Abstract
Autophagy allows cells to encapsulate parts of their cytosol into unique double-membrane structures. These autophagosomes mature to fuse with lysosomes and deliver the enclosed contents for degradation. Three recent papers, including one by Takáts et al. (2013. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201211160), have taken different routes to discover a role for Syntaxin 17 in the maturation of autophagosomes.
Autophagy contributes to cellular homeostasis and is a defense mechanism for metazoan cells stressed by starvation or by intracellular flotsam and jetsam—mitochondria gone bad, aggregates of misfolded proteins, or pathogens that escaped from phagosomes (Kroemer et al., 2010). All of these conditions elicit a stereotypical response that starts with the formation of a phagophore (also called an isolation membrane). This flat membrane cisternae expands to encapsulate the offending target or, in response to starvation, to engulf part of the cytosol. The nascent autophagosome continues to grow until its edges merge, and the resulting double-membraned autophagosome sequesters the cargo from the remaining cytosol.
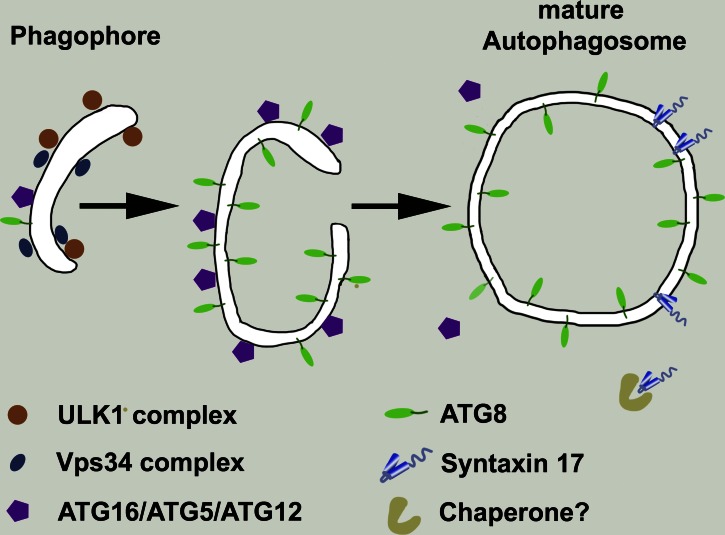
Most proteins necessary for autophagy, termed ATG proteins, were first described in yeast (Klionsky and Ohsumi, 1999), and much effort has gone into describing the interactions between these proteins and the order of their actions on maturing autophagosomes in yeast and metazoan cells (Mizushima et al., 2011). Two multiprotein complexes participate in the initial formation of the phagophore: a first containing the protein kinase ULK1 and the accessory subunits ATG13, ATG101, and FIP200 and a second including Beclin1, ATG14L, and the lipid kinase Vps34. Further phagophore growth depends on two conjugation systems involving the small, ubiquitin-like ATG3 and ATG8 (or LC3) proteins. Among these, ATG8 serves multiple roles as receptor for different cargoes and during the transport of autophagosomes. Although other ATG proteins dissociate from the early autophagosome before its closure, ATG8 persists and serves as a marker for mature autophagosomes (Fig. 1).
Figure 1.
Stage-specific markers of autophagosome maturation. Progression of autophagosome maturation is accompanied by the dissociation of the early phagophore components, including the ULK1 complex, which also contains ATG13, ATG101, FIP200, and the Vps34 complex with the Beclin1 and Atg14L subunits. Later, the complex of ATG5, ATG12, and ATG16 dissociates. Syntaxin 17 is inserted into autophagosomes after only lipidated ATG8 remains (Itakura et al., 2012; Takáts et al., 2013). It is not known whether a chaperone participates in maintaining Syntaxin 17 in the cytosol.
It is less clear how these mature autophagosomes complete their task to fuse specifically with lysosomes and deliver their content for degradation. For autophagosomes in yeast, their fusion with vacuoles involves the SNARE proteins Vti1, Ykt6, Vam3, and Vam7 (Fischer von Mollard and Stevens, 1999; Dilcher et al., 2001; Ishihara et al., 2001; Ohashi and Munro, 2010), but the latter two have no clear homologues in metazoan cells. Furthermore, the interpretation of genetic analysis is complicated by the indirect roles other membrane fusion events may exert. For example, in metazoan cells, unlike in yeast, autophagosomes fuse not only with lysosomes but also with late endosomes to form so-called amphisomes. Their formation is necessary for normal delivery of autophagic cargo to lysosomes (Filimonenko et al., 2007; Rusten et al., 2007). Thus, mutations effecting the formation or function of late endosomes may have secondary effects on autophagic flux measured by the degradation of ATG8 or cargo. Therefore, the search was on to identify SNARE proteins that localize to autophagosomes and are necessary for their fusion with lysosomes. The recent efforts of three different research teams converged on one candidate—Syntaxin 17 (see Takáts et al. in this issue; Itakura et al., 2012; Hamasaki et al., 2013)
In this issue of JCB, Juhász and coworkers (Takáts et al., 2013) report the result of an unbiased RNAi screen to systematically test all SNAREs in the Drosophila melanogaster genome for a role in starvation-induced autophagy. In that system, knockdown of Syntaxin 17, Usnp, or Vamp7, three SNAREs that they show can physically interact with each other, interfered with autophagosome maturation. For a more rigorous test of its requirement in autophagy, they generated the first syx17 (syntaxin 17) mutant animal model. Whereas these syx17-null flies were viable, although at a reduced rate, they exhibited severe neurodegeneration. At first glance, this was similar to the consequences of loss of the core autophagy genes ATG5 or ATG7 in mice or flies (Hara et al., 2006; Komatsu et al., 2006; Juhász et al., 2007). However, in contrast to those mutants, which block autophagy early before the appearance of autophagosomes, syx17 mutant neurons accumulated mature autophagosomes, indicating a requirement for their fusion with lysosomes. Consistent with such a late function, endogenous Syntaxin 17 was only detected on mature autophagosomes after they lost their early markers (Fig. 1). Together, these data point to Syntaxin 17 as the long-sought autophagosomal SNARE protein.
A role for Syntaxin 17 in autophagy had also emerged in two other recent studies (Itakura et al., 2012; Hamasaki et al., 2013), which both were based on the notion that the ER is an important source of autophagosome membranes and that ER membranes may bring along the sought-after autophagosomal SNARE. They observed that, among the SNARE proteins enriched in the ER, Syntaxin 17 was also found on autophagosomes. Furthermore, knockdown of Syntaxin 17 in both studies reduced autophagic flux. Although agreeing on the key observation of an important role of Syntaxin 17 in autophagy, the studies differed on some significant details.
In imaging experiments focused on the origin of autophagosome membranes, Yoshimori and coworkers observed a surprising correlation between the emergence of phagophores and the sites of contact between the ER and mitochondria (Hamasaki et al., 2013). Strikingly, GFP-tagged Syntaxin 17 was enriched at these ER–mitochondria contact sites, as was the phagophore marker Atg14. Furthermore, knockdown of Syntaxin 17 interfered with autophagic flux, leading the authors to suggest a role of ER-derived Syntaxin 17 early in the formation of autophagosomes (Hamasaki et al., 2013).
The study by Mizushima and coworkers followed the redistribution of overexpressed GFP-tagged SNAREs, which in fed mammalian cells are enriched in the ER. They found Syntaxin 17, but not Syntaxin 18, localized to autophagosomes upon starvation (Itakura et al., 2012). When more closely examining its distribution along the pathway of autophagosome maturation using an extensive set of markers, they did not detect GFP-Syntaxin 17 on emerging phagophores or early autophagosomes (Fig. 1). Instead, GFP-Syntaxin 17 appeared only on late autophagosomes after they were sealed. In a further parallel to the results in Drosophila (Takáts et al., 2013), Itakura et al. (2012) found that Syntaxin 17 knockdown in mammalian cells did not block autophagosome formation but did block their fusion with lysosomes.
If, therefore, mammalian and Drosophila Syntaxin 17 are not transferred directly form ER membranes to phagophores, the question arises how Syntaxin 17 arrives at autophagosomes. An important hint comes from structural considerations. Among SNARE proteins, only Syntaxin 17 has two transmembrane domains characterized by relative low hydrophobicity and a glycine zipper. Replacement of glycine with leucine residues did not interfere with its insertion into ER membranes but prevented Syntaxin 17 from reaching autophagosomes. Furthermore, in fed cells, substantial amounts of Syntaxin 17 were found in the cytosol after cell fractionation (Itakura et al., 2012). Together, these findings point to a model in which Syntaxin 17 is stored in the cytosol until it is transferred to autophagosomes upon their induction. Nothing is currently known about this transfer; obvious questions include a possible energy requirement for the transfer and the identities of any chaperone or autophagosomal receptor that may facilitate this novel mechanism to direct vesicles toward their proper fusion target.
Acknowledgments
The author is supported by grants from the National Eye Institute (EY10199 and EY021922).
References
- Dilcher M., Köhler B., von Mollard G.F. 2001. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 276:34537–34544 10.1074/jbc.M101551200 [DOI] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500 10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G., Stevens T.H. 1999. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 10:1719–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Mizushima N. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Juhász G., Erdi B., Sass M., Neufeld T.P. 2007. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21:3061–3066 10.1101/gad.1600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1–32 10.1146/annurev.cellbio.15.1.1 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J.-i., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., Levine B. 2010. Autophagy and the integrated stress response. Mol. Cell. 40:280–293 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Munro S. 2010. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. 21:3998–4008 10.1091/mbc.E10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.-P., Brech A., Bilder D., Stenmark H. 2007. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17:1817–1825 10.1016/j.cub.2007.09.032 [DOI] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga A., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., Juhász G. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]