Lysosomal degradation and recycling of sequestered autophagosome content is crucial to maintain proper functioning of the fly nervous system.
Abstract
During autophagy, phagophores capture portions of cytoplasm and form double-membrane autophagosomes to deliver cargo for lysosomal degradation. How autophagosomes gain competence to fuse with late endosomes and lysosomes is not known. In this paper, we show that Syntaxin17 is recruited to the outer membrane of autophagosomes to mediate fusion through its interactions with ubisnap (SNAP-29) and VAMP7 in Drosophila melanogaster. Loss of these genes results in accumulation of autophagosomes and a block of autolysosomal degradation during basal, starvation-induced, and developmental autophagy. Viable Syntaxin17 mutant adults show large-scale accumulation of autophagosomes in neurons, severe locomotion defects, and premature death. These mutant phenotypes cannot be rescued by neuron-specific inhibition of caspases, suggesting that caspase activation and cell death do not play a major role in brain dysfunction. Our findings reveal the molecular mechanism underlying autophagosomal fusion events and show that lysosomal degradation and recycling of sequestered autophagosome content is crucial to maintain proper functioning of the nervous system.
Introduction
Autophagy ensures degradation and recycling of intracellular material, including macromolecules and even whole organelles in eukaryotic cells. Autophagy has a role in a wide range of physiological and pathological settings, such as cellular adaptation to stress, starvation, and protection from aging, cancer, neurodegeneration, and invading pathogens (Mizushima et al., 2008). Cytoplasmic cargo sequestered into double-membrane autophagosomes by phagophore cisterns (also called isolation membranes) is transported to lysosomes for degradation and reuse to support biosynthetic and energy production pathways (Tooze and Yoshimori, 2010). Several different sources such as ER, Golgi, mitochondria, endosomes, and plasma membrane have been suggested to supply membranes for the phagophore (Axe et al., 2008; Tooze and Yoshimori, 2010). Regardless of the membrane source, all autophagosomes fuse with late endosomes and lysosomes to generate amphisomes and autolysosomes, respectively, whereas phagophores do not. Therefore, autophagosomes must gain competence for specific fusion through acquiring the required molecular machinery during their maturation process.
Vesicle fusion events are usually mediated by the action of different SNARE proteins that assemble into complexes in a combinatorial fashion. A SNARE complex contains four parallel SNARE helix bundles, supplied by a Qa SNARE and either separate Qb and Qc SNAREs or a SNAP protein that contains both Qb and Qc SNARE domains, all located on the membrane of the first vesicle, and an R SNARE anchored to the second vesicle (Hong, 2005). Yeast SNARE proteins Vam3, Vam7, Vti1, and Ykt6 have all been suggested to play a role in fusion of autophagosomes with the vacuole, the equivalent of metazoan lysosomes (Dilcher et al., 2001; Ishihara et al., 2001; Ohashi and Munro, 2010). Vam3 and Vam7 have no clear homologues in metazoa. Moreover, we and others showed that autophagosomes accumulate if formation of late endosomes is blocked in Drosophila melanogaster and mammals, indicating that amphisomes play a critical role in autophagosome clearance in these cells, unlike in yeast (Filimonenko et al., 2007; Rusten et al., 2007; Juhász et al., 2008). Mammalian VAMP7, VAMP8, and Vti1b were all suggested to be involved in autophagosomal fusion events (Fader et al., 2009; Furuta et al., 2010). The accumulation of both autophagosomes and autolysosomes in Vti1b knockout mice indicates that this gene product is likely to function in later steps of autophagy (Atlashkin et al., 2003). Ykt6 and VAMP7 have also been implicated in the formation of phagophores in yeast and mammalian cells, respectively (Moreau et al., 2011; Nair et al., 2011).
In a screen for SNAREs involved in autophagy, we identified Syx17 (Syntaxin17, Qa), the SNAP-29 homologue ubisnap (usnp, Qbc), and VAMP7 (CG1599, R) as required for autophagosome fusion events. We showed that Syx17 is recruited to the outer membrane of autophagosomes to acquire fusion competence, and loss of Syx17 results in neuronal dysfunction, locomotion defects, and early death of adult flies.
Results and discussion
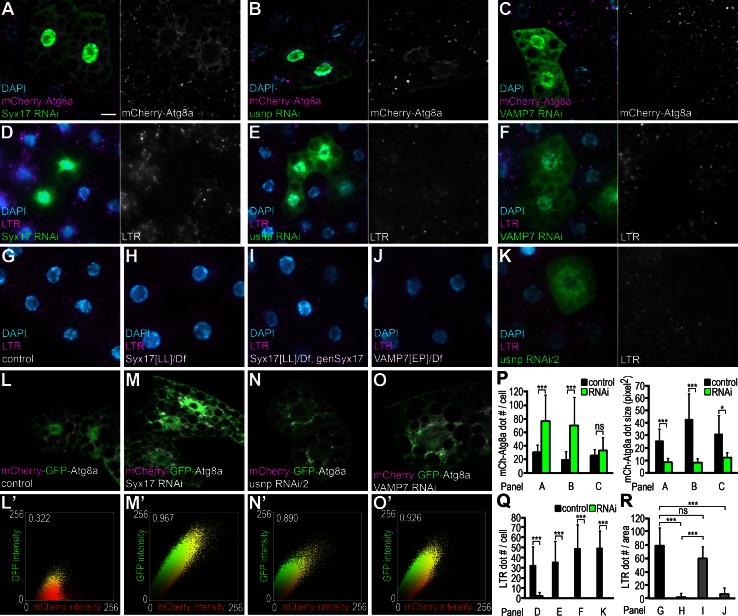
We performed a genetic screen for SNARE proteins involved in starvation-induced autophagy, by generating GFP-marked RNAi cell clones in mCherry-Atg8a–expressing Drosophila fat bodies. The mCherry-Atg8a reporter is bound to phagophores and autophagosomes through a lipid anchor on its C terminus. In addition, mCherry-Atg8a attached to the inner membrane of autophagosomes is selectively transported to autolysosomes, which are prominently labeled by this reporter as a result of large-scale accumulation of the protease- and low pH–resistant mCherry tag inside lysosomes (Kimura et al., 2007). Syx17, usnp, and VAMP7 knockdown cells showed a similar and very characteristic phenotype: small mCherry-Atg8a dots accumulated in the perinuclear region, unlike the evenly distributed, bigger, and brighter dots observed in neighboring control cells (Fig. 1, A–C and P; and Table S1 shows the results of our screen).
Figure 1.
Syx17, usnp, and VAMP7 are required for autophagy in Drosophila. (A–C) Syx17 (A), usnp (B), or VAMP7 (C) depletion in GFP-marked fat cell clones leads to formation of numerous small, mostly perinuclear mCherry-Atg8a dots, unlike the larger, brighter, evenly distributed punctae in surrounding control cells of starved larvae. (D–F) Knockdown of Syx17 (D), usnp (E), or VAMP7 (F) in LAMP1-GFP–marked cells blocks starvation-induced punctate LysoTracker red (LTR) staining. (G–J) Starvation leads to formation of LTR dots in control larvae (G). No LTR punctae form in starved Syx17 mutants (H), whereas LTR staining is restored in mutants expressing a Syx17 transgene (I). No LTR dots appear in VAMP7 mutants (J). (K) Expression of a second, independent usnp RNAi transgene also blocks LTR puncta formation. (L–O) Tandem-tagged mCherry-GFP-Atg8a is transported to autolysosomes that appear as mCherry-positive puncta (magenta in L) in control cells of starved larvae. Silencing of Syx17 (M), usnp (N), or VAMP7 (O) results in the formation of numerous dots positive for both mCherry and GFP (white). Dot plots in L′–O′ show intensity and colocalization profiles of mCherry and GFP dots. Pearson correlation coefficients shown at the top indicate strong colocalization of GFP and mCherry in M′–O′. (P–R) Quantification of data presented in A–C (P), D–F and K (Q), and G–J (R); n = 10 for all genotypes. mCh, mCherry. Error bars mark SDs. *, P < 0.05; ***, P < 0.001. Bar, 20 µm.
Depletion of Syx17, usnp, or VAMP7 resulted in a complete block of starvation-induced punctate LysoTracker staining, a dye commonly used to label autolysosomes in Drosophila fat body (Fig. 1, D–F and Q). Starvation-induced LysoTracker red (LTR) staining was also impaired in Syx17[LL] and VAMP7[EP] mutant larvae that carry transposon insertions in the coding sequences of these genes 22 and 24 nucleotides downstream of the translation start site, respectively (Fig. 1, G, H, J, and R), similar to an independent RNAi line targeting usnp (Fig. 1, K and Q). Transgenic expression of Syx17 restored punctate LysoTracker staining in Syx17 mutants, confirming that loss of Syx17 caused the mutant phenotype (Fig. 1, I and R). Tandem-tagged mCherry-GFP-Atg8a is commonly used for analyzing autophagic flux (Kimura et al., 2007). This reporter is normally transported to autolysosomes where GFP is rapidly quenched, but mCherry persists in control cells (Fig. 1, L and L′). In contrast, structures positive for both GFP and mCherry accumulated in the perinuclear region of Syx17, usnp, and VAMP7 RNAi cells, indicating a block of GFP inactivation (Fig. 1, M–O′). Developmental autophagy of the fat body at the onset of metamorphosis was also impaired in Syx17, usnp, and VAMP7 RNAi cells (Fig. S1, A–F, M, and N).
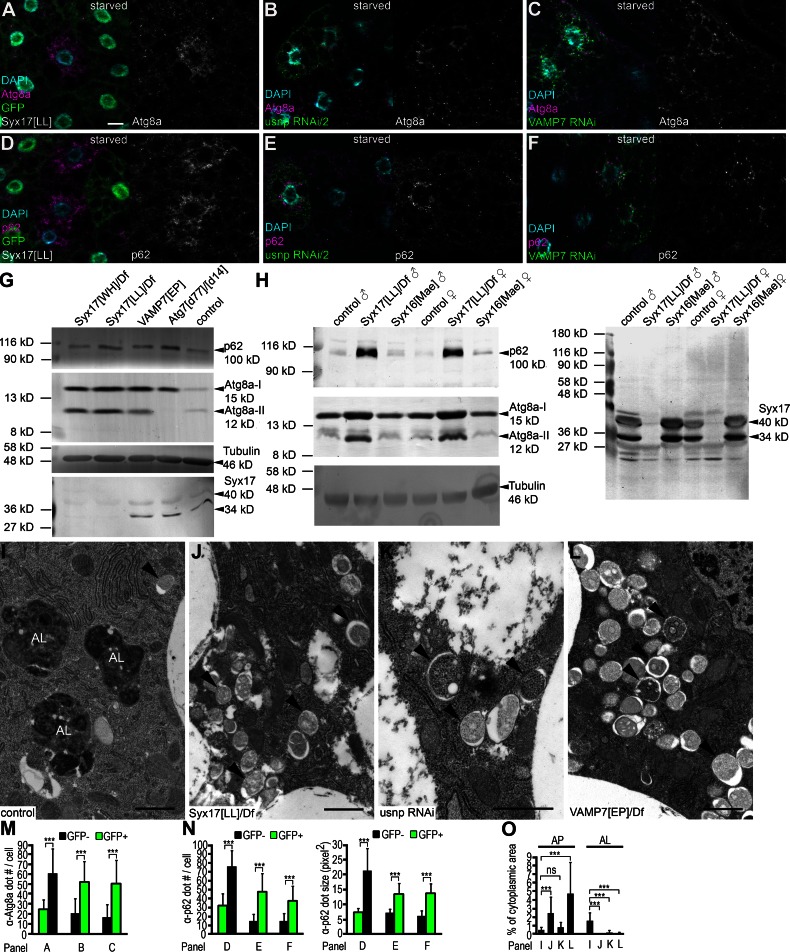
Immunostainings revealed accumulation of endogenous Atg8a-positive dots representing autophagosomes in Syx17, usnp, and VAMP7 loss-of-function cells compared with adjacent control fat body cells, both in starved (Fig. 2, A–C and M) and well-fed larvae (Fig. S1, G–I and O). Levels of the specific cargo p62 inversely correlate with autophagic degradation in flies and mammals (Bjørkøy et al., 2009; Pircs et al., 2012). p62 aggregates accumulated in Syx17, usnp, and VAMP7 loss-of-function cells both in starved (Fig. 2, D–F and N) and well-fed larvae (Fig. S1, J–L and P), indicating impaired autophagic breakdown. Western blots also revealed increased p62 and autophagosome-associated, lipidated Atg8a-II levels in starved Syx17 and VAMP7 mutant larvae compared with controls (Fig. 2 G). Larvae expressing usnp RNAi in all cells showed similar accumulation of Atg8a-II and p62 (Fig. S1 Q). Syx17 mutants are viable despite profound autophagy defects, similar to Atg7- and Atg8a-null mutants (Juhász et al., 2007; Simonsen et al., 2008). Levels of p62 and lipidated Atg8a-II also increased in well-fed Syx17 mutant adult flies compared with controls (Fig. 2 H). Western blots using our novel polyclonal rat and guinea pig anti-Syx17 antisera detected two bands near the predicted molecular weight of this protein, both of which were missing from Syx17 mutants (Fig. 2, G and H; and Fig. S1 R). Faint bands were visible in the independent Syx17 transposon insertion mutant line Syx17[WH] (Fig. 2 G). All these results indicated that autophagosomes cannot progress to autolysosomes in the absence of Syx17, usnp, and VAMP7. Ultrastructural analysis indeed revealed accumulation of double-membrane autophagosomes and impaired generation of autolysosomes in starved Syx17 mutant, usnp RNAi, and VAMP7 mutant fat body cells (Fig. 2, I–L and O).
Figure 2.
Autophagosomes accumulate upon loss of Syx17, usnp, or VAMP7. (A–C) Increased numbers of Atg8a-positive autophagosomes are seen in Syx17 mutant (A), usnp (B), and VAMP7 (C) RNAi cells in starved larvae. (D–F) p62 aggregates accumulate in Syx17 mutant (D), usnp (E), and VAMP7 (F) RNAi cells in starved larvae. Note that Syx17 mutant cells are marked by lack of GFP expression in A and D, whereas RNAi cells express LAMP1-GFP in B, C, E, and F. (G) Western blots show increased autophagosome-associated, lipidated Atg8a-II levels in starved Syx17 and VAMP7 mutant larvae compared with controls. Accumulation of p62 in Syx17 and VAMP7 mutants is comparable to Atg7 mutants that are unable to lipidate Atg8a. Both the 34- and 40-kD isoforms of Syx17 disappear in Syx17[LL] mutants based on rat anti-Syx17 immunoblots, whereas faint bands are visible in Syx17[WH] mutant larvae. (H) Atg8a-II and p62 accumulate in well-fed Syx17 mutant adults compared with controls or Syx16 mutants (an additional control). Both Syx17-specific bands are missing from Syx17 mutant adults. (I–L) Numerous large autolysosomes (AL) and few double-membrane autophagosomes (arrowheads) are visible in ultrastructural images of control fat body cells from starved animals (I). Loss of Syx17 (J), usnp (K), or VAMP7 (L) function leads to accumulation of autophagosomes and lack of autolysosomes. (M–O) Quantification of data presented in A–C (M), D–F (N), and I–L (O). AP, autophagosome. n = 10 for A–F, and n = 4 for I–L. Error bars mark SDs. ***, P < 0.001. Bars: (A–F) 20 µm; (I–L) 1 µm.
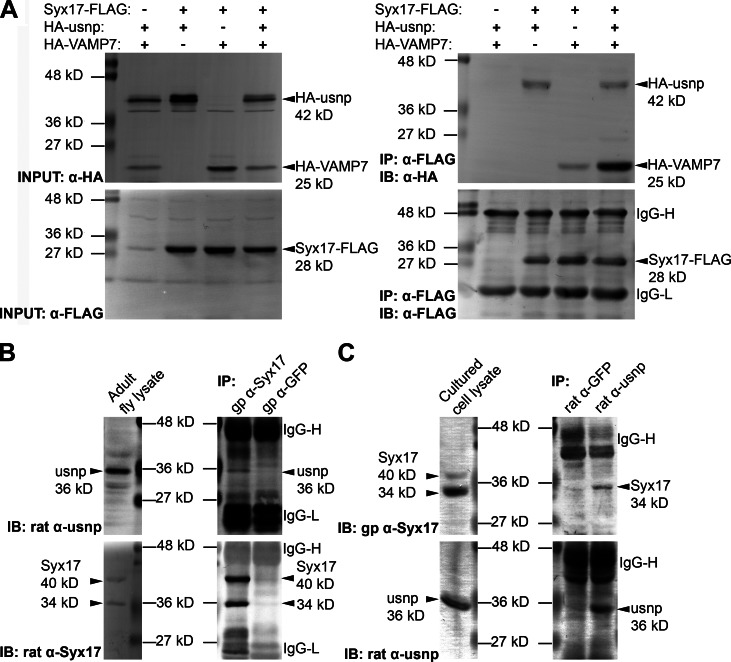
Our loss-of-function data suggested that Syx17, usnp, and VAMP7 function as part of the same SNARE complex. Human STX17 (Syntaxin17) was previously shown to bind to SNAP-29, but the significance of this interaction remained unknown (Steegmaier et al., 1998). We found that both HA-tagged usnp and VAMP7 coimmunoprecipitated with FLAG-tagged Syx17 in cultured Drosophila cells (Fig. 3 A). The amount of VAMP7 bound to beads increased when all three proteins were coexpressed, suggesting that Syx17 and usnp together bind more efficiently to VAMP7 than Syx17 alone (Fig. 3 A). We also produced anti-usnp antisera to detect interactions of endogenous proteins. These polyclonal rat antibodies specifically recognized endogenous usnp as a single 36-kD band, which was reduced by systemic expression of usnp RNAi transgenes in larvae (Fig. S1 Q). We could readily detect endogenous usnp in anti-Syx17 immunoprecipitates from lysates of adult flies (Fig. 3 B). Vice versa, endogenous Syx17 could be immunoprecipitated with anti-usnp antibodies from cultured cell lysates (Fig. 3 C).
Figure 3.
Syx17 binds to usnp and VAMP7. (A) Coimmunoprecipitation shows that Syx17-FLAG binds to both HA-usnp and HA-VAMP7 in cultured Drosophila cells. Note that usnp facilitates the interaction of overexpressed VAMP7 with Syx17 (both lacking transmembrane domains). (B) Endogenous usnp coimmunoprecipitates with endogenous Syx17. (C) Endogenous Syx17 coimmunoprecipitates with endogenous usnp. gp, guinea pig; IP, immunoprecipitation; IB, immunoblotting.
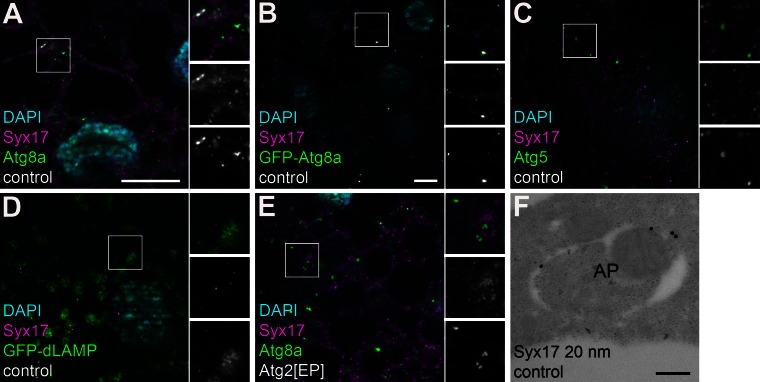
These results established the identity of autophagosome fusion-specific SNARE complex subunits but did not reveal which ones are present on autophagosomes. As ubisnap/SNAP-29 has both Qb and Qc SNARE domains but no transmembrane domain or lipidation site, it is likely recruited to its target membrane through interaction with Syx17, a Qa SNARE. Drosophila Syx17 localized to the ER (Fig. S2, A and B), similar to human STX17 (Hong, 2005). Drosophila VAMP7 is predicted to be present in late endosomes and lysosomes (Hong, 2005). We thus reasoned that Syx17 could also localize to autophagosomes to recruit usnp, and they mediate fusion of autophagosomes and endo/lysosomes by binding to VAMP7 located in the membranes of the degradative organelles. Endogenous Syx17 was indeed detected in 30.5% (61/200) of endogenous Atg8a-positive autophagosomes in starved animals and also colocalized with 43% (86/200) of GFP-Atg8a dots (Fig. 4, A and B). In contrast, Syx17 essentially did not colocalize with the phagophore marker Atg5 (2.5%, 5/200) or GFP-dLAMP–positive late endosomes and lysosomes (6.8%, 16/236; Fig. 4, C and D). Loss of Atg2 results in accumulation of stalled phagophores that already contain Atg8 homologues in worms and mammals (Lu et al., 2011; Velikkakath et al., 2012). Accordingly, Atg8a and Syx17 did not colocalize in Atg2 mutants (2.8%, 9/322; Fig. 4 E). Finally, immuno-EM showed the presence of endogenous Syx17 in the outer membrane of autophagosomes (Fig. 4 F). Thus, Syx17 is recruited to autophagosomes to promote their fusion with late endosomes and lysosomes through its interactions with usnp and VAMP7.
Figure 4.
Syx17 is recruited to completed autophagosomes. (A and B) Endogenous Syx17 colocalizes with both endogenous (A) and GFP-tagged (B) Atg8a in starved fat body cells. (C and D) No colocalization is observed between Syx17 and the phagophore marker Atg5 (C) or late endosomes and lysosomes, labeled by GFP-dLAMP (D). (E) Syx17 and Atg8a do not colocalize in Atg2 mutants that accumulate Atg8a-positive stalled phagophores. Insets show merged images (top), Syx17 channels (middle), and relevant green channels (bottom) enlarged from boxed areas in A–E. (F) Immunogold labeling reveals that Syx17 is associated with the outer membrane of autophagosomes (AP). Bars: (A and C–E) 20 µm; (B) 20 µm; (F) 100 nm.
Two papers relying on siRNA depletion of human STX17 were published while our manuscript was under review (Itakura et al., 2012; Hamasaki et al., 2013). Itakura et al. (2012) found that STX17, SNAP-29, and VAMP8 cooperate to mediate fusion of autophagosomes with late endosomes and lysosomes in mammalian cells. Also, GFP-STX17 localized to the outer membrane of autophagosomes. In contrast, Hamasaki et al. (2013) suggested that no autophagosomes form in STX17 siRNA–treated cells. Our results support the findings of the former work with the exception of VAMP8, but flies appear to have only VAMP7 and lack the closely related VAMP8. According to Itakura et al. (2012), a pool of free cytosolic STX17 may be directly inserted into the outer membrane of autophagosomes. This is likely mediated by its two glycine-rich transmembrane domains and the C-terminal region (Itakura et al., 2012). Drosophila Syx17 shows a similar domain structure to human STX17, so it may also be transported to autophagosomes by this pathway (Fig. S2 C).
Depletion of syntaxin5 was recently shown to cause defects in autophagic protein degradation caused by impaired trafficking of hydrolases from ER to Golgi and thus lysosomes (Renna et al., 2011). Importantly, syntaxin5 seemed to be dispensable for the fusion of autophagosomes with endosomes and lysosomes, indicating that it functions at a later step during autophagy than Syx17.
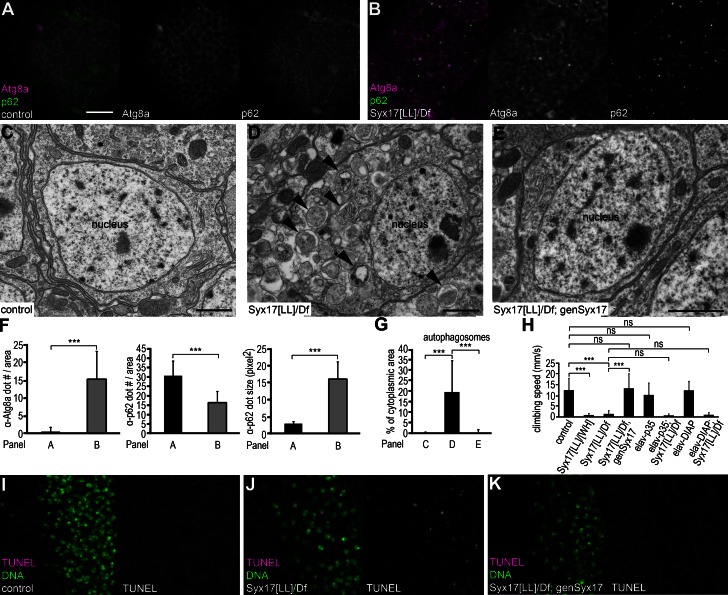
Continuous basal autophagy is critical for the homeostasis of quiescent, nondividing cells, such as neurons. We and others reported earlier that autophagy deficiency results in accumulation of protein aggregates and progressive neurodegeneration in flies and mice (Hara et al., 2006; Komatsu et al., 2006; Juhász et al., 2007). Syx17 mutant adults were viable but unable to fly or climb properly, and all died within 4 d of eclosion (n = 351). We detected large-scale accumulation of Atg8a-positive autophagosomes and p62 aggregates in Syx17 mutant brains (Fig. 5, A, B, and F). Neurons of well-fed Syx17 mutant adult flies contained vast numbers of autophagosomes: on average, 20% of total cytoplasm was enclosed within them (Fig. 5, D and G). In contrast, autophagic structures were rarely observed in ultrastructural images of control brains or mutants rescued by transgenic expression of Syx17 (Fig. 5, C, E, and G). Altogether 86% of cells (201/234, n = 4) contained autophagosomes in ultrastructural sections of mutant brains, in contrast with 1% of neurons in control flies (3/270, n = 4) or 2% in rescued flies (6/327, n = 4).
Figure 5.
Impaired autophagosome maturation leads to locomotion defects in 2-d-old adult flies. (A and B) Atg8a-positive autophagosomes and p62 aggregates accumulate in Syx17 mutant brains (B) compared with similarly aged controls (A). (C–E) No autophagosomes are found by EM in neurons of control adult flies (C). Large-scale accumulation of autophagosomes (arrowheads) is obvious in Syx17 mutant neurons (D). Autophagosome accumulation is rescued in Syx17 mutant neurons by transgenic expression of Syx17 (E). (F and G) Quantification of data presented in A and B (F) and C–E (G); n = 9 for A and B, and n = 4 for C–E. Error bars mark SDs. ***, P < 0.001. (H) Syx17 mutant adults perform poor compared with controls in climbing tests. Expression of Syx17 in mutants rescues locomotion defects. Neuron-specific expression of caspase inhibitors p35 or DIAP1 have no influence on climbing performance of adult flies, and these do not rescue the defects of Syx17 mutant adults. n = 90 for all genotypes. Error bars mark SDs. ***, P < 0.001. (I–K) TUNEL assays reveal apoptotic DNA fragmentation in Syx17 mutant brains (J) compared with controls (I), which is rescued by expression of Syx17 (K). Bars: (A, B, and I–K) 20 µm; (C–E) 1 µm.
2-d-old Syx17 mutants performed poorly in a climbing test (Fig. 5 H), an established measure of nervous system function (Juhász et al., 2007). Again, transgenic expression of Syx17 rescued this locomotion defect (Fig. 5 H). We showed earlier that loss of Atg7 leads to widespread apoptosis in brains of 30-d-old mutant adults (Juhász et al., 2007). On average, 25% of neurons were positive for active caspase 3 in the absence of Syx17 in mutant brains, unlike in similarly aged controls (Fig. S2, D–F). TUNEL assays showed that 20% of mutant cells contained fragmented DNA indicating cell death, whereas practically no TUNEL-positive neurons were identified in control or rescued 2-d-old adult brains (Fig. 5, I–K; and Fig. S2 G, quantification). This seemed specific to neurons, as no TUNEL-positive cells were detected in muscles of mutant adults (Fig. S2, H and I). Transgenic expression of the effector caspase inhibitor p35 in mutant neurons suppressed TUNEL staining (Fig. S2, G and J), but 85% of neurons (202/238, n = 3) still contained autophagosomes in brain sections similar to Syx17 mutants (Fig. S2 K). The potential contribution of caspase activation to mutant phenotypes was tested in epistasis analyses. Neuron-specific expression of p35 or the pancaspase inhibitor DIAP1 (Drosophila inhibitor of apoptosis protein 1) failed to rescue locomotion defects in 2-d-old Syx17 mutant adults (Fig. 5 H). In addition, although transgenic expression of Syx17 fully rescued the viability of Syx17 mutant adults (174/175 adults alive on day 4), all mutant flies expressing either p35 or DIAP1 still died within 4 d of eclosion (n = 90 and 105, respectively). These results suggest that neuronal caspase activation may not be a major cause for the locomotion defects and premature death of Syx17 mutant adult flies.
Collectively, we showed that a complex of SNARE proteins Syx17, usnp, and VAMP7 is required for autophagosome clearance and that autophagosomes gain competence to fuse with late endosomes and lysosomes by recruiting Syx17. The locomotion defects and early death of viable Syx17 mutants indicate that lysosomal degradation and recycling of sequestered autophagosomal cargo is crucial to maintain organismal health and proper brain functions in Drosophila.
Materials and methods
Fly strains and genetics
Flies were maintained on standard yeast/cornmeal/agar media. In climbing tests, a total of 90 adults per genotype were recorded and scored in cohorts of three to six flies, after tapping them down in a plastic graduated cylinder (Juhász et al., 2007). In clonal analyses, RNAi cells were generated spontaneously in larvae carrying hs-Flp; upstream activation sequence (UAS)-Dcr2; Actin>CD2>Gal4 UAS-RNAi (and UAS-GFP or UAS-LAMP1-GFP as a knockdown cell marker), and mutant cell clones (marked by lack of GFP expression) were generated by heat shocking 2–4-h embryos of the genotype hs-Flp; ubi-GFP FRT2A/Syx17[LL] FRT2A in a 38°C water bath for 1 h (Juhász et al., 2007, 2008; Pircs et al., 2012). Expression of mCherry-Atg8a was driven by a fat body–specific r4 promoter in our screen (transgenic flies were provided by T. Neufeld, University of Minnesota, Minneapolis, MN; Pircs et al., 2012). We used the GFP knockin line dLAMP[CPTI001775] (Drosophila Genetic Resource Center) to label lysosomes, w[1118] as control, UAS-GFP-KDEL and Pdi[G00198] as ER reporters (Bloomington Drosophila Stock Center), Atg2[EP3697] (Berry and Baehrecke, 2007), Atg7[d77]/Atg7[d14] (Juhász et al., 2007) mutants, and SNARE loss-of-function strains listed in Table S1. Knockdown of usnp was induced by Actin-Gal4 for Western blots and collagen-Gal4 for EM in L3 stage larvae, and overexpression of UAS-p35 or UAS-DIAP1 in adult neurons was mediated by elav-Gal4 (all obtained from Bloomington Drosophila Stock Center).
Histology and imaging
LTR stainings were performed by incubating dissected L3 stage fat bodies in 100 nM LTR (Invitrogen) for 5 min. For immunofluorescent labeling, bisected larvae were fixed overnight in 3.7% paraformaldehyde at 4°C and blocked in PBS with 0.1% Triton X-100, 0.05% sodium deoxycholate, and 3% goat serum for 3 h followed by overnight incubations with primary and secondary antibodies in blocking buffer at 4°C (Juhász et al., 2008; Pircs et al., 2012). We used chicken anti-GFP (1:1,500; Invitrogen), rabbit anti-Atg5 (1:100; Sigma-Aldrich), rabbit anti-Atg8a (1:500; provided by K. Kohler, Eidgenössische Technische Hochschule Zürich, Zurich, Switzerland; Barth et al., 2011), rabbit anti-p62 (1:2,000; Pircs et al., 2012), rabbit anti–active caspase 3 (1:300; Cell Signaling Technology), rat anti-Atg8a (1:300), and rat anti-Syx17 (1:300; this study) primary and Alexa Fluor 488 anti–chicken, Alexa Fluor 488 anti–rabbit, Alexa Fluor 568 anti–rat, and Alexa Fluor 647 anti–rabbit (all 1:1,500; Invitrogen) secondary antibodies. For TUNEL stainings, adult heads and half-thoraces were fixed in 3.7% paraformaldehyde overnight at 4°C and embedded into paraffin following standard protocols. Sections were processed using In Situ Cell Death Detection Kit, tetramethylrhodamine red (Roche) with SYTOX green DNA stain (Juhász et al., 2007). Images were obtained on a microscope (Axio Imager.M2; Carl Zeiss) equipped with a grid confocal unit (ApoTome.2; Carl Zeiss) at room temperature, using Plan-Neofluar 20×, 0.5 NA (air), 40×, 0.75 NA (air), and 100×, 1.3 NA (oil) objectives, a camera (AxioCam MRm; Carl Zeiss), and AxioVision software (Carl Zeiss). Microscope settings were identical for experiments of the same kind. Primary images were processed in AxioVision and Photoshop (Adobe) to produce final figures. Note that Alexa Fluor 568 or 647 channels are pseudocolored magenta.
Image analysis and statistics
Colocalization of puncta was counted manually on a computer situated in a darkroom, only taking into account overlapping structures with a similar shape and size in relevant fluorescent channels. For statistical analyses, original, unmodified images were imported in ImageJ (National Institutes of Health), and the intensity threshold for the relevant channel was set so that as many dots as possible were selected without the merging of clearly separable adjacent dots. In clonal analyses, mutant or RNAi cells were then manually encircled based on the GFP channel, and the number and size of dots in the relevant channel within that area were recorded. Adjacent control cells were selected randomly from the same image and analyzed. When comparing images from different control and mutant animals, a 300 × 300–pixel area was randomly selected from each image and analyzed as in clonal experiments. Images taken from the number of animals (n) per genotype as indicated in figure legends were evaluated for all experiments. Data were then imported into SPSS Statistics (IBM) and tested for normality of data distribution, and then, p-values were calculated with the appropriate statistical tests: two-tailed, two-sample, unequal variance t test or U test for pairwise comparison of normal or nonnormal distribution data, respectively, and analysis of variance or nonparametric Kruskal-Wallis tests for multiple comparisons of normal or nonnormal distribution data, respectively. For autophagic flux measurements with mCherry-GFP-Atg8a, primary images were imported into ImageJ and analyzed automatically using the colocalization plugin with the intensity correlation tool. Clones from 5–10 animals per genotype were evaluated with similar results, and one representative dot plot is shown in Fig. 1 (L′–O′) for each.
Molecular cloning and generation of polyclonal antibodies
The genomic region containing Drosophila Syx17 was amplified using primers 5′-GGCGCGCCGCATGCGGCCGCTTAGACGTAAGCGCACCACCGC-3′ and 5′-GCATGCTAGCGGCCGCGTGACCAACGAGAACACGGGC-3′ and cloned into pCasper5 as an SphI fragment. Transgenic flies genSyx17 were generated following standard procedures (BestGene, Inc.). Syx17 coding sequences were amplified by PCR using primers 5′-ATGACGCGCGATGAGAAACTGCC-3′ and 5′-ATCCTTCTGGCTTCTCTTTTAGCTCCAGTCTCTC-3′, phosphorylated, blunt cloned into XmnI–EcoRV-digested dephosphorylated pENTR1A (Invitrogen), and subsequently recombined into pDEST17 (Invitrogen). N-terminally His-tagged protein was expressed in the Escherichia coli Rosetta strain (EMD Millipore) and purified using Ni-agarose beads (QIAGEN). Recombinant protein was used to immunize rats and guinea pigs following standard procedures with Freund’s adjuvants (Sigma-Aldrich). Syx17 pENTR1A was recombined into pTWF (Drosophila Genomics Resource Center) to yield UAS-Syx17-FLAG, in which 3×FLAG replaces the two transmembrane domains and the extreme C-terminal region of Syx17. usnp and VAMP7 coding sequences were amplified using primers 5′-GTAGCGGCCGCGGGTACCGGAATGGCCCATAACTACCTGCAGC-3′/5′-TATGGTACCGCGGCCGCTACTTCTTCAGAAGCTTGCTCATGTCC-3′ and 5′-GTAGCGGCCGCGGGTACCGGACCGATACTATATAGTGTGATATCGCGGG-3′/5′-ATATGGTACCGCGGCCGCTAGACGCGGATGTTCTTCCAAAA-3′, respectively, and cloned into pUAST-3×HA to generate UAS-HA-usnp and UAS-HA-VAMP7. Note that the transmembrane domain is removed from tagged VAMP7. The aforementioned primers were used to blunt clone usnp coding sequences into pENTR1A followed by recombination of the resulting entry clone into pDEST17, expression and purification of the recombinant protein, and immunization of rats as for Syx17.
Cell culture and coimmunoprecipitations
Embryonic hemocyte-derived D.Mel-2 cells were maintained in Express-Five Serum-Free medium (Invitrogen) and transfected with UAS constructs and metallothionein-Gal4 plasmid using TransIT-2020 reagent (Mirus Bio LLC). 48 h later, protein expression was induced by adding 1 mM CuSO4 for overnight incubation. Cultured cells were collected, washed twice in PBS, lysed on ice in lysis buffer (0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, and 20 mM Tris-HCl, pH 7.5) containing complete protease and phosphatase inhibitor cocktails (Sigma-Aldrich), and spun for 10 min at 10,000 g in a centrifuge (5430R; Eppendorf) at 4°C followed by the addition of anti-FLAG slurry (Sigma-Aldrich) to the cleared supernatant. After incubation at 4°C for 2 h, beads were collected by centrifugation at 5,000 g for 30 s at 4°C followed by extensive washes in lysis buffer and finally boiling in 30 µl Laemmli sample buffer. Coimmunoprecipitations were repeated using a different DNA clone for all constructs, with similar results. For endogenous interaction experiments, either 100 mg of adult flies was starved for 2 h or 150 mg of cultured cells was washed for 2 × 10 min in PBS and homogenized for 2 × 10 s on ice in 1 ml lysis buffer containing 1% Triton X-100, using an homogenizer (Ultra-Turret T10; IKA) equipped with a disperser (S10N-5G; IKA). Lysates were cleared by centrifugation at 30,130 g for 10 min at 4°C (adult supernatants were spun once more to completely get rid of fat and unbroken cuticle pieces) followed by incubation with 3 µl rat anti-usnp (this study) or anti-GFP (Pircs et al., 2012), or guinea pig anti-Syx17 (this study) or anti-GFP (raised in collaboration with J. Mihály and I. Andó, Biological Research Center, Szeged, Hungary) antisera for 2 h at 4°C. Antibody–antigen complexes were collected by addition of 20 µl protein A (for guinea pig sera) or protein G (for rat sera) agarose 50% slurry for 1 h at 4°C and processed for Western blots as in co-overexpression experiments. Immunoprecipitations were performed from both cultured cells and adult flies, and representative examples are shown.
Transmission EM and immuno-EM
Dissected tissues were fixed in 3.2% paraformaldehyde, 0.5% glutaraldehyde, 1% sucrose, and 0.028% CaCl2 in 0.1 N sodium cacodylate, pH 7.4, overnight at 4°C, postfixed in 0.5% osmium tetroxide for 1 h, and embedded into Durcupan (Fluka) according to the manufacturer’s recommendations (Juhász et al., 2007, 2008). 70-nm sections were stained in Reynold’s lead citrate and viewed on a transmission electron microscope (JEM-1011; JEOL) equipped with camera (Morada; Olympus) and iTEM software (Olympus). A total of 17–29 randomly taken 10,000× magnification images of sections from four animals were evaluated per genotype by manually encircling relevant structures in Photoshop and calculating their percentage of area relative to total cytoplasm. P-values were calculated as described in Image analysis and statistics. For immuno-EM, samples were fixed as for conventional EM and embedded in London Resin white resin (Sigma-Aldrich) without postfixation. 90-nm sections were cut and incubated with rat anti-Syx17 (1:30) in PBS with 3% milk overnight at 4°C followed by biotin-conjugated anti–rat (1:100; Jackson ImmunoResearch Laboratories, Inc.) for 1 h in PBS with 1.5% milk at 25°C and anti-biotin conjugated to 20-nm gold for 5 h in TBS with 1.5% milk and 0.25% Tween 20 at 4°C (1:100; British Biocell; Juhász et al., 2007, 2008).
Western blots
Equal amounts of proteins per sample were separated by denaturing SDS-PAGE and processed for Western blots as previously described (Juhász et al., 2007; Pircs et al., 2012). Mouse anti-tubulin (1:1,000; AA4.3-s; Developmental Studies Hybridoma Bank), mouse anti-FLAG (1:2,000; M2; Sigma-Aldrich), rabbit anti-HA (1:2,000; Sigma-Aldrich), rabbit anti-p62 (1:8,000; Pircs et al., 2012), rabbit anti-Atg8a (1:5,000), rat anti-Syx17 (1:5,000; this study), guinea pig anti-Syx17 (1:5,000; this study), rat anti-usnp (1:3,000; this study) primary and alkaline phosphatase–conjugated anti–guinea pig, anti–rat (1:5,000; Sigma-Aldrich), anti–rabbit, and anti–mouse (1:5,000; EMD Millipore) secondary antibodies were used in TBS, pH 7.4, with 0.1% Tween 20 and 0.25% casein followed by colorimetric detection with nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl phosphate (Sigma-Aldrich).
Online supplemental material
Fig. S1 shows that Syx17, usnp, and VAMP7 are required for developmental and basal autophagy. Fig. S2 shows additional Syx17 localization, sequence alignment, and interaction data. Table S1 lists Drosophila homologues of human and yeast SNARE proteins, RNAi and mutant lines for these genes used in this study, and results of our small-scale RNAi screen. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211160/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201211160.dv.
Supplementary Material
Acknowledgments
We thank Sarolta Pálfia, Zsófia Kováts, Eszter Papp, and Eszter Vágó for technical assistance, public repositories, and colleagues listed in the Materials and methods section for reagents,
We thank the Wellcome Trust (087518/Z/08/Z), the Hungarian Scientific Research Fund (K83509), and the Hungarian Academy of Sciences (BO/00552/11) for support.
Footnotes
Abbreviations used in this paper:
- LTR
- LysoTracker red
- UAS
- upstream activation sequence
References
- Atlashkin V., Kreykenbohm V., Eskelinen E.L., Wenzel D., Fayyazi A., Fischer von Mollard G. 2003. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol. Cell. Biol. 23:5198–5207 10.1128/MCB.23.15.5198-5207.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth J.M., Szabad J., Hafen E., Köhler K. 2011. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 18:915–924 10.1038/cdd.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D.L., Baehrecke E.H. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 131:1137–1148 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Pankiv S., Øvervatn A., Brech A., Johansen T. 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 452:181–197 10.1016/S0076-6879(08)03612-4 [DOI] [PubMed] [Google Scholar]
- Dilcher M., Köhler B., von Mollard G.F. 2001. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 276:34537–34544 10.1074/jbc.M101551200 [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D.G., Mestre M.B., Colombo M.I. 2009. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta. 1793:1901–1916 10.1016/j.bbamcr.2009.09.011 [DOI] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179:485–500 10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N., Fujita N., Noda T., Yoshimori T., Amano A. 2010. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol. Biol. Cell. 21:1001–1010 10.1091/mbc.E09-08-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hong W. 2005. SNAREs and traffic. Biochim. Biophys. Acta. 1744:120–144 10.1016/j.bbamcr.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Mizushima N. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Juhász G., Erdi B., Sass M., Neufeld T.P. 2007. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21:3061–3066 10.1101/gad.1600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Hill J.H., Yan Y., Sass M., Baehrecke E.H., Backer J.M., Neufeld T.P. 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181:655–666 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F., Lin L., Kovács A.L., Yu L., Zhang H. 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 21:343–357 10.1016/j.devcel.2011.06.024 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K., Ravikumar B., Renna M., Puri C., Rubinsztein D.C. 2011. Autophagosome precursor maturation requires homotypic fusion. Cell. 146:303–317 10.1016/j.cell.2011.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W.L., Griffith J., Nag S., Wang K., Moss T., et al. 2011. SNARE proteins are required for macroautophagy. Cell. 146:290–302 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Munro S. 2010. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. 21:3998–4008 10.1091/mbc.E10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircs K., Nagy P., Varga A., Venkei Z., Erdi B., Hegedus K., Juhasz G. 2012. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS ONE. 7:e44214 10.1371/journal.pone.0044214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna M., Schaffner C., Winslow A.R., Menzies F.M., Peden A.A., Floto R.A., Rubinsztein D.C. 2011. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J. Cell Sci. 124:469–482 10.1242/jcs.076489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., Stenmark H. 2007. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17:1817–1825 10.1016/j.cub.2007.09.032 [DOI] [PubMed] [Google Scholar]
- Simonsen A., Cumming R.C., Brech A., Isakson P., Schubert D.R., Finley K.D. 2008. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 4:176–184 [DOI] [PubMed] [Google Scholar]
- Steegmaier M., Yang B., Yoo J.S., Huang B., Shen M., Yu S., Luo Y., Scheller R.H. 1998. Three novel proteins of the syntaxin/SNAP-25 family. J. Biol. Chem. 273:34171–34179 10.1074/jbc.273.51.34171 [DOI] [PubMed] [Google Scholar]
- Tooze S.A., Yoshimori T. 2010. The origin of the autophagosomal membrane. Nat. Cell Biol. 12:831–835 10.1038/ncb0910-831 [DOI] [PubMed] [Google Scholar]
- Velikkakath A.K., Nishimura T., Oita E., Ishihara N., Mizushima N. 2012. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell. 23:896–909 10.1091/mbc.E11-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.