Abstract
The muscular dystrophies are a group of heterogeneous genetic diseases characterized by progressive degeneration and weakness of skeletal muscle. Since the discovery of the first muscular dystrophy gene encoding dystrophin, a large number of genes have been identified that are involved in various muscle-wasting and neuromuscular disorders. Human genetic studies complemented by animal model systems have substantially contributed to our understanding of the molecular pathomechanisms underlying muscle degeneration. Moreover, these studies have revealed distinct molecular and cellular mechanisms that link genetic mutations to diverse muscle wasting phenotypes.
Skeletal muscle is a highly specialized organ system evolved for locomotion and energy metabolism in multicellular organisms. Deterioration of muscle cell integrity as a result of genetic mutations leads to progressive muscle wasting with detrimental consequences including early death. Typically, mutations in different genes tend to target distinct muscle groups, on the basis of which these conditions have been subdivided into several groups. However, mutations in different regions of the same protein, or even identical mutations, have also been implicated in clinically distinct phenotypes. Gene mapping efforts in families with affected individuals have been instrumental in elucidating the underlying cellular and molecular processes perturbed in most of these diseases. Mutations in a wide range of proteins, including many structural proteins and enzymes that posttranslationally modify some of these proteins have been implicated in muscular dystrophy. A comprehensive review of all types of muscular dystrophies and myopathies is beyond the scope of this review. A revised list of causative genes and reclassification is annually reported by the journal Neuromuscular Disorders (Kaplan and Hamroun, 2012). Here we will discuss distinct disease mechanisms of several well-understood muscular dystrophies and highlight several recently identified genes that will likely uncover novel cellular mechanisms underlying disease.
The structure of skeletal muscle cells
The myofiber is the functional unit of skeletal muscle and is a multinucleated tubular structure formed from the fusion of multiple mononucleated muscle cells (myoblasts). Myoblasts are generated by asymmetric division and differentiation of muscle stem cells, called satellite cells, that are located between the muscle cell membrane (sarcolemma) and basal lamina (Mauro, 1961). In addition to typical cellular organelles, the cytoplasm (sarcoplasm) of a myofiber contains a regular array of contractile units (sarcomeres) comprised of actin-containing thin filaments and myosin-containing thick filaments that, along with additional structural and regulatory proteins, are arranged longitudinally as myofibrils. The peripheral myofibrils are connected to the sarcolemma along the Z-disks via interactions with subsarcolemmal protein complexes called costameres. These structures transmit contractile forces from sarcomeres of one myofiber to another, which prevents sarcolemma ruptures by synchronizing contraction of myofibers within a muscle. Sarcolemma is firmly attached to the basal lamina, which consists of ECM proteins. Failure to attach properly results in sarcolemmal disruption, which is the predominant underlying cause of several forms of muscular dystrophies. Cross-section of healthy muscle reveals myofibers that are roughly equal in diameter with multiple nuclei pushed to the periphery of the cell. However, dystrophic muscle displays centrally located nuclei and myofibers of variable size, which are caused by successive rounds of degeneration and regeneration. Extensive damage to muscle activates satellite cells to promote muscle regeneration. Although these cells are replenished by self-renewal, recurrent degeneration and regeneration of skeletal muscle in disease state depletes the satellite cell pool and fails to further regenerate the muscle contributing to muscle wasting (Collins et al., 2005). Furthermore, atrophied muscle is gradually substituted by fibrous and fatty tissues, which is one of the hallmarks of muscular dystrophy. Disruption of sarcolemma causes leakage of muscle proteins such as creatine kinase (CK) into the serum. Thus, highly elevated CK concentration in circulating blood serum is used as an informative biomarker for degenerative processes occurring in muscle. However, the precise diagnosis of muscular dystrophies needs to be determined by a combination of protein testing with immunostaining or immunoblotting on a muscle biopsy followed by targeted genetic testing. Even though the end point of these conditions is severe muscle loss and accumulation of fibrotic and fatty tissues, different molecular pathomechanisms have been implicated in distinct types of muscle disease.
Cellular and molecular mechanisms of muscle degeneration
Disruption of cytoskeleton–ECM connection.
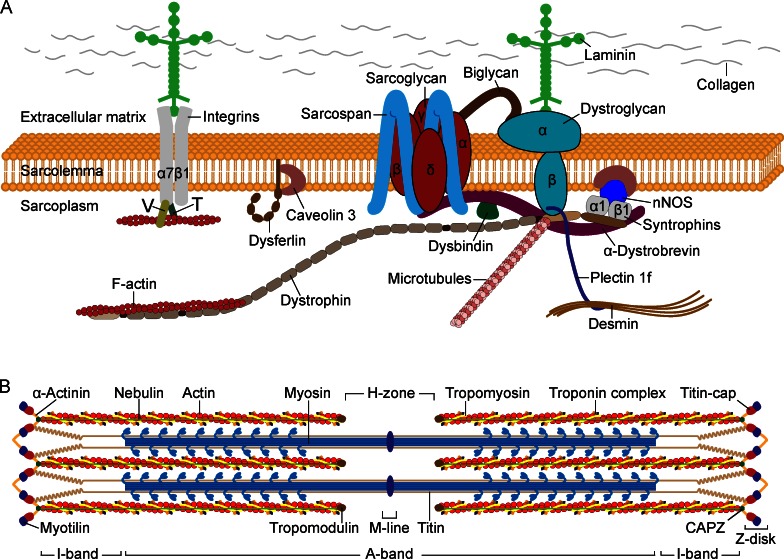
The most common and severely debilitating neuromuscular disorder, Duchenne muscular dystrophy (DMD), affects ∼1 in 3,500 males. It is manifested by rapidly progressive proximal muscle wasting starting around 3 years of age, culminating with respiratory insufficiency and cardiac failure that leads to premature death by the mid-20s. The allelic disorder Becker muscular dystrophy (BMD) is less common and milder, with late disease onset and relatively advanced survival age. Both diseases are caused by mutations in the DMD gene, the largest gene in the human genome, located on the X chromosome, which encodes the 427-kD protein dystrophin (Monaco et al., 1986; Burghes et al., 1987; Hoffman et al., 1987; Koenig et al., 1987). DMD is caused by recessive, frameshifting deletions and duplications or nonsense mutations that lead to complete loss of or expression of nonfunctional dystrophin in myofibers, whereas mutations causing BMD produce semifunctional dystrophin (Hoffman et al., 1988; Monaco et al., 1988; Koenig et al., 1989). Dystrophin is an intracellular, rod-shaped protein with four major functional domains. This protein is localized to the inner surface of the sarcolemma, with a high abundance at costameres. The N terminal and a portion of the middle rod domain interact with the cytoskeletal filamentous actin (F-actin), whereas the C-terminal domain interacts with multiple proteins to assemble the dystrophin-associated protein complex (DAPC), a group of proteins that span the sarcolemma of the skeletal and cardiac muscle (Ervasti et al., 1990; Yoshida and Ozawa, 1990; Fig. 1 A). This protein complex, in addition to dystrophin, encompasses intracellular (α1- and β1-syntrophin, α-dystrobrevin, and nNOS), transmembrane (β-dystroglycan, α-, β-, γ-, and δ-sarcoglycan, and sarcospan), and extracellular proteins (α-dystroglycan and laminin-2). The core component of the DAPC, dystroglycan, is encoded by a single gene and posttranslationally cleaved to yield two noncovalently associated subunits. The transmembrane subunit β-dystroglycan binds to dystrophin and other cytolinker proteins at the intracellular periphery of sarcolemma, whereas the cell-surface subunit α-dystroglycan binds to β-dystroglycan and to the ECM proteins such as laminin-2; together, these proteins connect the intracellular cytoskeleton through sarcolemma with the basement membrane (Ibraghimov-Beskrovnaya et al., 1992). The integrity of this protein complex is crucial for the contracting myofiber to withstand the mechanical stress generated by sarcomeres and to prevent its fragile sarcolemma from contraction-induced injuries (Petrof et al., 1993). Disruption of the DAPC, as a result of structural or posttranslational defects in one of the components building this protein complex, weakens the sarcolemma and causes different types of muscular dystrophy depending on the altered protein. The subcomplex of integral proteins sarcoglycans and sarcospan within the DAPC provides additional mechanical support to the DAPC and thereby to the sarcolemma (Yoshida et al., 1994). Mutations in genes encoding the α, β, γ, and δ subunits of the sarcoglycan complex cause sarcoglycanopathies, a subtype of recessively inherited limb-girdle muscular dystrophies (LGMDs; Table 1). In general, structural defects in one sarcoglycan subunit lead to reduction or complete loss of the other subunits, destabilizing the entire protein complex, which consequently weakens the sarcolemma. LGMDs are the most heterogeneous subgroup of muscular dystrophies that predominantly involve the proximal limb-girdle musculature, mainly those around hips and shoulders. Besides sarcoglycanopathies, there are several additional LGMDs with distinct molecular underpinnings that can arise as a result of mutations in proteins unrelated to the DAPC, each type categorized based on the mode of inheritance and altered gene product.
Figure 1.
Sarcolemmal proteins and sarcomere structure. (A) The DAPC is a multimeric protein complex that connects the intracellular cytoskeleton of a myofiber to the ECM, which is composed of laminin, collagen, and other proteins. The muscle-specific laminin is composed of α2, β1, and γ1 chains. The α2 subunit directly interacts with glycosylated α-dystroglycan, which in turn interacts with the transmembrane β-dystroglycan. The dystrophin protein has four functional domains including the N-terminal, a long middle rod, cysteine-rich, and C-terminal domains. The central rod domain consists of 24 spectrin-like repeats arranged head-to-tail and interspersed by four flexible hinges. The N-terminal and the spectrin-like repeats bind to F-actin of the cytoskeleton, but not to the α-actin of thin sarcomeric filaments. The cysteine-rich domain binds to β-dystroglycan and the adjacent C-terminal domain binds to α-dystrobrevin and syntrophin. The cytolinker protein plectin binds β-dystroglycan and dystrophin and connects desmin IFs with the DAPC. Microtubules also interact with dystrophin. The four subunits of the sarcoglycan complex interact with each other and with the transmembrane protein sarcospan. The small leucine-rich repeat proteoglycan biglycan in the ECM binds to α- and γ-sarcoglycan and α-dystroglycan. Syntrophins bind to dystrophin, α-dystrobrevin, nNOS, and caveolin-3. The α7β1 integrin dimer binds laminin extracellularly and actin intracellularly via the vinculin (V) and talin (T) proteins. (B) The basic contractile unit of skeletal muscle, the sarcomere, is composed of thin and thick filaments predominantly composed of actin and myosin, respectively. Thin filaments of adjacent sarcomeres are anchored at the Z-disk, which defines the lateral borders of the sarcomere. Myosin has a long, fibrous tail and a globular head, which interacts with actin to produce muscle contraction.
Table 1.
Genes associated with muscular dystrophies
| Disease | Inheritance | Gene | Protein | References |
| DMD, Becker muscular dystrophy | XR | DMD | Dystrophin | Monaco et al., 1986; Burghes et al., 1987 |
| Emery-Dreifuss muscular dystrophy | XR | EMD | Emerin | Bione et al., 1994; |
| XR | FHL1 | Four and a half LIM domains 1 | Gueneau et al., 2009 | |
| AD/AR | LMNA | Lamin A/C | Bonne et al., 1999 | |
| LGMD1A | AD | MYOT | Myotilin | Hauser et al., 2000 |
| LGMD1B | AD | LMNA | Lamin A/C | Muchir et al., 2000 |
| LGMD1C | AD | CAV3 | Caveolin-3 | Minetti et al., 1998 |
| LGMD1D | AD | DES | Desmin | Greenberg et al., 2012 |
| LGMD1E | AD | DNAJB6 | DnaJ (Hsp40) homolog, subfamily B, member 6 | Sarparanta et al., 2012 |
| LGMD2A | AR | CAPN3 | Calpain 3 | Richard et al., 1995 |
| LGMD2B, Miyoshi myopathy, distal anterior compartment myopathy | AR | DYSF | Dysferlin | Bashir et al., 1998; Liu et al., 1998 |
| LGMD2C | AR | SGCG | γ-Sarcoglycan | Noguchi et al., 1995 |
| LGMD2D | AR | SGCA | α-Sarcoglycan | Roberds et al., 1994 |
| LGMD2E | AR | SGCB | β-Sarcoglycan | Bönnemann et al., 1995; Lim et al., 1995 |
| LGMD2F | AR | SGCD | δ-Sarcoglycan | Nigro et al., 1996 |
| LGMD2G | AR | TCAP | Titin-cap | Moreira et al., 2000 |
| LGMD2H | AR | TRIM32 | Tripartite motif containing 32 | Frosk et al., 2002 |
| LGMD2J | AR | TTN | Titin | Hackman et al., 2002 |
| Tibial MD | AD | |||
| LGMD2L | AR | ANO5 | Anoctamin 5 | Bolduc et al., 2010 |
| LGMD2Q | AR | PLEC | Plectin | Gundesli et al., 2010 |
| MDDGA1, MDDGC1 (LGMD2K)a | AR | POMT1 | Protein-O-mannosyltransferase 1 | Beltrán-Valero de Bernabe et al., 2002 |
| MDDGA2, MDDGC2 (LGMD2N) | AR | POMT2 | Protein-O-mannosyltransferase 2 | van Reeuwijk et al., 2005 |
| MDDGA3, MDDGC3 (LGMD2O) | AR | POMGNT1 | Protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase | Yoshida et al., 2001 |
| MDDGA4, MDDGC4 (LGMD2M) | AR | FKTN | Fukutin | Kobayashi et al., 1998 |
| MDDGA5, MDDGC5 (LGMD2I) | AR | FKRP | Fukutin related protein | Brockington et al., 2001 |
| MDDGA6 | AR | LARGE | like-glycosyltransferase | Longman et al., 2003 |
| MDDGA7 | AR | ISPD | Isoprenoid synthase domain containing | Roscioli et al., 2012; Willer et al., 2012 |
| MDDGA8 | AR | GTDC2 | Glycosyltransferase-like domain containing 2 | Manzini et al., 2012 |
| MDDGA | AR | B3GNT1 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 | Buysse et al., 2013 |
| MDDGA | AR | B3GALNT2 | β-1,3-N-acetylgalactosaminyltransferase 2 | Stevens et al., 2013 |
| MDDGC9 (LGMD) | AR | DAG1 | Dystroglycan | Hara et al., 2011 |
| DM1 | AD | DMPK | CTGexp in 3′ UTR | Brook et al., 1992; Fu et al., 1992; Mahadevan et al., 1992 |
| DM2 | AD | CNBP | CCTGexp in intron 1 | Liquori et al., 2001 |
| FSHD1 | AD | DUX4 | Double homeobox 4 | Kowaljow et al., 2007; Lemmers et al., 2010 |
| FSHD2 | AD, Digenic | DUX4, SMCHD1 | Double homeobox 4, structural maintenance of chromosomes flexible hinge domain containing 1 | Lemmers et al., 2012 |
AD, autosomal dominant; AR, autosomal recessive; XR, X-linked recessive; LGMD, limb-girdle muscular dystrophy (LGMD1, autosomal dominant; LGMD2, autosomal recessive); MDDG, muscular dystrophy-dystroglycanopathy; DM, myotonic dystrophy; FSHD, facioscapulohumeral muscular dystrophy; exp, expansion.
Alternative or previous nomenclature is provided in parentheses.
Aberrant glycosylation of α-dystroglycan.
During posttranslational maturation, the mucin domain of α-dystroglycan undergoes extensive O-glycosylation at serine/threonine residues catalyzed by multiple enzymes residing in the Golgi apparatus and sarcoplasmic reticulum in a stepwise manner. The branched O-linked carbohydrate moieties are required for high-affinity binding to G domain–containing proteins of the ECM such as laminin 2, agrin, and perlecan (Barresi and Campbell, 2006). Hypoglycosylation of α-dystroglycan diminishes its binding capacity to the ECM proteins and destabilizes the sarcolemma. Functional impairment of several known or putative glycosyltransferases as a result of genetic mutations causes them to fail to properly glycosylate the α-dystroglycan protein and leads to a clinically heterogeneous group of congenital muscular dystrophies (CMDs): Walker-Warburg syndrome (WWS), muscle-eye-brain disease (MEB), and Fukuyama CMD (Michele et al., 2002; Moore et al., 2002). In addition to hypotonia and severe muscle weakness, present at birth or thereafter, the affected individuals suffer from severe brain and eye malformations. To date, recessive mutations in 10 genes have been identified in ∼50% of individuals with CMD (Table 1), and the number of causative genes is increasing rapidly with the advent of whole-exome sequencing. Mutations in these genes produce a wide spectrum of clinical severity that lead to recent reclassification of the most severe forms of CMD—WWS, MEB, and Fukuyama CMD—as muscular dystrophy-dystroglycanopathy type A (MDDGA). An intermediate form, with or without cognitive impairment, is designated as MDDGB, and the least severe form with no cognitive impairment and manifested as later onset LGMD is designated as MDDGC (Table 1). However, in the majority of cases the causative gene is difficult to predict from the clinical phenotype. Because these disorders are not caused by mutations in the dystroglycan gene itself, but rather by insufficient posttranslational glycosylation of the protein, they are often classified as secondary dystroglycanopathies. A missense change of a highly conserved amino acid (Thr192Met) in dystroglycan was recently identified in an individual affected with LGMD and cognitive impairment, the first reported case of a primary dystroglycanopathy within this disease classification (Hara et al., 2011). Functional studies, including a mutation knock-in mouse model, determined that this mutation interferes with LARGE-mediated maturation of phosphorylated O-mannosyl glycans on α-dystroglycan, thereby decreasing its binding efficiency to laminin.
Loss of integrin-mediated linkage between cytoskeleton and ECM.
Similar to the DAPC, the α7β1 integrin heterodimer connects the actin cytoskeleton to the ECM (Fig. 1 A). Mutations in ITGA7 encoding the α7 chain of α7β1 integrin cause CMD (Hayashi et al., 1998). In developing myoblasts, α7β1 integrin plays a role in cell adhesion, proliferation, and differentiation. In mature myofibers, it is expressed throughout the sarcolemma to reinforce its attachment to the basal lamina, and in particular these proteins are concentrated at costameres, neuromuscular junctions (NMJs), and myotendinous junctions (MTJs). Targeted deletion of the α7 chain causes progressive muscle degeneration with specific impairment of function at the MTJ (Mayer et al., 1997). Loss of the DAPC in DMD muscle increases the amount α7β1 integrin at the sarcolemma, likely to compensate for the weakened link between the sarcolemma and basal lamina. Furthermore, transgenic overexpression of α7β1 integrin reduced sarcolemma damage and significantly ameliorated disease symptoms in mdx mice, the most widely used animal model for DMD with a nonsense mutation in Dmd (Burkin et al., 2001; Liu et al., 2012). The integrin linkage system appears to functionally complement the DAPC-mediated linkage between the cytoskeleton and the ECM. Therefore, enhancing sarcolemmal expression of the integrin heterodimers provides a potential therapeutic approach for DMD and other muscular dystrophies caused by loss of the DAPC.
Disruption of the ECM.
The ECM is a meshwork of glycoproteins, collagen, and proteoglycans that provides an attachment and signaling scaffold for myofibers. Mutations in several genes encoding the structural components of the ECM are responsible for CMDs. The most common form of CMD, merosin-deficient or type 1A (MDC1A), is caused by the deficiency of laminin-α2 chain (LAMA2; Helbling-Leclerc et al., 1995). Secondary loss of laminin-α2 is common to severe forms of CMD caused by mutations in other genes (Brockington et al., 2001). Laminin-α2 is one of the three subunits of muscle-specific laminin that makes distinct interacting complexes with the DAPC through glycosylated α-dystroglycan and with the α7β1 integrin dimer. These protein complexes are secreted from myofibers and interact with entactin, which binds to collagens to build the ECM. In addition to destabilized sarcolemma in laminin-deficient muscle, mitochondrial apoptosis appears to contribute substantially to disease progression in MDCA1. Detachment of cell membrane from the ECM induces a particular form of apoptotic pathway called anoikis. Inhibition of muscle cell death by transgenic overexpression of the antiapoptosis protein Bcl-2, or through targeted ablation of the pro-apoptosis protein Bax in Lama2−/− mice, alleviated the dystrophic phenotype and extended lifespan of these animals severalfold (Girgenrath et al., 2004). Therefore antiapoptosis therapies could potentially benefit MDCA1 and likely other neuromuscular disorders where apoptosis plays an important role in disease pathogenesis, such as those that result from defective collagen VI. Different types of mutations in three genes (COL6A1, COL6A2, and COL6A3) encoding the subunits of collagen VI cause two clinically distinct myopathies: Ullrich CMD and Bethlem myopathy (Bönnemann, 2011).
Impaired cell signaling at the sarcolemma.
The intracellular module of the DAPC that includes α-dystrobrevin, syntrophins, and neuronal nitric oxide synthase (nNOS) regulates the signal transduction cascade at the sarcolemma. The nNOS enzyme produces nitric oxide (NO) from l-arginine, which in turn induces guanylyl cyclase to synthesize cyclic GMP, a signaling molecule broken down by cyclic nucleotide phosphodiesterases (PDEs). cGMP functions as a potent vasodilator by inhibiting the vasoconstrictor response to α-adrenergic receptor activation. Muscle contractions activate nNOS and the resultant NO augments local blood flow to meet the increased energy demand of contracting muscle (Kobzik et al., 1994). nNOS is anchored to the sarcolemma via the PDZ domain of the adaptor protein syntrophin, which binds directly to the C-terminal domain of dystrophin. To ensure rapid diffusion of NO to blood vessels, sarcolemmal localization of nNOS is essential for its function as a vasodilator in skeletal muscle. Complete loss of dystrophin dissociates nNOS from the sarcolemma (Brenman et al., 1995), causing functional ischemia and exercise-induced fatigue in DMD patients (Sander et al., 2000). Interestingly, a recent study demonstrated that not only the DMD muscle, but muscles from other types of muscular dystrophies, including those that do not directly involve the DAPC, tend to lose nNOS from the sarcolemma, which implies that many muscular dystrophies may share common mechanisms of exercise-induced fatigue (Kobayashi et al., 2008). Furthermore, using nNOS-null mice, this study demonstrated that exercise-induced fatigue as a result of nNOS loss is distinct from general muscle weakness pertinent to muscle wasting diseases. Treatment of the mdx and Sgca−/− mice, which lack α-sarcoglycan, with the phosphodiesterase 5A inhibitor sildenafil citrate to boost cGMP signaling significantly improved vasodilation and reduced exercise-induced fatigue, which supports the therapeutic benefits of vasomodulation in muscular dystrophies (Kobayashi et al., 2008). Furthermore, syntrophins also bind the dystrophin-related protein α-dystrobrevin, which in turn interacts with dystrophin as well as with sarcoglycans. Mice null for α-dystrobrevin exhibit muscular dystrophy and cardiomyopathy, not because of disruption of the DAPC and sarcolemma but as a result of compromised signaling at the sarcolemma (Grady et al., 1999). In muscle from these mice, nNOS is dislocated from the DAPC and the production of NO is reduced, leading to muscle and heart abnormalities. Recent studies demonstrate that nNOS can also bind directly to the dystrophin protein via a 10–amino acid fragment within spectrin-like repeat 17 (Lai et al., 2009, 2013). In fact, direct interaction with dystrophin appears to be critical for the recruitment of nNOS to the sarcolemma. Although minidystrophin, a truncated version of dystrophin without these specific spectrin-like repeats engineered for antiviral gene therapy, reverses dystrophic pathology in the mdx mouse, it is unable to reduce exercise-induced fatigue; therefore, inclusion of spectrin-like repeats required for nNOS interaction with minidystrophin should be considered in gene replacement therapies. In conclusion, these studies underscore the importance of nNOS signaling in the functioning and maintenance of skeletal muscle.
Cytoskeletal defects in muscular dystrophy.
The main intracellular cytoskeletal component that transmits the mechanical force through the DAPC to ECM is the microfilament network consisting of F-actin (Ervasti and Campbell, 1993; Rybakova et al., 2000). The N-terminal actin-binding domain and spectrin-like repeats within the rod domain of dystrophin bind to actin filaments. However, several recent studies revealed that dystrophin interacts with the other two elements of the cytoskeleton; intermediate filaments (IFs) and microtubules. Multiple members of the IF network are expressed in striated muscle; among these desmin is the predominantly expressed IF protein in myofibers. Desmin connects Z-disks of neighboring myofibrils and anchors myofibrils to intracellular organelles, such as mitochondria and the nucleus, and to the sarcolemma, to maintain a spatial organization between myofibrils and other structural components of the myofiber. Mutations in desmin are associated with the autosomal dominant LGMD1D with dilated cardiomyopathy, with characteristic desmin-positive protein aggregates within the sarcoplasma (Greenberg et al., 2012). The linkage between desmin and the DAPC appears to be mediated by the giant cytolinker protein plectin, mutations in which also underlie muscular dystrophy with skin blistering (epidermolysis bullosa simplex; Smith et al., 1996). Plectin is expressed as multiple isoforms through alternative splicing of its variable N-terminal domain. The C-terminal domain of plectin-1f isoform binds desmin while the actin-binding and plakin domains bind dystrophin and β-dystroglycan at costameres, respectively (Rezniczek et al., 2007). A homozygous 9bp deletion mutation in the first plectin-1f isoform-encoding exon of PLEC was identified in progressive LGMD2Q without skin manifestation, demonstrating a unique role of plectin isoform 1f in skeletal muscle function (Gundesli et al., 2010). Enlarged distance between the sarcolemma and myofibrils and misaligned Z lines in patient muscles suggest that lack of this particular isoform fails to hold these structures together. Other cytolinker proteins such as syncoilin and synemin are expressed in skeletal muscle and have been suggested to mediate interaction of the desmin IF cytoskeleton network with the DAPC (Bellin et al., 2001; Poon et al., 2002). Furthermore, dystrophin directly interacts and stabilizes the organization of the third component of the cellular cytoskeleton—microtubules (Prins et al., 2009)—and microtubule disorganization has been proposed to contribute to disease pathogenesis in DMD. Brief stretching of muscle from mdx mice induces microtubule-dependent activation of NADPH oxidase, which in turn produces reactive oxygen species and increases sarcolemmal Ca+2 influx (Khairallah et al., 2012). Collectively, in contrast to the initial assumption that the F-actin is the only cytoskeletal element interacting with the DAPC, it is now well established that IFs as well as microtubules interact with the DAPC.
Defective sarcolemma repair.
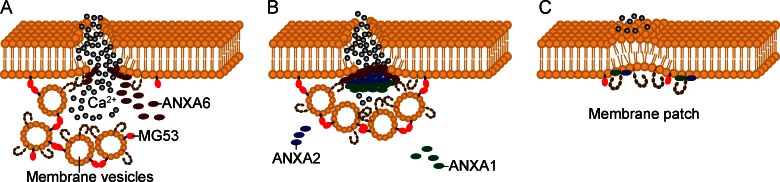
Frequent mechanical stress and the large size of individual muscle fibers render the fragile sarcolemma susceptible to microinjuries. In healthy muscle these contraction-induced membrane injuries are efficiently resealed by a Ca2+-dependent repair pathway. The dynamic repair process performed by fusion of membrane-bound intracellular vesicles with the sarcolemma is orchestrated by intricate interactions between multiple proteins residing both on the sarcolemma and on these vesicles (Fig. 2 A). In contrast to myofiber degeneration that results from loss of connection between the cytoskeleton and the ECM, muscle degeneration can also arise from inefficient repair of naturally damaged sarcolemma. Dysferlin, a 230-kD transmembrane protein with six intracellular C2 domains, is a key component of the Ca2+-dependent sarcolemma repair pathway. It promotes the fusion of the intracellular vesicles with each other and with the sarcolemma at the injury site. Loss-of-function mutations in DYSF underlie three clinically distinct muscular dystrophies, collectively called dysferlinopathies: LGMD2B, Miyoshi myopathy, and distal anterior compartment myopathy (DACM; Bashir et al., 1998; Liu et al., 1998). Miyoshi myopathy prominently affects distal limb muscles, whereas LGMD2B is a proximal muscle disease. Interestingly, identical mutations in DYSF have been associated with both types (Weiler et al., 1999). Dysf−/− mice develop progressive muscular dystrophy, with intact DAPC, which indicates that loss of dysferlin does not destabilize the DAPC (Bansal et al., 2003). Because the intracellular vesicles fail to fuse with the damaged sarcolemma, there is a characteristic accumulation of subsarcolemmal vesicles in dysferlin-deficient muscle.
Figure 2.
Dysferlin-mediated sarcolemma repair. (A) Repetitive muscle contractions often cause membrane disruption. Mitsugumin 53 (MG53; red) located on intracellular membrane-bound vesicles oligomerizes when exposed to oxidized extracellular components (gray circles) and recruits dysferlin-carrying vesicles to the injury site. Simultaneously but independently of these vesicles, intracellular annexin 6 (ANXA6) accumulates at the membrane lesion. (B) Together with dysferlin, ANXA6 sequentially recruits ANXA2 and ANXA1 to the injury site. (C) Elevated intracellular Ca2+ concentration facilitates fusion of intracellular vesicles with each other and with the plasma membrane through interactions between dysferlin, annexins, and other proteins at the disrupted site, rapidly forming a membrane repair patch.
Dysferlin interacts with multiple proteins. Among these proteins, caveolin-3 (Matsuda et al., 2001) is the principle constituent of plasma membrane caveolae, small invaginations of the plasma membrane that are responsible for LGMD1C when mutated (Minetti et al., 1998). Mitsugumin 53 (MG53), a muscle-specific TRIM (Tri-partite motif) family protein encoded by the TRIM72 gene, is another important member of the sarcolemma membrane repair pathway. Although no mutations in TRIM72 have yet been identified in human muscular dystrophy, MG53 knockout mice develop progressive muscle degeneration, which was demonstrated to result from defective membrane repair (Cai et al., 2009a). Influx of oxidized extracellular environment through damaged sarcolemma into the sarcoplasma appears to oligomerize MG53 residing on intracellular vesicles, which in turn induces their translocation to and nucleation at the acute injury site. Subsequently, through functional interaction with MG53 and caveolin-3, dysferlin facilitates rapid membrane patch formation in the presence of elevated Ca2+ level, and disruption of one of these components can affect the localization and function of the other components (Cai et al., 2009b). Dysferlin also interacts with annexins A1 and A2 (Lennon et al., 2003). Annexins are a family of proteins that bind phospholipids in a Ca2+-dependent manner to assemble a docking platform for interacting proteins at the membrane. A recent study using high-resolution in vivo imaging in zebrafish demonstrated that after membrane injury, cytoplasmic annexin A6 accumulates at the membrane lesion, independently and simultaneously with dysferlin-docked intracellular vesicles, and then together with dysferlin sequentially recruit annexins A2 and A1 to the membrane repair site (Roostalu and Strähle, 2012; Fig. 2 A). In dysferlin-deficient muscles, the localization and distribution of annexin A1 and A2 at the sarcolemma is perturbed (Lennon et al., 2003). In conclusion, sarcolemmal repair involves sequential interaction between multiple proteins, and these proteins could be strong candidates for muscular dystrophies that have not yet been associated with a genetic mutation. Furthermore, variation within these proteins might also modulate phenotypic variability of dysferlin-associated muscle diseases.
Anoctamins are a family of transmembrane proteins with Ca2+-activated chloride channel activity (Tian et al., 2012). Loss-of-function mutations in ANO5, encoding anoctamin 5, were identified in families with LGMD2L and distal nondysferlin Miyoshi myopathy 3 (MMD3; Bolduc et al., 2010). Muscles from subjects with ANO5 mutations displayed multifocal sarcolemma lesions, but unlike in typical dysferlinopathies no subsarcolemmal vesicle accumulation was observed. However, fibroblasts from one MMD3 subject did show impaired membrane repair when the cell membrane was injured in vitro. The function of anoctamin 5 in skeletal muscle still remains unclear. Progressive loss of both proximal as well as distal muscles in different individuals, defective membrane repair in one subject, and predicted Ca2+-activated chloride channel activity point to its potential role in sarcolemma repair, likely through the dysferlin-dependent repair pathway described above. According to the proposed model, increased Ca2+ concentration within a damaged myofiber might activate the putative chloride ion channel ANO5 residing on intracellular vesicles and allow chloride transport into the vesicles to modify their conformation and facilitate recruitment to the injury site (Bolduc et al., 2010).
Disintegration of muscle sarcomeres.
The assembly and maintenance of myofibrils is regulated by multiple proteins that interact with the primary sarcomeric proteins actin and myosin (Fig. 1 B). Mutations in proteins expressed in sarcomeres have been associated with neuromuscular disorders, including several LGMDs, but mutations in sarcomeric proteins are more common in cardiomyopathies. Associated genes and molecular mechanisms of inherited cardiomyopathies are discussed in two recent reviews (Harvey and Leinwand, 2011; McNally et al., 2013). Interestingly, mutations in sarcomeric proteins causative of muscular dystrophy tend to affect distal muscles, i.e., those in hands, forearms, lower legs, and feet. The myotilin protein cross-links α-actinin, filamin-C, and F-actin, and promotes myofibrillogenesis at Z-disks (Salmikangas et al., 2003). Mutations in the MYOT gene were identified in families with LGMD1A (Hauser et al., 2000). Dominant mutations in the actin-binding domain of filamin-C that increase its binding affinity for actin have recently been associated with a distal myopathy (Duff et al., 2011). Nebulin spans the length of the thin filament interacting with its components actin, tropomyosin, tropomodulin, and CAPZ. It regulates the filament length by stabilizing and preventing actin depolymerization (Pappas et al., 2010). Mutations in nebulin are responsible for nemaline myopathy, a group of congenital muscle disorders characterized by generalized muscle weakness and rod-like nemaline bodies in myofibers (Wallgren-Pettersson et al., 2011). Titin extends from the opposing Z-disks of a sarcomere and overlaps in the middle of the sarcomere and is thought to serve as the molecular ruler that controls the length of sarcomere during contraction and relaxation. Mutations in titin have been associated with both muscular dystrophy and cardiomyopathy. Heterozygous mutations in the C-terminal domain of titin, located within the M-line of sarcomeres, cause the late-onset tibial muscular dystrophy, whereas homozygous mutations underlie the more severe LGMD2J (Hackman et al., 2002). Titin mutations associated with dilated cardiomyopathy are, however, overrepresented in its A-band region but are absent in the Z-disk and M-line regions (Herman et al., 2012). Furthermore, mutations in titin lead to secondary reduction of calpain 3, a muscle-specific, calcium-dependent protease that is mutated in LGMD2A (Richard et al., 1995). LGMD2G is associated with mutations in telethonin (titin-cap), which anchors two parallel titin molecules within a single sarcomere to the Z-disk (Moreira et al., 2000). Given the complexity of the organization and biogenesis of sarcomeres, mutations in many of the interacting proteins have been associated with several forms of muscular dystrophies. These disorders have been reviewed recently (Udd, 2009).
Toxic RNA induced muscle degeneration.
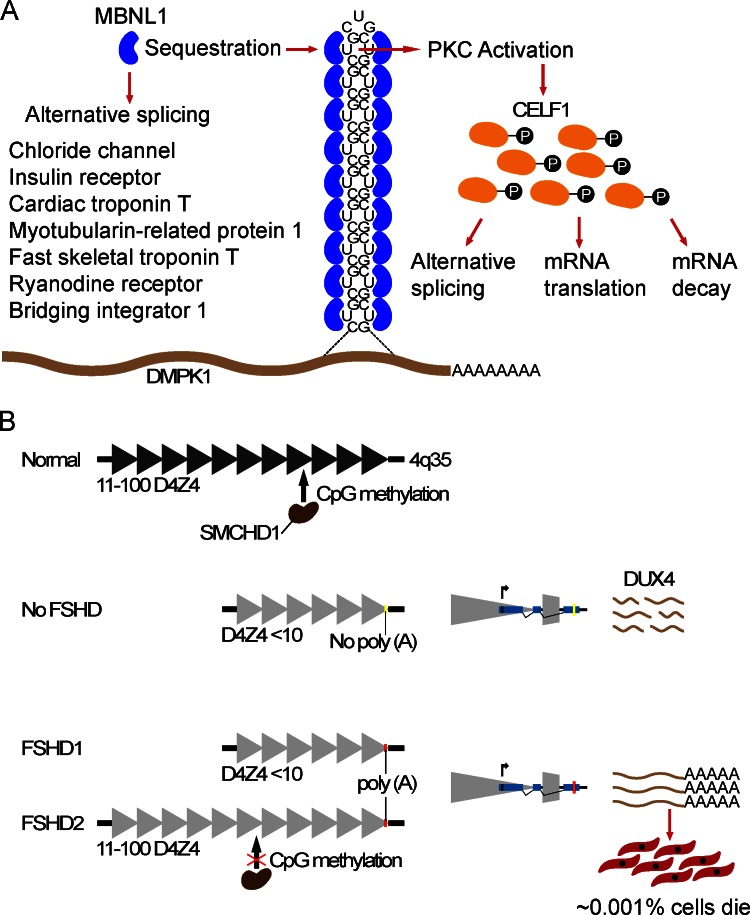
Unlike the majority of muscular dystrophies caused by mutations that alter the amino acid sequence, the primary cause of myotonic dystrophy (DM) is the accumulation of toxic RNA in cell nucleus. Myotonic dystrophy type 1 (DM1) is caused by a CTG trinucleotide repeat expansion (from 50 to >2,000) within the 3′ UTR of the dystrophia myotonica protein kinase (DMPK) gene (Brook et al., 1992; Fu et al., 1992; Mahadevan et al., 1992). DM1 is a multisystem disease that predominantly affects skeletal muscles and the central nervous system, and it is also considered the most common adult onset muscular dystrophy, with a prevalence of ∼1/8,000. It is characterized by myotonia (slow relaxation of the muscles after contraction), muscle degeneration, cataracts, cardiac arrhythmias, and cognitive impairment. The less frequent DM2 results from a CCTG repeat expansion (from 75 to >10,000) in intron 1 of the CCHC-type zinc finger nucleic acid binding protein (CNBP; Liquori et al., 2001). The mRNA products transcribed from the expanded alleles of these genes are retained within the nucleus, forming punctuated foci of RNP aggregates (Taneja et al., 1995; Davis et al., 1997; Mankodi et al., 2003). In both types of DM, mis-splicing of downstream target gene transcripts secondary to these repeat sequence amplifications underlie the molecular etiology of the disease. Muscleblind-like proteins (MBNL) are a highly conserved, developmentally regulated family of proteins that regulate alternative splicing of a large number of pre-mRNAs. The native cognate binding site of MBNL1 includes a CUG motif. Therefore, MBNL1 binds to expanded CUG and CCUG repeats of the DMPK1 and CNBP mRNAs, respectively, and becomes sequestered within the nucleus (Miller et al., 2000; Mankodi et al., 2001; Fig. 3 A). Sequestration of MBNL1 depletes its cellular abundance, which in turn leads to inappropriate splicing of many pre-mRNAs that depend on MBNL1 for their transition from an embryonic to postnatal splicing pattern (Lin et al., 2006). Two of the well-studied pre-mRNAs that are prominently affected and directly contribute to disease symptoms encode muscle-specific chloride channel, responsible for the hyperexcitability of muscle (myotonia; Charlet-B et al., 2002), and insulin receptor, which explains insulin resistance in DM patients (Savkur et al., 2001). Mouse models expressing CTG repeats from the 3′ UTR of the human skeletal α-actin transgene (HASLR; Mankodi et al., 2000) and null for muscleblind-like 1 (Mbnl1ΔE3/ΔE3; Kanadia et al., 2003) reproduce several human phenotypes including embryonic pattern of pre-mRNA splicing in adult tissues. Genome-wide gene expression and pre-mRNA splicing analyses in these two mouse models revealed >200 exons with altered splicing profiles (Du et al., 2010). More than 80% of the splicing differences observed in HASLR muscles were also seen in Mbnl1-deficient muscles, which suggests that loss of Mbnl1 likely accounts for the majority of abnormal splicing occurring in DM. Furthermore, expression levels of genes encoding the ECM proteins were overrepresented among the altered transcripts, highlighting common molecular defects with other types of muscular dystrophies. These genome-wide studies of mRNA expression and splicing events have identified potential candidate genes that might contribute to disease symptoms when inappropriately spliced. The specific roles of individual target genes in disease etiology remain to be established. In addition to MBNL1 depletion, expanded CUG repeats induce aberrant activation of PKC, which in turn phosphorylates and increases the activity of a second splicing regulatory factor CUGBP Elav like family 1 (CELF1), formerly called as CUGPB1 (Roberts et al., 1997; Philips et al., 1998; Kuyumcu-Martinez et al., 2007; Fig. 3 A). MBNL1 and CELF1 are thought to act as antagonistic splicing regulatory factors. CELF1 is also known to control mRNA translation and stability. Consequently, imbalance between these two genomic regulatory factors has a widespread effect on a large number of genes that result in systemic disease.
Figure 3.
Primary molecular mechanisms underlying toxic RNA and toxic transcription factor–induced muscular dystrophies. (A) The expanded CUG tract within the 3′ UTR of the DMPK mRNA folds into a double-stranded hairpin structure that resembles the cognate binding site of the muscleblind-like 1 (MBNL1) protein. MBNL1 binds the expanded RNA molecules and becomes sequestered within the nucleus, resulting in loss of its normal function in RNA splicing and enhancing formation of the foci that trap the expanded RNA in the nucleus. The nuclear accumulation of this RNA disrupts RNA-processing functions in the nucleus and cytoplasm, affecting the regulation of alternative splicing and translation of many pre-mRNAs. A handful of well-studied examples are listed in the figure. Hyperphosphorylation and up-regulation of CELF1 as a result of expanded CUG repeats affect alternative splicing, translation, and mRNA stability of its target genes. (B) Unaffected individuals carry 11–100 repeats (triangles) within the D4Z4 macrosatellite on the telomeric end of chromosome 4q35. Contraction of D4Z4 repeats to <10 repeats relaxes the chromatin and induces DUX4 expression from the distal-most repeat unit (shown separately). DUX4 expressed from the nonpermissive chromosomal allele that does not contain the poly(A) signal (yellow bar) does not get polyadenylated and is unstable, whereas polyadenylated transcripts expressed from the permissive allele (red bar) are stable and translate into toxic transcription factor. Black triangles depict condensed chromatin, whereas gray triangles depict relaxed chromatin as a result of hypomethylation. SMCHD1 regulates D4Z4 methylation. In FSHD2, mutated SMCH1 fails to methylate D4Z4 and suppress DUX4 expression.
Ectopic expression of a toxic transcription factor.
Facioscapulohumeral muscular dystrophy (FSHD) is an unusual and the third most common muscular dystrophy, characterized by the asymmetric and progressive weakness and atrophy of skeletal muscles of the face, scapula, and upper arms, frequently accompanied by retinal telangiectasia and/or hearing loss. The common form of FSHD, type 1, is caused by contractions of tandemly arrayed repeat elements of 3.3 kb within the highly polymorphic macrosatellite element D4Z4 on the distal end of chromosome 4q35 (Wijmenga et al., 1992; van Deutekom et al., 1993). FSHD1 is, however, associated with D4Z4 mutations that arise only on the permissive chromosomal allele 4qA, whereas deletions on the equally common allele in the general population 4qB do not present any pathogenic consequences (Lemmers et al., 2002). In unaffected individuals, the D4Z4 array usually consists of 11–100 repeats, whereas FSHD1 patients carry 1–10 repeats. Furthermore, each repeat unit of D4Z4 contains an open reading frame encoding the transcription factor double homeobox 4 (DUX4). Reduction of the D4Z4 size to a pathogenic threshold of <10 repeats relaxes the chromatin state within D4Z4 and induces the expression of the otherwise presumably repressed DUX4 retrogene. The expression of DUX4 embedded in the last repeat extends into the adjacent pLAM sequence, which contains a polymorphic canonical polyadenylation signal. Transcripts expressed from the permissive allele (4qA) are polyadenylated and stabilized, consequently producing protein, whereas the nonpolyadenylated transcripts transcribed from the nonpermissive allele (4qB) are degraded (Lemmers et al., 2010). The double homeobox domain containing transcription factor DUX4 in turn induces muscle cell death (Kowaljow et al., 2007; Snider et al., 2010; Fig. 3 B). The exact mechanisms of its toxicity to muscle cells are not clear yet. Furthermore, DUX4 is spliced into two isoforms, the full-length protein (DUX4-fl), which was shown to be expressed exclusively in ∼1/1,000 cultured FSHD myoblasts, causing cell death shortly after translation, and a shorter nontoxic isoform expressed in both affected and unaffected cells and muscles (Snider et al., 2010).
Later findings challenged the notion that the expression of DUX4 is restricted to affected cells and muscles, by demonstrating that DUX4-fl is also expressed in unaffected cells and muscles, albeit at lower frequency, which is suggestive of potential disease modifiers (Jones et al., 2012). DUX4-fl was shown to induce a large number of genes that are normally expressed in stem cells and germline (Geng et al., 2012). However, the significance of these genes in disease pathogenesis is elusive. Strikingly, injection of very low amounts of DUX4-fl mRNA into zebrafish embryos recapitulated several human phenotypes, including the extramuscular phenotypes such as eye and ear abnormalities and asymmetric involvement of affected organs (Mitsuhashi et al., 2013).
The less common type, FSHD2, which is clinically indistinguishable from FSHD1, was previously linked to hypomethylation of D4Z4 chromatin (van Overveld et al., 2003). However, the genetic determinant of this phenomenon was not known until recently. Whole-exome sequencing in FSHD2 families identified heterozygous mutations in the SMCHD1 gene that co-segregate with D4Z4 hypomethylation of the permissive allele in affected family members (Lemmers et al., 2012). This study demonstrated that in addition to the haploinsufficiency of SMCHD1, inheritance of the polyadenylated DUX4 is required for disease manifestation. Consequently, these findings converge the underlying disease mechanisms of FSHD1 and FSHD2. SMCHD1 encodes for the SMCHD1 (structural maintenance of chromosomes flexible hinge domain containing 1) protein and is required for maintenance of the genome-wide hypermethylation of D4Z4, and likely other repeat elements. It has yet to be tested whether genetic variants in SMCHD1 also modify the disease phenotype of FSHD1, such as the highly variable age of onset, mutation penetrance, and extramuscular involvement.
Conclusion
Remarkable progress has been made over the last two decades toward deciphering the cellular and molecular underpinnings of neuromuscular disorders. Better understanding of the molecular mechanisms has paved the way for the development of targeted therapeutic interventions for several muscular dystrophies, not only specific for each type of disease, but also specific for individuals with certain mutations. Progress in antisense oligonucleotide-mediated exon skipping to restore the open reading frame of mutated dystrophin or nonsense mutation read-through approaches developed to treat DMD has already started showing encouraging outcomes in clinical trials. Furthermore, recent advances in improved diagnostics, in particular using next-generation sequencing technologies, now allow the rapid (literally within hours) screening of newborns for potentially pathogenic genetic lesions. Early identification of mutations causative of muscular dystrophies will enable clinicians to take proactive measures to halt or reverse muscle degeneration.
Acknowledgments
The work in our laboratory is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development grant (5U54HD060848) to the Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center for FSHD, by research (MDA177911) and development (MDA202863) grants from the Muscular Dystrophy Association, and by a grant from the Bernard F. and Alva B. Gimbel Foundation.
We apologize to our colleagues whose work we could not cite because of space limitations.
Footnotes
Abbreviations used in this paper:
- CK
- creatine kinase
- CMD
- congenital muscular dystrophy
- DAPC
- dystrophin-associated protein complex
- DM
- myotonic dystrophy
- DMD
- Duchenne muscular dystrophy
- F-actin
- filamentous actin
- FSHD
- facioscapulohumeral muscular dystrophy
- IF
- intermediate filament
- LGMD
- limb-girdle muscular dystrophy
- MDDG
- muscular dystrophy-dystroglycanopathy
- nNOS
- neuronal nitric oxide synthase
- NO
- nitric oxide
References
- Bansal D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., Campbell K.P. 2003. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 423:168–172 10.1038/nature01573 [DOI] [PubMed] [Google Scholar]
- Barresi R., Campbell K.P. 2006. Dystroglycan: from biosynthesis to pathogenesis of human disease. J. Cell Sci. 119:199–207 10.1242/jcs.02814 [DOI] [PubMed] [Google Scholar]
- Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., et al. 1998. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 20:37–42 10.1038/1689 [DOI] [PubMed] [Google Scholar]
- Bellin R.M., Huiatt T.W., Critchley D.R., Robson R.M. 2001. Synemin may function to directly link muscle cell intermediate filaments to both myofibrillar Z-lines and costameres. J. Biol. Chem. 276:32330–32337 10.1074/jbc.M104005200 [DOI] [PubMed] [Google Scholar]
- Beltrán-Valero de Bernabé D., Currier S., Steinbrecher A., Celli J., van Beusekom E., van der Zwaag B., Kayserili H., Merlini L., Chitayat D., Dobyns W.B., et al. 2002. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am. J. Hum. Genet. 71:1033–1043 10.1086/342975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S., Maestrini E., Rivella S., Mancini M., Regis S., Romeo G., Toniolo D. 1994. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8:323–327 10.1038/ng1294-323 [DOI] [PubMed] [Google Scholar]
- Bolduc V., Marlow G., Boycott K.M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., et al. 2010. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86:213–221 10.1016/j.ajhg.2009.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21:285–288 10.1038/6799 [DOI] [PubMed] [Google Scholar]
- Bönnemann C.G. 2011. The collagen VI-related myopathies: muscle meets its matrix. Nat. Rev. Neurol. 7:379–390 10.1038/nrneurol.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönnemann C.G., Modi R., Noguchi S., Mizuno Y., Yoshida M., Gussoni E., McNally E.M., Duggan D.J., Angelini C., Hoffman E.P. 1995. Beta-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat. Genet. 11:266–273 10.1038/ng1195-266 [DOI] [PubMed] [Google Scholar]
- Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. 1995. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 82:743–752 10.1016/0092-8674(95)90471-9 [DOI] [PubMed] [Google Scholar]
- Brockington M., Blake D.J., Prandini P., Brown S.C., Torelli S., Benson M.A., Ponting C.P., Estournet B., Romero N.B., Mercuri E., et al. 2001. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am. J. Hum. Genet. 69:1198–1209 10.1086/324412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. 1992. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 68:799–808 10.1016/0092-8674(92)90154-5 [DOI] [PubMed] [Google Scholar]
- Burghes A.H., Logan C., Hu X., Belfall B., Worton R.G., Ray P.N. 1987. A cDNA clone from the Duchenne/Becker muscular dystrophy gene. Nature. 328:434–437 10.1038/328434a0 [DOI] [PubMed] [Google Scholar]
- Burkin D.J., Wallace G.Q., Nicol K.J., Kaufman D.J., Kaufman S.J. 2001. Enhanced expression of the α7β1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J. Cell Biol. 152:1207–1218 10.1083/jcb.152.6.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buysse K., Riemersma M., Powell G., van Reeuwijk J., Chitayat D., Roscioli T., Kamsteeg E.-J., van den Elzen C., van Beusekom E., Blaser S., et al. 2013. Missense mutations in β-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) cause Walker-Warburg syndrome. Hum. Mol. Genet. 22:1746–1754 10.1093/hmg/ddt021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., Ko J.K., Lin P., Thornton A., Zhao X., et al. 2009a. MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 11:56–64 10.1038/ncb1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Weisleder N., Ko J.K., Komazaki S., Sunada Y., Nishi M., Takeshima H., Ma J. 2009b. Membrane repair defects in muscular dystrophy are linked to altered interaction between MG53, caveolin-3, and dysferlin. J. Biol. Chem. 284:15894–15902 10.1074/jbc.M109.009589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet-B N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A. 2002. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 10:45–53 10.1016/S1097-2765(02)00572-5 [DOI] [PubMed] [Google Scholar]
- Collins C.A., Olsen I., Zammit P.S., Heslop L., Petrie A., Partridge T.A., Morgan J.E. 2005. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 122:289–301 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. 1997. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA. 94:7388–7393 10.1073/pnas.94.14.7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Cline M.S., Osborne R.J., Tuttle D.L., Clark T.A., Donohue J.P., Hall M.P., Shiue L., Swanson M.S., Thornton C.A., Ares M., Jr 2010. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17:187–193 10.1038/nsmb.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff R.M., Tay V., Hackman P., Ravenscroft G., McLean C., Kennedy P., Steinbach A., Schöffler W., van der Ven P.F., Fürst D.O., et al. 2011. Mutations in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am. J. Hum. Genet. 88:729–740 10.1016/j.ajhg.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti J.M., Campbell K.P. 1993. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 122:809–823 10.1083/jcb.122.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. 1990. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 345:315–319 10.1038/345315a0 [DOI] [PubMed] [Google Scholar]
- Frosk P., Weiler T., Nylen E., Sudha T., Greenberg C.R., Morgan K., Fujiwara T.M., Wrogemann K. 2002. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 70:663–672 10.1086/339083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.H., Pizzuti A., Fenwick R.G., Jr, King J., Rajnarayan S., Dunne P.W., Dubel J., Nasser G.A., Ashizawa T., de Jong P., et al. 1992. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 255:1256–1258 10.1126/science.1546326 [DOI] [PubMed] [Google Scholar]
- Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., Tapscott S.J. 2012. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 22:38–51 10.1016/j.devcel.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenrath M., Dominov J.A., Kostek C.A., Miller J.B. 2004. Inhibition of apoptosis improves outcome in a model of congenital muscular dystrophy. J. Clin. Invest. 114:1635–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady R.M., Grange R.W., Lau K.S., Maimone M.M., Nichol M.C., Stull J.T., Sanes J.R. 1999. Role for alpha-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat. Cell Biol. 1:215–220 10.1038/12034 [DOI] [PubMed] [Google Scholar]
- Greenberg S.A., Salajegheh M., Judge D.P., Feldman M.W., Kuncl R.W., Waldon Z., Steen H., Wagner K.R. 2012. Etiology of limb girdle muscular dystrophy 1D/1E determined by laser capture microdissection proteomics. Ann. Neurol. 71:141–145 10.1002/ana.22649 [DOI] [PubMed] [Google Scholar]
- Gueneau L., Bertrand A.T., Jais J.P., Salih M.A., Stojkovic T., Wehnert M., Hoeltzenbein M., Spuler S., Saitoh S., Verschueren A., et al. 2009. Mutations of the FHL1 gene cause Emery-Dreifuss muscular dystrophy. Am. J. Hum. Genet. 85:338–353 10.1016/j.ajhg.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundesli H., Talim B., Korkusuz P., Balci-Hayta B., Cirak S., Akarsu N.A., Topaloglu H., Dincer P. 2010. Mutation in exon 1f of PLEC, leading to disruption of plectin isoform 1f, causes autosomal-recessive limb-girdle muscular dystrophy. Am. J. Hum. Genet. 87:834–841 10.1016/j.ajhg.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackman P., Vihola A., Haravuori H., Marchand S., Sarparanta J., De Seze J., Labeit S., Witt C., Peltonen L., Richard I., Udd B. 2002. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 71:492–500 10.1086/342380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y., Balci-Hayta B., Yoshida-Moriguchi T., Kanagawa M., Beltrán-Valero de Bernabé D., Gündeşli H., Willer T., Satz J.S., Crawford R.W., Burden S.J., et al. 2011. A dystroglycan mutation associated with limb-girdle muscular dystrophy. N. Engl. J. Med. 364:939–946 10.1056/NEJMoa1006939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P.A., Leinwand L.A. 2011. The cell biology of disease: cellular mechanisms of cardiomyopathy. J. Cell Biol. 194:355–365 10.1083/jcb.201101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M.A., Horrigan S.K., Salmikangas P., Torian U.M., Viles K.D., Dancel R., Tim R.W., Taivainen A., Bartoloni L., Gilchrist J.M., et al. 2000. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum. Mol. Genet. 9:2141–2147 10.1093/hmg/9.14.2141 [DOI] [PubMed] [Google Scholar]
- Hayashi Y.K., Chou F.L., Engvall E., Ogawa M., Matsuda C., Hirabayashi S., Yokochi K., Ziober B.L., Kramer R.H., Kaufman S.J., et al. 1998. Mutations in the integrin alpha7 gene cause congenital myopathy. Nat. Genet. 19:94–97 10.1038/ng0598-94 [DOI] [PubMed] [Google Scholar]
- Helbling-Leclerc A., Zhang X., Topaloglu H., Cruaud C., Tesson F., Weissenbach J., Tomé F.M., Schwartz K., Fardeau M., Tryggvason K., Guicheney P. 1995. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat. Genet. 11:216–218 10.1038/ng1095-216 [DOI] [PubMed] [Google Scholar]
- Herman D.S., Lam L., Taylor M.R., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E., et al. 2012. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366:619–628 10.1056/NEJMoa1110186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E.P., Brown R.H., Jr, Kunkel L.M. 1987. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 51:919–928 10.1016/0092-8674(87)90579-4 [DOI] [PubMed] [Google Scholar]
- Hoffman E.P., Fischbeck K.H., Brown R.H., Johnson M., Medori R., Loike J.D., Harris J.B., Waterston R., Brooke M., Specht L., et al. 1988. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N. Engl. J. Med. 318:1363–1368 10.1056/NEJM198805263182104 [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O., Ervasti J.M., Leveille C.J., Slaughter C.A., Sernett S.W., Campbell K.P. 1992. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 355:696–702 10.1038/355696a0 [DOI] [PubMed] [Google Scholar]
- Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P., Jr, et al. 2012. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 21:4419–4430 10.1093/hmg/dds284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. 2003. A muscleblind knockout model for myotonic dystrophy. Science. 302:1978–1980 10.1126/science.1088583 [DOI] [PubMed] [Google Scholar]
- Kaplan J.-C., Hamroun D. 2012. The 2013 version of the gene table of monogenic neuromuscular disorders (nuclear genome). Neuromuscul. Disord. 22:1108–1135 10.1016/j.nmd.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Khairallah R.J., Shi G., Sbrana F., Prosser B.L., Borroto C., Mazaitis M.J., Hoffman E.P., Mahurkar A., Sachs F., Sun Y., et al. 2012. Microtubules underlie dysfunction in duchenne muscular dystrophy. Sci. Signal. 5:ra56 10.1126/scisignal.2002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Nakahori Y., Miyake M., Matsumura K., Kondo-Iida E., Nomura Y., Segawa M., Yoshioka M., Saito K., Osawa M., et al. 1998. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 394:388–392 10.1038/28653 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y.M., Rader E.P., Crawford R.W., Iyengar N.K., Thedens D.R., Faulkner J.A., Parikh S.V., Weiss R.M., Chamberlain J.S., Moore S.A., Campbell K.P. 2008. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 456:511–515 10.1038/nature07414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobzik L., Reid M.B., Bredt D.S., Stamler J.S. 1994. Nitric oxide in skeletal muscle. Nature. 372:546–548 10.1038/372546a0 [DOI] [PubMed] [Google Scholar]
- Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. 1987. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 50:509–517 10.1016/0092-8674(87)90504-6 [DOI] [PubMed] [Google Scholar]
- Koenig M., Beggs A.H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C.R., Lindlöf M., Kaariainen H., et al. 1989. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 45:498–506 [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Mattéotti C., Arias C., Corona E.D., Nuñez N.G., Leo O., et al. 2007. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 17:611–623 10.1016/j.nmd.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. 2007. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 28:68–78 10.1016/j.molcel.2007.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Thomas G.D., Yue Y., Yang H.T., Li D., Long C., Judge L., Bostick B., Chamberlain J.S., Terjung R.L., Duan D. 2009. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J. Clin. Invest. 119:624–635 10.1172/JCI36612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Zhao J., Yue Y., Duan D. 2013. α2 and α3 helices of dystrophin R16 and R17 frame a microdomain in the α1 helix of dystrophin R17 for neuronal NOS binding. Proc. Natl. Acad. Sci. USA. 110:525–530 10.1073/pnas.1211431109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. 2002. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 32:235–236 10.1038/ng999 [DOI] [PubMed] [Google Scholar]
- Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. 2010. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 329:1650–1653 10.1126/science.1189044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R., et al. 2012. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 44:1370–1374 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon N.J., Kho A., Bacskai B.J., Perlmutter S.L., Hyman B.T., Brown R.H., Jr 2003. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 278:50466–50473 10.1074/jbc.M307247200 [DOI] [PubMed] [Google Scholar]
- Lim L.E., Duclos F., Broux O., Bourg N., Sunada Y., Allamand V., Meyer J., Richard I., Moomaw C., Slaughter C., et al. 1995. Beta-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat. Genet. 11:257–265 10.1038/ng1195-257 [DOI] [PubMed] [Google Scholar]
- Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A. 2006. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 15:2087–2097 10.1093/hmg/ddl132 [DOI] [PubMed] [Google Scholar]
- Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P. 2001. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 293:864–867 10.1126/science.1062125 [DOI] [PubMed] [Google Scholar]
- Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B., et al. 1998. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20:31–36 10.1038/1682 [DOI] [PubMed] [Google Scholar]
- Liu J., Milner D.J., Boppart M.D., Ross R.S., Kaufman S.J. 2012. β1D chain increases α7β1 integrin and laminin and protects against sarcolemmal damage in mdx mice. Hum. Mol. Genet. 21:1592–1603 10.1093/hmg/ddr596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman C., Brockington M., Torelli S., Jimenez-Mallebrera C., Kennedy C., Khalil N., Feng L., Saran R.K., Voit T., Merlini L., et al. 2003. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of alpha-dystroglycan. Hum. Mol. Genet. 12:2853–2861 10.1093/hmg/ddg307 [DOI] [PubMed] [Google Scholar]
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O’Hoy K., et al. 1992. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 255:1253–1255 10.1126/science.1546325 [DOI] [PubMed] [Google Scholar]
- Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. 2000. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 289:1769–1773 10.1126/science.289.5485.1769 [DOI] [PubMed] [Google Scholar]
- Mankodi A., Urbinati C.R., Yuan Q.P., Moxley R.T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M.S., Thornton C.A. 2001. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 10:2165–2170 10.1093/hmg/10.19.2165 [DOI] [PubMed] [Google Scholar]
- Mankodi A., Teng-Umnuay P., Krym M., Henderson D., Swanson M., Thornton C.A. 2003. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann. Neurol. 54:760–768 10.1002/ana.10763 [DOI] [PubMed] [Google Scholar]
- Manzini M.C., Tambunan D.E., Hill R.S., Yu T.W., Maynard T.M., Heinzen E.L., Shianna K.V., Stevens C.R., Partlow J.N., Barry B.J., et al. 2012. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am. J. Hum. Genet. 91:541–547 10.1016/j.ajhg.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda C., Hayashi Y.K., Ogawa M., Aoki M., Murayama K., Nishino I., Nonaka I., Arahata K., Brown R.H., Jr 2001. The sarcolemmal proteins dysferlin and caveolin-3 interact in skeletal muscle. Hum. Mol. Genet. 10:1761–1766 10.1093/hmg/10.17.1761 [DOI] [PubMed] [Google Scholar]
- Mauro A. 1961. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9:493–495 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U., Saher G., Fässler R., Bornemann A., Echtermeyer F., von der Mark H., Miosge N., Pöschl E., von der Mark K. 1997. Absence of integrin alpha 7 causes a novel form of muscular dystrophy. Nat. Genet. 17:318–323 10.1038/ng1197-318 [DOI] [PubMed] [Google Scholar]
- McNally E.M., Golbus J.R., Puckelwartz M.J. 2013. Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Invest. 123:19–26 10.1172/JCI62862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michele D.E., Barresi R., Kanagawa M., Saito F., Cohn R.D., Satz J.S., Dollar J., Nishino I., Kelley R.I., Somer H., et al. 2002. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 418:417–422 10.1038/nature00837 [DOI] [PubMed] [Google Scholar]
- Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. 2000. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 19:4439–4448 10.1093/emboj/19.17.4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti C., Sotgia F., Bruno C., Scartezzini P., Broda P., Bado M., Masetti E., Mazzocco M., Egeo A., Donati M.A., et al. 1998. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat. Genet. 18:365–368 10.1038/ng0498-365 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi H., Mitsuhashi S., Lynn-Jones T., Kawahara G., Kunkel L.M. 2013. Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 22:568–577 10.1093/hmg/dds467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A.P., Neve R.L., Colletti-Feener C., Bertelson C.J., Kurnit D.M., Kunkel L.M. 1986. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 323:646–650 10.1038/323646a0 [DOI] [PubMed] [Google Scholar]
- Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. 1988. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 2:90–95 10.1016/0888-7543(88)90113-9 [DOI] [PubMed] [Google Scholar]
- Moore S.A., Saito F., Chen J., Michele D.E., Henry M.D., Messing A., Cohn R.D., Ross-Barta S.E., Westra S., Williamson R.A., et al. 2002. Deletion of brain dystroglycan recapitulates aspects of congenital muscular dystrophy. Nature. 418:422–425 10.1038/nature00838 [DOI] [PubMed] [Google Scholar]
- Moreira E.S., Wiltshire T.J., Faulkner G., Nilforoushan A., Vainzof M., Suzuki O.T., Valle G., Reeves R., Zatz M., Passos-Bueno M.R., Jenne D.E. 2000. Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat. Genet. 24:163–166 10.1038/72822 [DOI] [PubMed] [Google Scholar]
- Muchir A., Bonne G., van der Kooi A.J., van Meegen M., Baas F., Bolhuis P.A., de Visser M., Schwartz K. 2000. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 9:1453–1459 10.1093/hmg/9.9.1453 [DOI] [PubMed] [Google Scholar]
- Nigro V., de Sá Moreira E., Piluso G., Vainzof M., Belsito A., Politano L., Puca A.A., Passos-Bueno M.R., Zatz M. 1996. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the delta-sarcoglycan gene. Nat. Genet. 14:195–198 10.1038/ng1096-195 [DOI] [PubMed] [Google Scholar]
- Noguchi S., McNally E.M., Ben Othmane K., Hagiwara Y., Mizuno Y., Yoshida M., Yamamoto H., Bönnemann C.G., Gussoni E., Denton P.H., et al. 1995. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 270:819–822 10.1126/science.270.5237.819 [DOI] [PubMed] [Google Scholar]
- Pappas C.T., Krieg P.A., Gregorio C.C. 2010. Nebulin regulates actin filament lengths by a stabilization mechanism. J. Cell Biol. 189:859–870 10.1083/jcb.201001043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. 1993. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. USA. 90:3710–3714 10.1073/pnas.90.8.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips A.V., Timchenko L.T., Cooper T.A. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 280:737–741 10.1126/science.280.5364.737 [DOI] [PubMed] [Google Scholar]
- Poon E., Howman E.V., Newey S.E., Davies K.E. 2002. Association of syncoilin and desmin: linking intermediate filament proteins to the dystrophin-associated protein complex. J. Biol. Chem. 277:3433–3439 10.1074/jbc.M105273200 [DOI] [PubMed] [Google Scholar]
- Prins K.W., Humston J.L., Mehta A., Tate V., Ralston E., Ervasti J.M. 2009. Dystrophin is a microtubule-associated protein. J. Cell Biol. 186:363–369 10.1083/jcb.200905048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezniczek G.A., Konieczny P., Nikolic B., Reipert S., Schneller D., Abrahamsberg C., Davies K.E., Winder S.J., Wiche G. 2007. Plectin 1f scaffolding at the sarcolemma of dystrophic (mdx) muscle fibers through multiple interactions with β-dystroglycan. J. Cell Biol. 176:965–977 10.1083/jcb.200604179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. 1995. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 81:27–40 10.1016/0092-8674(95)90368-2 [DOI] [PubMed] [Google Scholar]
- Roberds S.L., Leturcq F., Allamand V., Piccolo F., Jeanpierre M., Anderson R.D., Lim L.E., Lee J.C., Tomé F.M., Romero N.B., et al. 1994. Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell. 78:625–633 10.1016/0092-8674(94)90527-4 [DOI] [PubMed] [Google Scholar]
- Roberts R., Timchenko N.A., Miller J.W., Reddy S., Caskey C.T., Swanson M.S., Timchenko L.T. 1997. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc. Natl. Acad. Sci. USA. 94:13221–13226 10.1073/pnas.94.24.13221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu U., Strähle U. 2012. In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell. 22:515–529 10.1016/j.devcel.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Roscioli T., Kamsteeg E.J., Buysse K., Maystadt I., van Reeuwijk J., van den Elzen C., van Beusekom E., Riemersma M., Pfundt R., Vissers L.E., et al. 2012. Mutations in ISPD cause Walker-Warburg syndrome and defective glycosylation of α-dystroglycan. Nat. Genet. 44:581–585 10.1038/ng.2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova I.N., Patel J.R., Ervasti J.M. 2000. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 150:1209–1214 10.1083/jcb.150.5.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmikangas P., van der Ven P.F., Lalowski M., Taivainen A., Zhao F., Suila H., Schröder R., Lappalainen P., Fürst D.O., Carpén O. 2003. Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum. Mol. Genet. 12:189–203 10.1093/hmg/ddg020 [DOI] [PubMed] [Google Scholar]
- Sander M., Chavoshan B., Harris S.A., Iannaccone S.T., Stull J.T., Thomas G.D., Victor R.G. 2000. Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 97:13818–13823 10.1073/pnas.250379497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarparanta J., Jonson P.H., Golzio C., Sandell S., Luque H., Screen M., McDonald K., Stajich J.M., Mahjneh I., Vihola A., et al. 2012. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet. 44:450–455 10.1038/ng.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R.S., Philips A.V., Cooper T.A. 2001. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29:40–47 10.1038/ng704 [DOI] [PubMed] [Google Scholar]
- Smith F.J., Eady R.A., Leigh I.M., McMillan J.R., Rugg E.L., Kelsell D.P., Bryant S.P., Spurr N.K., Geddes J.F., Kirtschig G., et al. 1996. Plectin deficiency results in muscular dystrophy with epidermolysis bullosa. Nat. Genet. 13:450–457 10.1038/ng0896-450 [DOI] [PubMed] [Google Scholar]
- Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., Miller D.G. 2010. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6:e1001181 10.1371/journal.pgen.1001181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens E., Carss K.J., Cirak S., Foley A.R., Torelli S., Willer T., Tambunan D.E., Yau S., Brodd L., Sewry C.A., et al. 2013. Mutations in B3GALNT2 cause congenital muscular dystrophy and hypoglycosylation of α-dystroglycan. Am. J. Hum. Genet. 92:354–365 10.1016/j.ajhg.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. 1995. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128:995–1002 10.1083/jcb.128.6.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Schreiber R., Kunzelmann K. 2012. Anoctamins are a family of Ca2+-activated Cl- channels. J. Cell Sci. 125:4991–4998 10.1242/jcs.109553 [DOI] [PubMed] [Google Scholar]
- Udd B. 2009. Genetics and pathogenesis of distal muscular dystrophies. Adv. Exp. Med. Biol. 652:23–38 10.1007/978-90-481-2813-6_3 [DOI] [PubMed] [Google Scholar]
- van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. 1993. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 2:2037–2042 10.1093/hmg/2.12.2037 [DOI] [PubMed] [Google Scholar]
- van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. 2003. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 35:315–317 10.1038/ng1262 [DOI] [PubMed] [Google Scholar]
- van Reeuwijk J., Janssen M., van den Elzen C., Beltran-Valero de Bernabé D., Sabatelli P., Merlini L., Boon M., Scheffer H., Brockington M., Muntoni F., et al. 2005. POMT2 mutations cause alpha-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J. Med. Genet. 42:907–912 10.1136/jmg.2005.031963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallgren-Pettersson C., Sewry C.A., Nowak K.J., Laing N.G. 2011. Nemaline myopathies. Semin. Pediatr. Neurol. 18:230–238 10.1016/j.spen.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Weiler T., Bashir R., Anderson L.V., Davison K., Moss J.A., Britton S., Nylen E., Keers S., Vafiadaki E., Greenberg C.R., et al. 1999. Identical mutation in patients with limb girdle muscular dystrophy type 2B or Miyoshi myopathy suggests a role for modifier gene(s). Hum. Mol. Genet. 8:871–877 10.1093/hmg/8.5.871 [DOI] [PubMed] [Google Scholar]
- Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. 1992. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 2:26–30 10.1038/ng0992-26 [DOI] [PubMed] [Google Scholar]
- Willer T., Lee H., Lommel M., Yoshida-Moriguchi T., de Bernabe D.B., Venzke D., Cirak S., Schachter H., Vajsar J., Voit T., et al. 2012. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat. Genet. 44:575–580 10.1038/ng.2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Ozawa E. 1990. Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. 108:748–752 [DOI] [PubMed] [Google Scholar]
- Yoshida M., Suzuki A., Yamamoto H., Noguchi S., Mizuno Y., Ozawa E. 1994. Dissociation of the complex of dystrophin and its associated proteins into several unique groups by n-octyl beta-D-glucoside. Eur. J. Biochem. 222:1055–1061 10.1111/j.1432-1033.1994.tb18958.x [DOI] [PubMed] [Google Scholar]
- Yoshida A., Kobayashi K., Manya H., Taniguchi K., Kano H., Mizuno M., Inazu T., Mitsuhashi H., Takahashi S., Takeuchi M., et al. 2001. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev. Cell. 1:717–724 10.1016/S1534-5807(01)00070-3 [DOI] [PubMed] [Google Scholar]