STV-SV40NLS (green), and Tpr (purple) during mitosis. (B) IF microscopy for Ran (red), GFP-STV-SV40NLS (green), and Tpr (purple) during mitosis. Bars, 20 µm.
Tpr contains a bipartite NLS recognized by KPNA2 during nuclear import
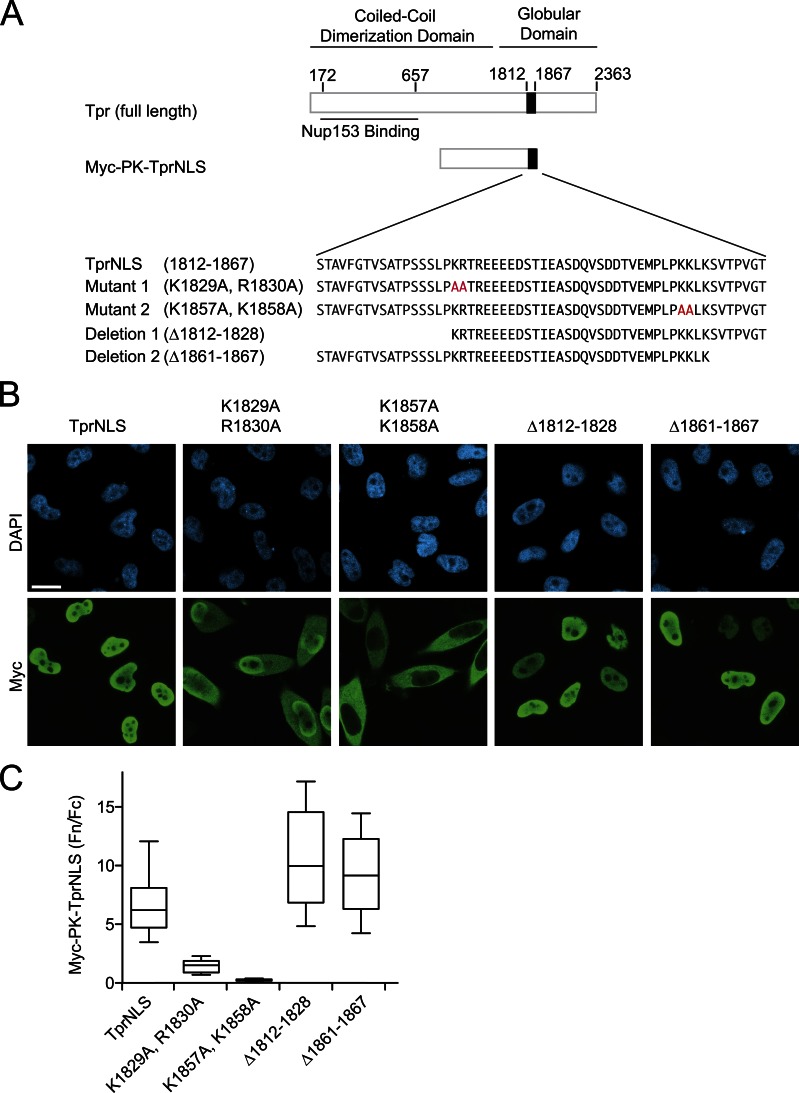
To gain insight into how different cargoes can display different sensitivities to the interphase Ran gradient, we explored whether there are any special features of the Tpr import pathway that explain its sensitivity to Progerin (Fig. 2). We examined the TprNLS defined by Cordes et al. (1998), which is relatively large (56 amino acids) compared with most other NLSs (Robbins et al., 1991). Within the 56–amino acid NLS are two clusters of basic amino acids (1,829–1,832 and 1,857–1,860) separated by a spacer sequence, an arrangement found in bipartite NLSs. Using Myc-tagged pyruvate kinase (PK; Myc-PK) as a fusion partner, we mutated the basic clusters and found that both contribute to the import efficiency of the TprNLS, with a larger contribution by the second cluster. We then engineered deletions on the N- and C-terminal sides of each basic cluster and determined that residues 1,812–1,828 and 1,861–1,867 are dispensable for activity of the import signal (Fig. 5, A–C). Our data suggest that the minimal import signal for Tpr is a 32–amino acid, bipartite NLS.
Figure 5.
Tpr contains a 32–amino acid bipartite NLS. (A) Diagram depicting major domains and amino acid substitutions engineered within the NLS of Tpr. NLS function of the mutants and deletions was determined using fusions to pyruvate kinase (Myc-PK-TprNLS). Red letters highlight alanine substitutions. (B and C) IF microscopy and Fn/Fc values of TprNLS mutants and deletions. Bar, 20 µm. Whiskers are 10th and 90th percentiles. Box lines are 25th, median, and 75th percentiles.
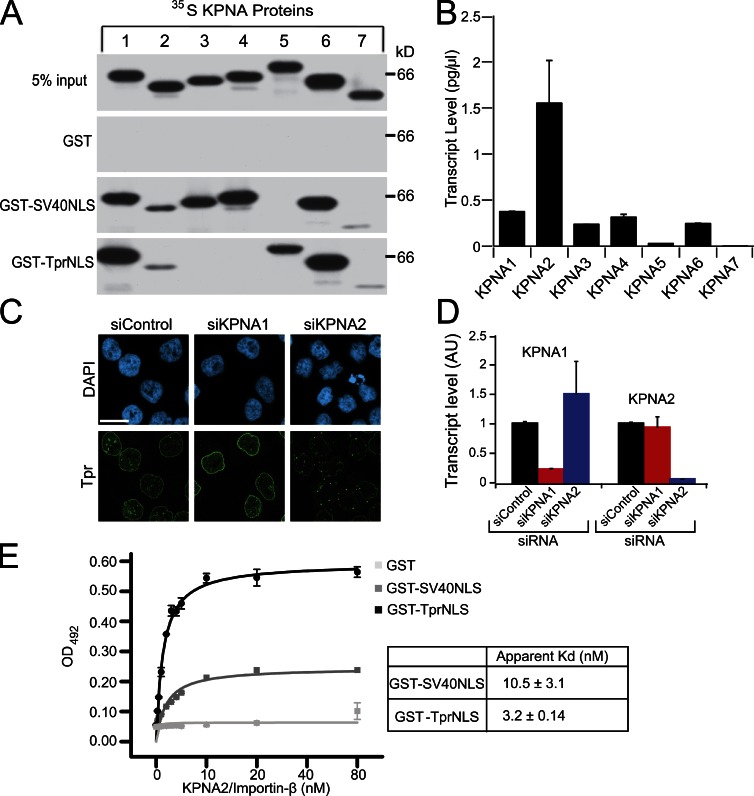
Although bipartite NLSs are common to nuclear-localized proteins, certain members of the Importin-α/Karyopherin-α (KPNA) family of nuclear import receptors display NLS binding preferences, suggesting some cargoes might have dedicated receptors (Köhler et al., 1999; Kelley et al., 2010). Thus, we considered it possible that Progerin selectively prevents Tpr import by inhibiting a specific transport receptor. We analyzed which KPNA proteins bind the TprNLS and mediate its import in cultured cells. The seven KPNA receptors were generated as 35S-labeled proteins and combined with GST fusion proteins in binding assays with the TprNLS and, as a control, the SV40NLS. KPNA1 and KPNA6 displayed the most robust binding to the TprNLS, whereas less binding was observed with KPNA2, KPNA5, and KPNA7 (Fig. 6 A). By quantitative PCR, transcripts for five KPNA isoforms were detected in HeLa cells, with KPNA2 as the most abundant isoform (Fig. 6 B). We applied KPNA1 and KPNA2 siRNA to HeLa cells and observed that depletion of KPNA2, but not KPNA1, results in a Tpr import defect and its accumulation in cytoplasmic aggregates (Fig. 6, C and D). Although KPNA2 displays weaker binding to the TprNLS compared with the other KPNA proteins, it appears to be responsible for Tpr import in HeLa cells. We found that KPNA2 has a slightly higher affinity for the TprNLS (3 nM) compared with SV40NLS (10 nM; Fig. 6 E). Thus, in a hypothetical scenario of Progerin-induced loss of KPNA2 function, SV40NLS-dependent import should be more sensitive than TprNLS-dependent import. Interestingly, KPNA specificity for SV40NLS binding overlaps the TprNLS in terms of receptor binding but also included binding to KPNA3 and KPNA4 (Fig. 6 A). The possibility that Progerin-insensitive SV40NLS-dependent import results from utilization of different KPNA proteins is addressed in subsequent experiments.
Figure 6.
Tpr import is mediated by the KPNA2/Importin-β heterodimer. (A) TprNLS and SV40NLS binding was analyzed using 35S-labeled KPNA isoforms and GST-immobilized signals. (B) Transcript levels (picograms per microliter) of KPNA isoforms in HeLa cells assayed by quantitative real-time PCR. (C) IF microscopy of Tpr (green) in HeLa cells treated with siRNA to KPNA1 and KPNA2. Bar, 20 µm. (D) RT-PCR analysis of KPNA1 and KPNA2 transcript levels in HeLa cells depleted of each isoform. AU, arbitrary unit. (E) Recombinant KPNA2/Importin-β binding to GST-TprNLS, GST-SV40NLS, and GST measured by ELISA. The apparent Kd for KPNA2 binding to GST-TprNLS is 3.2 ± 0.14 nM and GST-SV40NLS is 10.5 ± 3.1 nM (the mean Kd ± SEM calculated from two experiments). The data were fit with a one-site binding function in OriginPro. siControl, control siRNA. Error bars represent the standard deviation of duplicate wells.
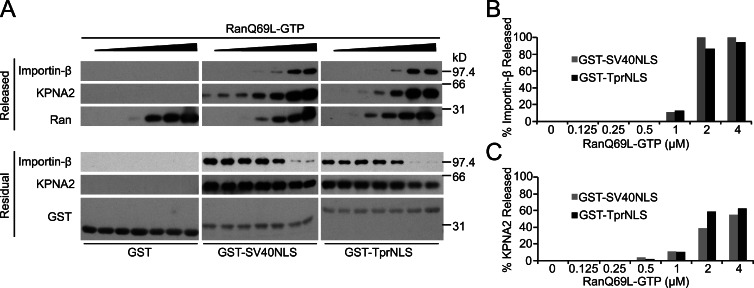
RanGTP is required to disassemble import complexes in the terminal step of nuclear import (Pemberton and Paschal, 2005). Progerin expression reduces the concentration of nuclear Ran and reduces the mobility of the Ran nucleotide exchange factor RCC1 (Kelley et al., 2011), observations consistent with the idea that Progerin-expressing cells have reduced levels of nuclear RanGTP. We therefore tested the sensitivities of TprNLS and SV40NLS to RanGTP-induced dissociation of import complexes containing these cargoes, reasoning that the particular NLS might dictate the concentration of Ran required for efficient dissociation as the final step of nuclear import. This hypothesis is in alignment with the observation that import mediated by different NLSs display different sensitivities to NTF2 levels in permeabilized cell assays (Hu and Jans, 1999). We found that KPNA2/Importin-β complexes with TprNLS and with SV40NLS were disassembled with similar concentrations of RanGTP. Importin-β, which is the direct target of RanGTP in this assay, was released almost quantitatively from both types of import complexes (Fig. 7 B). KPNA2 release was less complete (∼50%; Fig. 7 C), which could reflect the reduced efficiency of this reaction in the absence of CAS (Sun et al., 2008). Thus, although the Tpr and SV40 NLSs display threefold differences in the apparent affinity for KPNA binding, import receptor dissociation from these signals occurs with similar concentrations of RanGTP.
Figure 7.
Ran-dependent disassembly of import complexes containing the TprNLS and SV40NLS. (A) RanQ69L-GTP induced dissociation of import factors preassembled on immobilized SV40NLS and TprNLS. The released (top) and residual fractions (bottom) of KPNA2, Importin-β, Ran, and GST target proteins were analyzed by immunoblotting. (B and C) Quantitation of the released fraction of Importin-β and KPNA2. Bar graphs are from the blotting data in A and are representative of three experiments.
SV40NLS does not rescue the Tpr import defect
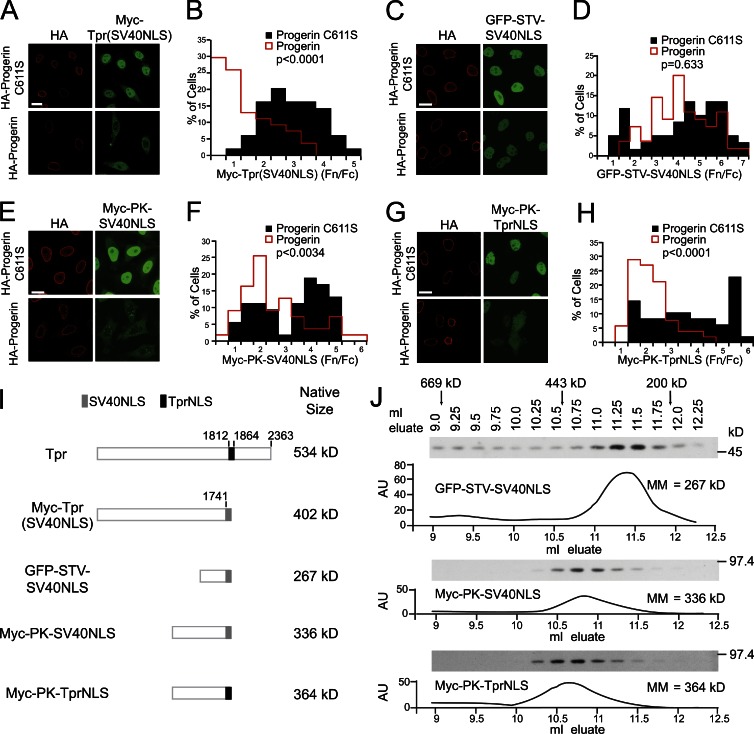
The fact that TprNLS-mediated import depends on the most abundant import receptor, KPNA2, and shows typical RanGTP-stimulated release from KPNA2, implies that another mechanism is responsible for the inhibitory effects of Progerin on Tpr import. We reasoned that if a region of Tpr outside of its NLS mediates the inhibitory effects of Progerin, a form of Tpr in which the SV40NLS has been substituted for the endogenous NLS (Cordes et al., 1998) should still display an import defect. Alternatively, if the inhibitory effect was linked to the TprNLS itself, substituting the SV40NLS could rescue the Tpr import defect caused by Progerin. We found that nuclear import of Myc-Tpr(SV40NLS) was strongly inhibited by Progerin expression (Fig. 8, A and B, P < 0.0001). This contrasted sharply with the Progerin-resistant import of GFP-STV-SV40NLS (Fig. 8, C and D, P = 0.633). Thus, Progerin inhibition of Tpr import is associated with a feature of Tpr apart from its endogenous NLS.
Figure 8.
Progerin inhibition of Tpr import is size dependent but not signal specific. (A–H) IF microscopy and quantitative analysis of Tpr and reporter proteins cotransfected with HA-Progerin C611S and HA-Progerin. (A and B) Myc-Tpr, in which the endogenous TprNLS was replaced with SV40NLS, cotransfected with HA-Progerin C611S (n = 49) or HA-Progerin (n = 54). P < 0.0001. The IF microscopy is from a single experiment and is representative of three experiments. (C and D) GFP-STV-SV40NLS cotransfected with HA-Progerin C611S (n = 59) or HA-Progerin (n = 55). P = 0.633. The IF microscopy is from a single experiment and is representative of 10 experiments. (E and F) Myc-PK-SV40NLS cotransfected with HA-Progerin C611S (n = 53) or HA-Progerin (n = 55). P < 0.0034. (G and H) Myc-PK-TprNLS cotransfected with HA-Progerin C611S (n = 52) or HA-Progerin (n = 48). P < 0.0001. Images shown are of severely affected cells. The IF microscopy is from a single experiment and is representative of two experiments. Bars, 20 µm. (I) Diagrams of Tpr and reporter proteins with apparent native sizes. (J) Gel filtration (Superdex 200) chromatography of reporter proteins. Elution positions of reporter proteins were determined by immunoblotting and compared with protein standards. Data are representative of two experiments for each reporter. AU, arbitrary unit; MM, molecular mass.
To further test whether a feature of Tpr outside of its NLS is required for the inhibitory effect of Progerin, we constructed TprNLS and SV40NLS fusions with Myc-PK and analyzed the effect of Progerin on reporter protein localization. Progerin inhibited nuclear import of both Myc-PK-TprNLS and Myc-PK-SV40NLS (P < 0.0001 and P < 0.0034, respectively; Fig. 8, E–H). This was unexpected because of the aforementioned result (Fig. 8, A and B) indicating that the inhibitory effect of Progerin is NLS independent and because SV40NLS fused to the GFP reporter was insensitive to Progerin (Fig. 8, C and D). Our results could be reconciled, however, if nuclear import defects caused by Progerin are related to the size of the protein cargo. This would explain why Tpr import cannot be rescued by the SV40NLS and why import mediated by SV40NLS fused to Myc-PK, which assembled into a tetramer with an apparent size of >300 kD, is inhibited by Progerin. We used gel filtration chromatography to compare the apparent sizes of the reporter proteins with the predicted sizes of Tpr and Myc-Tpr(SV40NLS). Reporter proteins that displayed Progerin sensitivity have molecular masses of >300 kD (Fig. 8, I and J), and Tpr and Myc-Tpr(SV40NLS) (dimers) have sizes in the 400–500-kD range. Thus, the size of the protein cargo, irrespective of the NLS, can determine the sensitivity to Progerin.
To further understand the characteristics of proteins whose nuclear import might be affected by Progerin, we analyzed the subcellular localization of reporter proteins with molecular masses smaller than Tpr. This set included proteins with well-described nuclear localization pathways (GFP-Jun and GFP-telomerase), as well as proteins from the LIFEdb (life database) library (Bannasch et al., 2004; Mehrle et al., 2006) whose import pathways are not well defined. Coexpressing HA-Progerin did not have a significant effect on any of the “intermediate-sized” proteins tested (n = 33 proteins; Table S1 and examples in Fig. S1).
Large cargo import is correlated with nuclear Ran
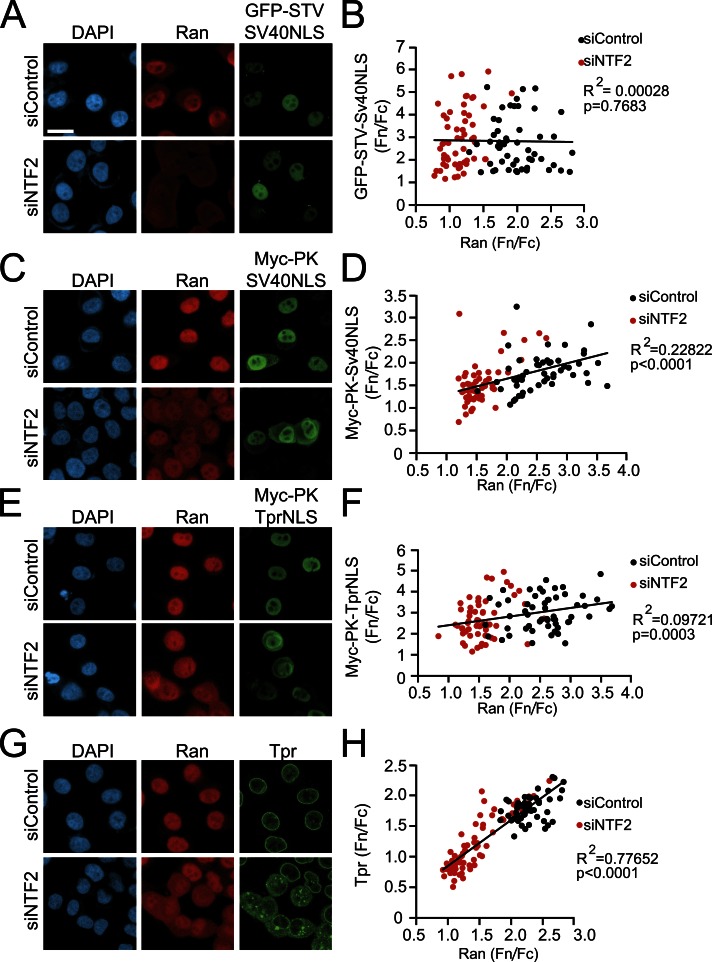
In the context of nuclear transport assays performed in digitonin-permeabilized cells, Lyman et al. (2002) found that large protein cargoes require the addition of higher concentrations of RanGTP for efficient import. Given that the Ran gradient is disrupted in HGPS cells (Fig. 3 A) and the defect in Tpr import can be attributed to its large size (Fig. 8), we examined the Ran sensitivity of nuclear import for different NLS reporter proteins in cells. This was achieved by depleting the Ran import factor NTF2. The Fn/Fc ratio of GFP-STV-SV40NLS reporter protein, which is not affected by Progerin (Fig. 8, C and D), was not significantly correlated with the Ran Fn/Fc ratio (Fig. 9 B, Spearman P = 0.7683). Conversely, the Fn/Fc ratios of both Myc-PK-SV40NLS and Myc-PK-TprNLS were correlated with Ran Fn/Fc (Fig. 9, D and F; Spearman P < 0.0001 and P = 0.0003, respectively). Endogenous Tpr is highly correlated with Ran Fn/Fc (Fig. 9, G and H, Spearman P < 0.0001). Thus, NLS-containing reporter proteins that are sensitive to Progerin expression are also sensitive to the Ran gradient as indicated by a distribution that correlates with the Ran Fn/Fc.
Figure 9.
Large NLS-containing reporter proteins exhibit a strong dependence on the Ran protein gradient for import. HeLa cells were treated with NTF2 siRNA to disrupt the Ran gradient, and Fn/Fc values of the indicated proteins were plotted as a function of Ran Fn/Fc. (A and B) GFP-STV-SV40NLS in control siRNA (siControl; n = 79) and siNTF2 cells (n = 56). Spearman P = 0.7683 for collated data. (C and D) Myc-PK-SV40NLS in control siRNA (n = 50) and siNTF2 cells (n = 57). Spearman P < 0.0001 for collated control siRNA and siNTF2 data. (E and F) Myc-PK-TprNLS in control siRNA (n = 56) and siNTF2 cells (n = 58). Spearman P = 0.0003 for collated control siRNA and siNTF2 data. (G and H) Endogenous Tpr distribution in control siRNA (n = 54) and siNTF2 cells (n = 70) cells. Spearman P < 0.0001 for collated control siRNA and siNTF2 data. All data are representative of at least two experiments. The data were fitted with trend lines in Excel. Bar, 20 µm. Bar applies to all IF images.
Large nuclear complexes have Progerin-induced import defects and depend on the Ran gradient
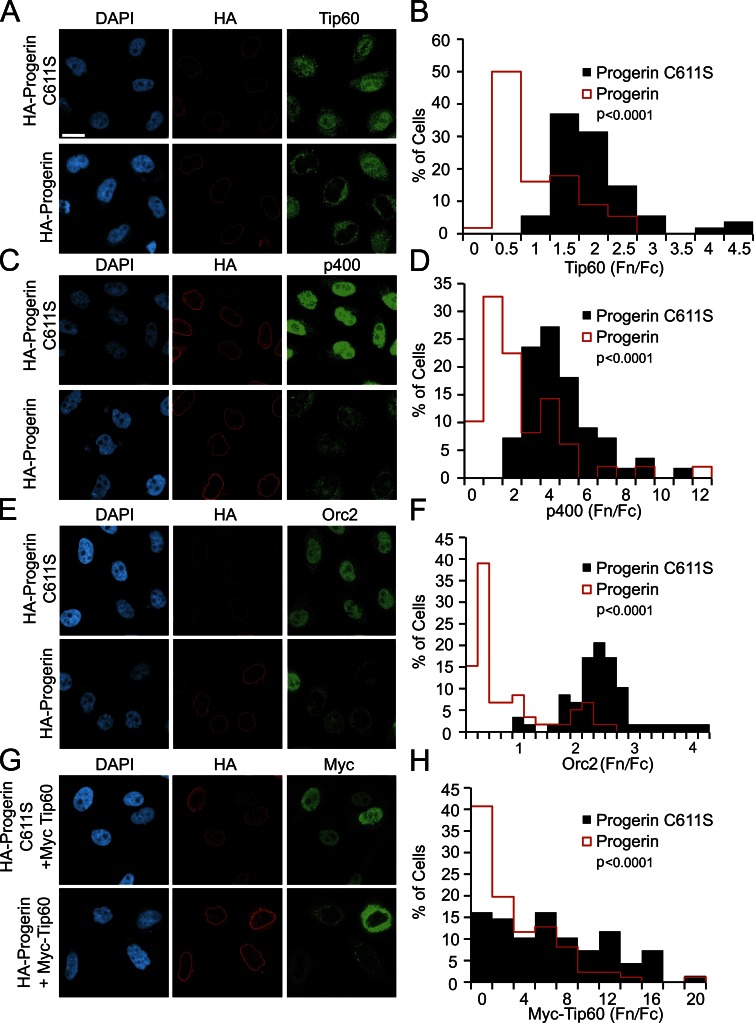
If Progerin inhibits nuclear import of Tpr principally because of cargo size and an associated requirement for Ran, by extension, nuclear transport of other large nuclear complexes could be defective in Progerin-expressing cells. We tested this idea by examining the localization of three nuclear proteins known to exist in high molecular mass complexes: Tip60 (histone acetyltransferase complex component; DNA repair complex), p400 (E1A binding complex), and Orc2 (origin of replication complex). Using antibodies that recognize the endogenous proteins, we found that HA-Progerin, but not the C611S form of Progerin, causes nuclear import defects of these proteins (Fig. 10, A–F, P < 0.0001 for each). Total cellular p400 and Orc2 signals also appear to decrease with HA-Progerin expression (Fig. 10 C), suggesting that protein degradation might be associated with the import defects. A Myc-tagged form of Tip60 also displayed a robust import defect in response to Progerin expression (Fig. 10, G and H, P < 0.0001). Nuclear levels of endogenous Tip60, p400, and Orc2 were each correlated with Ran Fn/Fc in cells depleted of the Ran import factor NTF2 (Fig. S2), underscoring that these complexes undergo Ran-sensitive nuclear import.
Figure 10.
Defective import of protein complexes in cells expressing Progerin. (A–H) IF microscopy and histograms of endogenous and transfected protein Fn/Fc levels in HeLa cells expressing Progerin. (A and B) Tip60 in HA-Progerin C611S (n = 54)– and HA-Progerin (n = 56)–transfected cells. P < 0.0001. (C and D) p400 in HA-Progerin C611S (n = 55)– and HA-Progerin (n = 49)–transfected cells. P < 0.0001. (E and F) Orc2 in HA-Progerin C611S (n = 58)– and HA-Progerin (n = 59)–transfected cells. P < 0.0001. (G and H) Myc-Tip60 in HA-Progerin C611S (n = 68) and HA-Progerin (n = 86). P < 0.0001. The IF microscopy is from a single experiment and is representative of at least two experiments. Bar, 20 µm. Bar applies to all IF images.
Discussion
Progerin expression drives complex cellular changes in HGPS that include alterations in nuclear morphology, reduced levels of epigenetic marks associated with gene repression, cell cycle perturbation, and signaling changes linked to DNA damage (Eriksson et al., 2003; Csoka et al., 2004; Goldman et al., 2004; Liu et al., 2006; Shumaker et al., 2006; Dechat et al., 2007; Taimen et al., 2009). Although there is little insight into the mechanisms that link Progerin to these phenotypic changes, experimental approaches that interfere with farnesylation of Progerin or inhibit proteolytic processing of pre–lamin A have made it clear that the biological effects of Progerin are strictly dependent on its stable attachment to the nuclear membrane (Capell et al., 2005; Mallampalli et al., 2005; Young et al., 2005; Caron et al., 2007). Because the inner nuclear membrane and associated nuclear lamina are key architectural elements in the nucleus, current models suggest Progerin exerts biological effects of the cell, at least in part, by changing chromatin organization at the nuclear periphery (Burke and Stewart, 2006).
In previous work, we showed Progerin disrupts the nuclear/cytoplasmic distribution of Ran, resulting in defective import of Tpr, the nucleoporin that forms the basket on the nuclear side of the NPC (Krull et al., 2004). Depending on the model system, Tpr function has been linked to a variety of nuclear activities, including the cell cycle, RNA export, tethering of deSUMOylating enzymes, and chromatin organization (Bangs et al., 1998; Lee et al., 2008; Krull et al., 2010; Nakano et al., 2010; Chow et al., 2012; Rajanala and Nandicoori, 2012). Given that Tpr function in human cells is probably important for several nuclear pathways, loss of the NPC basket might contribute to HGPS phenotypes by multiple mechanisms.
Toward the goal of understanding how Progerin induces cellular changes in HGPS, we set out to understand the molecular basis of the Tpr nuclear import defect. We observed that SV40NLS-mediated import was unaffected by Progerin, despite the fact that the same cells showed strong Tpr import defects (Fig. 2). Based on this finding, we hypothesized the dominant-negative effects of Progerin would involve inactivation of the TprNLS, its import receptor, or some cargo-specific feature. To that end, we analyzed the import signal and receptors that recognize the signal, both biochemically and in cultured cells. We determined that Tpr contains a bipartite NLS that can bind KPNA1, KPNA2, KPNA5, and KPNA6. Interestingly, although KPNA1 and KPNA6 display the most robust binding to the TprNLS, siRNA experiments demonstrated that KPNA2 together with Importin-β is responsible for Tpr import (Fig. 6 and Fig. S3). Because KPNA2 is the most abundant import receptor, yet global NLS-mediated import is not inhibited by Progerin, our data suggested that the defect in Tpr import is not caused by inactivation of its receptor.
Despite primary sequence differences between the TprNLS and SV40NLS, both signals bind KPNA2 with nanomolar affinity, and both undergo RanGTP-stimulated release from KPNA2 in vitro (Fig. 7). These data suggested that the features that make Tpr import sensitive to Progerin are not specific to the TprNLS, which we explored further in signal swapping experiments. Appending the SV40NLS to the first 1,741 amino acids of Tpr was insufficient to rescue Tpr import in the presence of Progerin (Fig. 8), indicating the defect is associated with a feature outside of the TprNLS. We then turned to PK as a reporter protein, which has been used widely as a fusion partner for nuclear transport signals because it lacks endogenous transport signals. Using signal fusions with PK, which assembles as a tetramer, we found that nuclear transport mediated by the TprNLS and the SV40NLS was inhibited by Progerin. By gel filtration, these fusion proteins are >300 kD. The SV40NLS fused to a GFP reporter displayed an apparent size of 260 kD, and its import was unaffected by Progerin. Thus, our data indicate that the Progerin-induced defect in Tpr import occurs because of the size of the cargo not because of the signal or receptor that defines the pathway. Additional support for this conclusion came from testing the effect of Progerin on the localization of a small set of nuclear proteins. Of these proteins, only those known to assemble into large multisubunit complexes displayed clear import defects in the presence of Progerin in our assays (Fig. 10 and Fig. S1).
Tpr might be one of the largest protein cargoes that undergoes nuclear import in the cell, given that its coiled-coil interactions generate a 535-kD dimer that has an overall length exceeding 100 nm (Hase et al., 2001). Work from other groups has shown that cargo size can, in fact, have a significant influence on nuclear import. In real-time tracking experiments, the frequency of “aborted attempts” at nuclear import of NLS-containing quantum dots was increased threefold by increasing the cargo diameter from 15 to 40 nm (Lowe et al., 2010). Nuclear import mediated by the Importin-β binding and M9 signals was dependent on protein size, with large protein fusions (500 and 669 kD) undergoing less efficient import than smaller fusions (60–125 kD; Lyman et al., 2002). The principle that size matters in nuclear transport likely extends to intermediate-sized cargoes as well. Using assays capable of detecting small kinetic differences in nuclear transport, it was shown that there is an inverse correlation between nuclear import efficiency and reporter protein size in the range of 38–88 kD (Ribbeck and Görlich, 2002).
The Tpr import defect in HGPS occurs because the Ran gradient is disrupted. Independent of Progerin expression, disruption of the Ran gradient is sufficient to inhibit Tpr import, and rescue of the Ran gradient in Progerin-expressing cells is sufficient to rescue Tpr import (Kelley et al., 2011). Nuclear import of Tpr shows a steep dependence on the nuclear/cytoplasmic distribution of Ran, explaining why it is particularly sensitive to Progerin. This sensitivity to the Ran gradient is a property shared by other large molecular mass protein cargoes (Fig. S2 and Fig. S4, model) and provides a plausible explanation for how Progerin can reduce the import of proteins whose common property is their large size. It should be noted that the extent to which Progerin causes defects in the Ran system and Tpr import varies between cells, both in patient cells and in our models that rely on ectopic expression. The penetrance of the dominant-negative effects might be related to Progerin expression levels or other features of the cell, such as passage number, that help determine the sensitivity to the effects of Progerin. The indication that the dominant-negative effects of Progerin in patients are strongly manifest in the vascular system already implies the susceptibility is somehow related to the cell or tissue type.
A question that remains is why large protein complexes are sensitive to the Ran gradient. Lyman et al. (2002) showed that nuclear import of large protein cargoes in permeabilized cells requires the addition of recombinant Ran and hydrolyzable GTP. This was taken as evidence that RanGTP modulates transport receptor interactions within the NPC that promote import. This view seems to present a paradox because RanGTP binding to import complexes can release cargo from its receptors in vitro. Whether Ran enhances large cargo import by acting directly on the transport complex or via another Ran-binding component within the NPC, such as cargo-free Importin-β, remains an important question. Our finding that nuclear localization of large protein complexes is highly sensitive to disruption of the Ran gradient provides a transport-based model that may help explain some of the complex changes in nuclear biology associated with HGPS.
Materials and methods
Cell culture
Primary human fibroblasts were obtained from the Coriell Cell Repositories. The AGO8469 cells (designated Normal 8469) are from an unaffected father of an HGPS patient. The AGO1972, AG11498, and AGO3199 cells (designated HGPS 1972, HGPS 1498, and HGPS 3199, respectively) are from HGPS patients. Primary human fibroblasts were grown in MEM (Gibco), containing 15% FBS (Hyclone), 1% MEM vitamin solution (HyClone), 1% glutathione (Gibco), and 1% penicillin/streptomycin (Gibco). Cells were passaged every 2–4 d. The patient cells used in our experiments (passage number 10–25) showed only modest defects in nuclear morphology. Adherent HeLa cells (American Type Culture Collection), including the HeLa cell line stably expressing GFP-STV-SV40NLS (Black et al., 1999), were grown in DMEM containing 10% FBS (Atlanta Biologicals). All cells were grown in 5% CO2 at 37°C.
IF microscopy and image analysis
IF microscopy was performed using standard methods as previously described (Kelley et al., 2011). Cells were grown on glass coverslips, washed with PBS, and fixed for 20 min with 3.75% formaldehyde. Cells were then washed three times with PBS and permeabilized in 0.2% Triton X-100 for 5 min. Cells were blocked for 1 h in blocking buffer (2% FBS and 2% BSA in PBS). Primary antibody was diluted in blocking buffer, and incubation was performed at room temperature for 2 h or overnight at 4°C. Rabbit antibodies to Tpr were generated by injecting a recombinant protein fragment that spans amino acids 1,649–1,912 (Cocalico Biologicals) and using the same fragment for antibody purification. Tpr knockdown by siRNA reduces the IF signal with this antibody (Fig. S5 A). Rabbit anti-p400 (E. Shibata, University of Virginia, Charlottesville, VA) was raised against a bacterially produced His6-tagged fragment of p400 (aa 2,185–2,487). Rabbit anti-Tip60 (Jha et al., 2010) was purified with peptide (sequence: CLHFTPKDWSKRGKW) coupled to SulfoLink Coupling gel (Thermo Fisher Scientific). Other primary antibodies used for IF were Nup153 mAb SA1 (B. Burke, University of Florida, Gainesville, FL; McMorrow et al., 1994), Ran mAb (catalog number 610341; BD), Myc mAb 9E10, Myc rabbit polyclonal (catalog number ab9106; Abcam), HA mAb 16B12 (Covance), HA rabbit polyclonal antibody Y-11 (Santa Cruz Biotechnology, Inc.), and Orc2 (BD). Secondary antibodies were diluted in blocking buffer, and incubation was performed at room temperature for 1 h. Secondary antibodies used were FITC-labeled donkey anti–mouse, Cy3-labeled goat anti–mouse, Cy3-labeled donkey anti–rabbit, Cy5-labeled donkey anti–rabbit, and Cy5-labeled donkey anti–mouse (antibodies all obtained from Jackson ImmunoResearch Laboratories, Inc.). Slides were mounted in Vectashield (Vector Laboratories) for imaging.
Images were acquired using confocal and wide-field microscopes. Most of the experiments were performed using a microscope (LSM 700; Carl Zeiss) equipped with a 40×, 1.3 NA oil immersion objective and ZEN software (Carl Zeiss). Fig. 1 and Fig. 3 were generated using images acquired with a microscope (LSM 510 UV; Carl Zeiss) using a 40×, 1.3 NA oil immersion objective and Axiovision software (Carl Zeiss). Fig. S1 and Fig. S3 contain images acquired on an upright microscope (Eclipse E800; Nikon) using a 40×, 1.0 NA oil immersion objective and were captured with a charge-coupled device camera (C4742-95; Hamamatsu Photonics) with OpenLab software (PerkinElmer). All imaging was performed at room temperature (∼24°C).
Quantitative analysis of IF images (ratios of the nuclear concentration to the cytoplasmic concentration [Fn/Fc ratios]) was measured as described previously (Kelley and Paschal, 2007; Kelley et al., 2011) using ImageJ (National Institutes of Health). Both nuclear and cytoplasmic regions of each cell were selected, and the mean IF was determined by ImageJ software. Fn/Fc ratios = (mean fluorescence of nucleus)/(mean fluorescence of cytoplasm). Statistical tests (Student’s t test and Spearman correlation) were performed using Prism (GraphPad Software). All graphs were made in Excel (Microsoft). Data were plotted as histograms, which help illustrate the range of Fn/Fc values within the experiment. All IF images shown were processed in Photoshop (Adobe). Adjustments to brightness and contrast were performed using the levels function. Individual channels were colored by overlaying the color onto the grayscale image as a separate layer. Images were collated in Illustrator (Adobe).
Plasmids and siRNA
The plasmid encoding HA-Progerin (Kelley et al., 2011) was engineered to encode to HA-Progerin C611S using the site-directed mutagenesis technique (QuikChange II; Agilent Technologies). C611 is the prenylation site in lamin A and Progerin. Because the serine substitution at this position abrogated the effects of Progerin on the Ran system, the C661S form of Progerin was used throughout the study as a control for any nonspecific effects of ectopic protein expression. HA-Progerin and HA-Progerin C611S have a pCDNA3 backbone (Ampr; Invitrogen). Plasmids encode KPNA1–7 from a parent vector of pCMVTNT (Promega) and are N-terminally T7 tagged as previously described (Kelley et al., 2010). Myc-Tpr(SV40NLS) and Myc-PK-TprNLS (parent vectors pRC/CMV and Ampr; Invitrogen) were gifts from V. Cordes (The Max Planck Institute for Biophysical Chemistry, Göttingen, Germany; Cordes et al., 1998). Myc-Tpr(SV40NLS) encodes the first 1,741 amino acids of Tpr followed by the SV40NLS sequence. Myc-PK-TprNLS was used to make Myc-PK-SV40NLS. In brief, the TprNLS was removed using Nhe1–Apa1, and annealed oligonucleotides containing the SV40NLS sequence were added. DNA transfection was performed using Transfectin according to the manufacturer’s directions (Invitrogen). In brief, HeLa cells were grown on glass coverslips for 24 h, transfected, and grown for an additional for 24 h before performing IF microscopy. Plasmids encoding intermediate-sized cargo tagged with GFP and YFP were cotransfected with HA-Progerin expressed from a bistronic plasmid that also encodes RFP. siRNAs for NTF2 were obtained from Santa Cruz Biotechnology, Inc. RT-PCR analysis and immunoblotting for the NTF2 knockdown are shown in Fig. S5 (B and C). siRNAs for KPNA1 and KPNA2 were SMARTpools (Thermo Fisher Scientific). siRNA for Tpr was as previously described (sequences 5′-GCACAACAGGATAAGGTTA-3′ and 5′-TAACCTTATCCTGTTGTGC-3′; Coyle et al., 2011). All siRNAs were transfected by using RNAiMAX (Invitrogen) according to the manufacturer’s instructions (reverse transfection protocol).
RT-PCR
The forward and reverse primers used for RT-PCR analysis of transcript levels are as follows: NTF2, 5′-TTCTGGAACGGAAGGCTAGA-3′ and 5′-ACCCAACTAGGCGCAATTTA-3′; KPNA1, 5′-TAGCAACATTTCTCCGCTTG-3′ and 5′-TCTCTGAATCCCGATGAGATG-3′; KPNA2, 5′-TGATTTTCCACATTGCTGCT-3′ and 5′-GATGATGCTACTTCTCCGCTG-3′; KPNA3, 5′-TTTTGTTCTTCCGCAGTTCC-3′ and 5′-CGCATCAAGAGCTTCAAGAAC-3′; KPNA4, 5′-CAACTTCATTTCGTTGTCTTCTC-3′ and 5′-CGGACAACGAGAAACTGGAC-3′; KPNA5, 5′-CGGCATTTCTTGTTGTTGTG-3′ and 5′-TGCTGGTGACAATGCAGAAT-3′; KPNA6, 5′-AATTGTCTTTCCCTGGGCTC-3′ and 5′-ATTGTCTACTGAAAGCTGCCG-3′; and KPNA7, 5′-CATCGAGAAGCACTTTGGTG-3′ and 5′-GGAGGTAGGGAGCTTGGCTA-3′. RT-PCR was performed using standard methods. For semiquantitative analysis of KPNA message levels, plasmid DNA was used to generate standard curves for each KPNA. Error bars shown are the standard deviation of duplicate wells.
Protein binding assays
ELISA was used to determine the apparent Kd of KPNA2 for GST-fused SV40NLS and TprNLS. GST, GST-SV40NLS, or GST-TprNLS was preabsorbed to 96-well plates at a concentration of 25 nM. The wells were blocked using 2% BSA, and indicated concentrations of KPNA2/Importin-β were added to wells and incubated overnight at 4°C. Binding was detected using the KPNA2 antibody (BD), and horseradish peroxidase–labeled secondary antibody was detected using o-phenylenediamine dihydrochloride (SIGMAFAST; Sigma-Aldrich). The apparent Kd was calculated using OriginPro 7.5 (OriginLab) by fitting the ELISA data to the equation specific binding = Bmax [L]/Kd + [L], in which [L] is the concentration of KPNA2/Importin-β, and Bmax is maximum binding.
For Ran-dependent disassembly of import complexes, GST, GST-SV40NLS, or GST-TprNLS was bound to glutathione–agarose beads overnight in PBS and protease inhibitors. KPNA2/Importin-β was then bound at an input concentration of 200 nM, and the beads were then washed with PBS. His-RanQ69L, prepared by standard methods, was loaded with GTP (Steggerda and Paschal, 2000). In brief, 300 µg RanQ69L was incubated in a 50-mM Hepes buffer, pH 7.4, containing 0.2 µM GTP, 2.5 mM DTT, 10 mM EDTA, and 2 mM ATP, for 20 min at 30°C. The loading reaction was stopped with 20 mM MgCl2, diluted 25-fold in PBS, and reconcentrated using a centrifugation filter column (Ultracel 10K; EMD Millipore) to remove unbound nucleotide. His-RanQ69L bound to GTP was incubated with the NLS receptor complexes for 1 h at room temperature. Beads were collected, and the unbound and bound fractions were analyzed by immunoblotting. The films (ECL detection) in Fig. 7 were quantitated using ImageJ.
In vitro translation of the KPNA isoforms and binding to GST, GST-SV40NLS, or GST-TprNLS was performed as previously described (Kelley et al., 2010). A reaction kit (TnT; Promega) was used to translate each KPNA isoform using 35S-labeled methionine. Each reaction was performed at 30°C for 1.5 h. Translated proteins were subjected to scintillation counting, and 300,000 counts per million of translated protein was added to each binding reaction in which GST, GST-TprNLS, or GST-SV40NLS was bound to glutathione–agarose beads. Binding was performed in PBS at 4°C for 3 h, the beads were washed with PBS, and bound fractions were loaded on a gel along with 5% input. Gels were treated with Autofluor (National Diagnostics) for 1 h, dried, and exposed to film for 24 h before developing.
Other biochemical methods
HeLa cells were transfected with the Myc or GFP constructs as indicated. Cell lysates were clarified at 100,000 g and chromatographed on a gel filtration column (Superdex 200; GE Healthcare) at a flow rate of 0.25 ml/min. Column fractions (0.25 ml each) were analyzed by immunoblotting and peaks were quantitated using ImageJ.
SDS-PAGE gel electrophoresis and immunoblotting were performed using standard methods. Antibodies used in immunoblotting, in addition to those mentioned previously, were Importin-β mAb (BD), Tubulin mAb (Sigma Aldrich), Jol-2 lamin A/C mAb (EMD Millipore), GFP mAb (Covance), KPNA2 mAb (BD), and NTF2 5e8 (Steggerda et al., 2000). Secondary horseradish peroxidase–coupled antibodies were detected using ECL.
Online supplemental material
Fig. S1 shows the distributions of three nuclear-localized cargoes (YFP–Tigger transposable element derived 6, YFP-REX2, and YFP–DEAD box polypeptide 49) that are unaffected by Progerin expression. Fig. S2 shows import defects of proteins that are components of large protein complexes (Tip60, p400, and Orc2) in response to Ran gradient disruption by NTF2 siRNA. Fig. S3 demonstrates that both KPNA2 and Importin-β from rabbit reticulocyte lysate bind to GST-TprNLS and that Importin-β knockdown is sufficient to mislocalize Tpr in HeLa cells. Fig. S4 is a cartoon summarizing the effects of Progerin on the Ran gradient and large protein cargo import. Fig. S5 demonstrates Tpr antibody specificity and NTF2 knockdown efficiency. Table S1 summarizes the 33 reporter proteins (small nuclear cargoes) that were tested and found to be unaffected by Progerin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212117/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201212117.dv.
Supplementary Material
Acknowledgments
We thank Drs. B. Burke and V. Cordes for their generous gifts of plasmids and antibodies as well as E. Shibata for the p400 antibody. We thank Dr. S. Wiemann (German Cancer Research Center, Heidelberg) for providing us with the LIFEdb library and C. Spillner for performing the initial library screen. We thank the Advanced Microscopy Core Facility at the University of Virginia for their assistance. We also thank A. Spencer and Dr. L. Ni for their technical assistance.
These studies were supported by National Institutes of Health award 1RO1AG040162 (to B.M. Paschal) and a National Science Foundation graduate fellowship award (to C.J. Snow).
Footnotes
Abbreviations used in this paper:
- Fn/Fc
- fluorescence nucleus/fluorescence cytoplasm
- HGPS
- Hutchinson-Gilford Progeria syndrome
- IF
- immunofluorescence
- KPNA
- Karyopherin-α
- NPC
- nuclear pore complex
- PK
- pyruvate kinase
- STV
- streptavidin
- Tpr
- translocated promoter region
References
- Antonin W., Ellenberg J., Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582:2004–2016 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- Bangs P., Burke B., Powers C., Craig R., Purohit A., Doxsey S. 1998. Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell Biol. 143:1801–1812 10.1083/jcb.143.7.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch D., Mehrle A., Glatting K.H., Pepperkok R., Poustka A., Wiemann S. 2004. LIFEdb: a database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 32(Suppl. 1):D505–D508 10.1093/nar/gkh022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I., Frosst P.D., Gerace L. 2009. Karyopherin binding interactions and nuclear import mechanism of nuclear pore complex protein Tpr. BMC Cell Biol. 10:74 10.1186/1471-2121-10-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 354:80–82 10.1038/354080a0 [DOI] [PubMed] [Google Scholar]
- Black B.E., Lévesque L., Holaska J.M., Wood T.C., Paschal B.M. 1999. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol. Cell. Biol. 19:8616–8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Enarson P., Chowdhury S., Salina D., Raharjo W.H., Burke B. 1999a. Function and assembly of nuclear pore complex proteins. Biochem. Cell Biol. 77:321–329 10.1139/o99-038 [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W.H., Bastos R., Lohka M., Burke B. 1999b. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264 [DOI] [PubMed] [Google Scholar]
- Burke B., Ellenberg J. 2002. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 3:487–497 10.1038/nrm860 [DOI] [PubMed] [Google Scholar]
- Burke B., Stewart C.L. 2006. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu. Rev. Genomics Hum. Genet. 7:369–405 10.1146/annurev.genom.7.080505.115732 [DOI] [PubMed] [Google Scholar]
- Capell B.C., Erdos M.R., Madigan J.P., Fiordalisi J.J., Varga R., Conneely K.N., Gordon L.B., Der C.J., Cox A.D., Collins F.S. 2005. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 102:12879–12884 10.1073/pnas.0506001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron M., Auclair M., Donadille B., Béréziat V., Guerci B., Laville M., Narbonne H., Bodemer C., Lascols O., Capeau J., Vigouroux C. 2007. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ. 14:1759–1767 10.1038/sj.cdd.4402197 [DOI] [PubMed] [Google Scholar]
- Chow K.H., Elgort S., Dasso M., Ullman K.S. 2012. Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus. 3:349–358 10.4161/nucl.20822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett A.H., Silver P.A. 1996. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J. Biol. Chem. 271:18477–18484 10.1074/jbc.271.31.18477 [DOI] [PubMed] [Google Scholar]
- Cordes V.C., Hase M.E., Müller L. 1998. Molecular segments of protein Tpr that confer nuclear targeting and association with the nuclear pore complex. Exp. Cell Res. 245:43–56 10.1006/excr.1998.4246 [DOI] [PubMed] [Google Scholar]
- Coyle J.H., Bor Y.C., Rekosh D., Hammarskjold M.L. 2011. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA. 17:1344–1356 10.1261/rna.2616111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka A.B., English S.B., Simkevich C.P., Ginzinger D.G., Butte A.J., Schatten G.P., Rothman F.G., Sedivy J.M. 2004. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 3:235–243 10.1111/j.1474-9728.2004.00105.x [DOI] [PubMed] [Google Scholar]
- D’Angelo M.A., Hetzer M.W. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456–466 10.1016/j.tcb.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Shimi T., Adam S.A., Rusinol A.E., Andres D.A., Spielmann H.P., Sinensky M.S., Goldman R.D. 2007. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc. Natl. Acad. Sci. USA. 104:4955–4960 10.1073/pnas.0700854104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 423:293–298 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 90:1051–1060 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F.S. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 101:8963–8968 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Panté N., Kutay U., Aebi U., Bischoff F.R. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584–5594 [PMC free article] [PubMed] [Google Scholar]
- Hase M.E., Cordes V.C. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell. 14:1923–1940 10.1091/mbc.E02-09-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase M.E., Kuznetsov N.V., Cordes V.C. 2001. Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol. Biol. Cell. 12:2433–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Jans D.A. 1999. Efficiency of importin alpha/beta-mediated nuclear localization sequence recognition and nuclear import. Differential role of NTF2. J. Biol. Chem. 274:15820–15827 10.1074/jbc.274.22.15820 [DOI] [PubMed] [Google Scholar]
- Jha S., Vande Pol S., Banerjee N.S., Dutta A.B., Chow L.T., Dutta A. 2010. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol. Cell. 38:700–711 10.1016/j.molcel.2010.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J.B., Paschal B.M. 2007. Hyperosmotic stress signaling to the nucleus disrupts the Ran gradient and the production of RanGTP. Mol. Biol. Cell. 18:4365–4376 10.1091/mbc.E07-01-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J.B., Talley A.M., Spencer A., Gioeli D., Paschal B.M. 2010. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 11:63 10.1186/1471-2121-11-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J.B., Datta S., Snow C.J., Chatterjee M., Ni L., Spencer A., Yang C.S., Cubeñas-Potts C., Matunis M.J., Paschal B.M. 2011. The defective nuclear lamina in Hutchinson-gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol. Cell. Biol. 31:3378–3395 10.1128/MCB.05087-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M., Speck C., Christiansen M., Bischoff F.R., Prehn S., Haller H., Görlich D., Hartmann E. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Thyberg J., Björkroth B., Rackwitz H.R., Cordes V.C. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 15:4261–4277 10.1091/mbc.E04-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Dörries J., Boysen B., Reidenbach S., Magnius L., Norder H., Thyberg J., Cordes V.C. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29:1659–1673 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Sterling H., Burlingame A., McCormick F. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22:2926–2931 10.1101/gad.1677208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M., Maffini S., Orr B., Ding Y., Florindo Cláudia, Sunkel C.E., Tavares A., Johansen J., Johansen K.M., Maiato H. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184:647–657 10.1083/jcb.200811012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Rusinol A., Sinensky M., Wang Y., Zou Y. 2006. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J. Cell Sci. 119:4644–4649 10.1242/jcs.03263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe A.R., Siegel J.J., Kalab P., Siu M., Weis K., Liphardt J.T. 2010. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 467:600–603 10.1038/nature09285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R.J., Trujillo M.A., Denham K.S., Wenger L., Sinensky M. 1992. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. USA. 89:3000–3004 10.1073/pnas.89.7.3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman S.K., Guan T., Bednenko J., Wodrich H., Gerace L. 2002. Influence of cargo size on Ran and energy requirements for nuclear protein import. J. Cell Biol. 159:55–67 10.1083/jcb.200204163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallampalli M.P., Huyer G., Bendale P., Gelb M.H., Michaelis S. 2005. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 102:14416–14421 10.1073/pnas.0503712102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMorrow I., Bastos R., Horton H., Burke B. 1994. Sequence analysis of a cDNA encoding a human nuclear pore complex protein, hnup153. Biochim. Biophys. Acta. 1217:219–223 10.1016/0167-4781(94)90040-X [DOI] [PubMed] [Google Scholar]
- Mehrle A., Rosenfelder H., Schupp I., del Val C., Arlt D., Hahne F., Bechtel S., Simpson J., Hofmann O., Hide W., et al. 2006. The LIFEdb database in 2006. Nucleic Acids Res. 34(Suppl. 1):D415–D418 10.1093/nar/gkj139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Funasaka T., Hashizume C., Wong R.W. 2010. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J. Biol. Chem. 285:10841–10849 10.1074/jbc.M110.105890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut M.E., Mizzen C.A., Stukenberg T., Allis C.D., Macara I.G. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 292:1540–1543 10.1126/science.292.5521.1540 [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki H., Nishimoto T. 1989. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109:1389–1397 10.1083/jcb.109.4.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M., Fritze C., Guan T., Gerace L. 1997. High levels of the GTPase Ran/TC4 relieve the requirement for nuclear protein transport factor 2. J. Biol. Chem. 272:21534–21539 10.1074/jbc.272.34.21534 [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Paschal B.M. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 6:187–198 10.1111/j.1600-0854.2005.00270.x [DOI] [PubMed] [Google Scholar]
- Qi H., Rath U., Wang D., Xu Y.Z., Ding Y., Zhang W., Blacketer M.J., Paddy M.R., Girton J., Johansen J., Johansen K.M. 2004. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell. 15:4854–4865 10.1091/mbc.E04-07-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanala K., Nandicoori V.K. 2012. Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS ONE. 7:e29921 10.1371/journal.pone.0029921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M., Blobel G. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 83:683–692 10.1016/0092-8674(95)90181-7 [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664–2671 10.1093/emboj/21.11.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky G., Kent H.M., Stewart M., Görlich D. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17:6587–6598 10.1093/emboj/17.22.6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilworth S.M., Laskey R.A., Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 64:615–623 10.1016/0092-8674(91)90245-T [DOI] [PubMed] [Google Scholar]
- Ryan K.J., McCaffery J.M., Wente S.R. 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160:1041–1053 10.1083/jcb.200209116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S., et al. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 103:8703–8708 10.1073/pnas.0602569103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M., Fantle K., Trujillo M., McLain T., Kupfer A., Dalton M. 1994. The processing pathway of prelamin A. J. Cell Sci. 107:61–67 [DOI] [PubMed] [Google Scholar]
- Smith A., Brownawell A., Macara I.G. 1998. Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 8:1403–1406 10.1016/S0960-9822(98)00023-2 [DOI] [PubMed] [Google Scholar]
- Stade K., Ford C.S., Guthrie C., Weis K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 90:1041–1050 10.1016/S0092-8674(00)80370-0 [DOI] [PubMed] [Google Scholar]
- Steggerda S.M., Paschal B.M. 2000. The mammalian Mog1 protein is a guanine nucleotide release factor for Ran. J. Biol. Chem. 275:23175–23180 10.1074/jbc.C000252200 [DOI] [PubMed] [Google Scholar]
- Steggerda S.M., Black B.E., Paschal B.M. 2000. Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol. Biol. Cell. 11:703–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Sun C., Yang W., Tu L.C., Musser S.M. 2008. Single-molecule measurements of importin alpha/cargo complex dissociation at the nuclear pore. Proc. Natl. Acad. Sci. USA. 105:8613–8618 10.1073/pnas.0710867105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taimen P., Pfleghaar K., Shimi T., Möller D., Ben-Harush K., Erdos M.R., Adam S.A., Herrmann H., Medalia O., Collins F.S., et al. 2009. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. USA. 106:20788–20793 10.1073/pnas.0911895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Plessmann U., Traub P. 1989. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 257:411–414 10.1016/0014-5793(89)81584-4 [DOI] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2:a000562 10.1101/cshperspect.a000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., Ostlund C., Wang Y. 2010. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000760 10.1101/cshperspect.a000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S.G., Fong L.G., Michaelis S. 2005. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 46:2531–2558 10.1194/jlr.R500011-JLR200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.